Radiomics, a Promising New Discipline: Example of Hepatocellular Carcinoma
Abstract
:1. Introduction
2. What Is Radiomics?
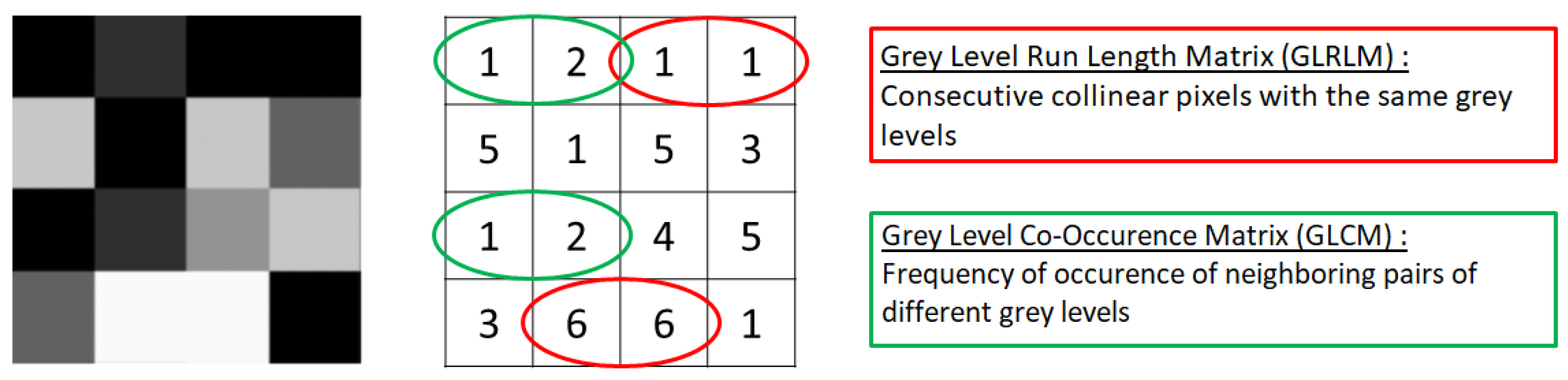
Characterization of Intra-Tumoral Heterogeneity
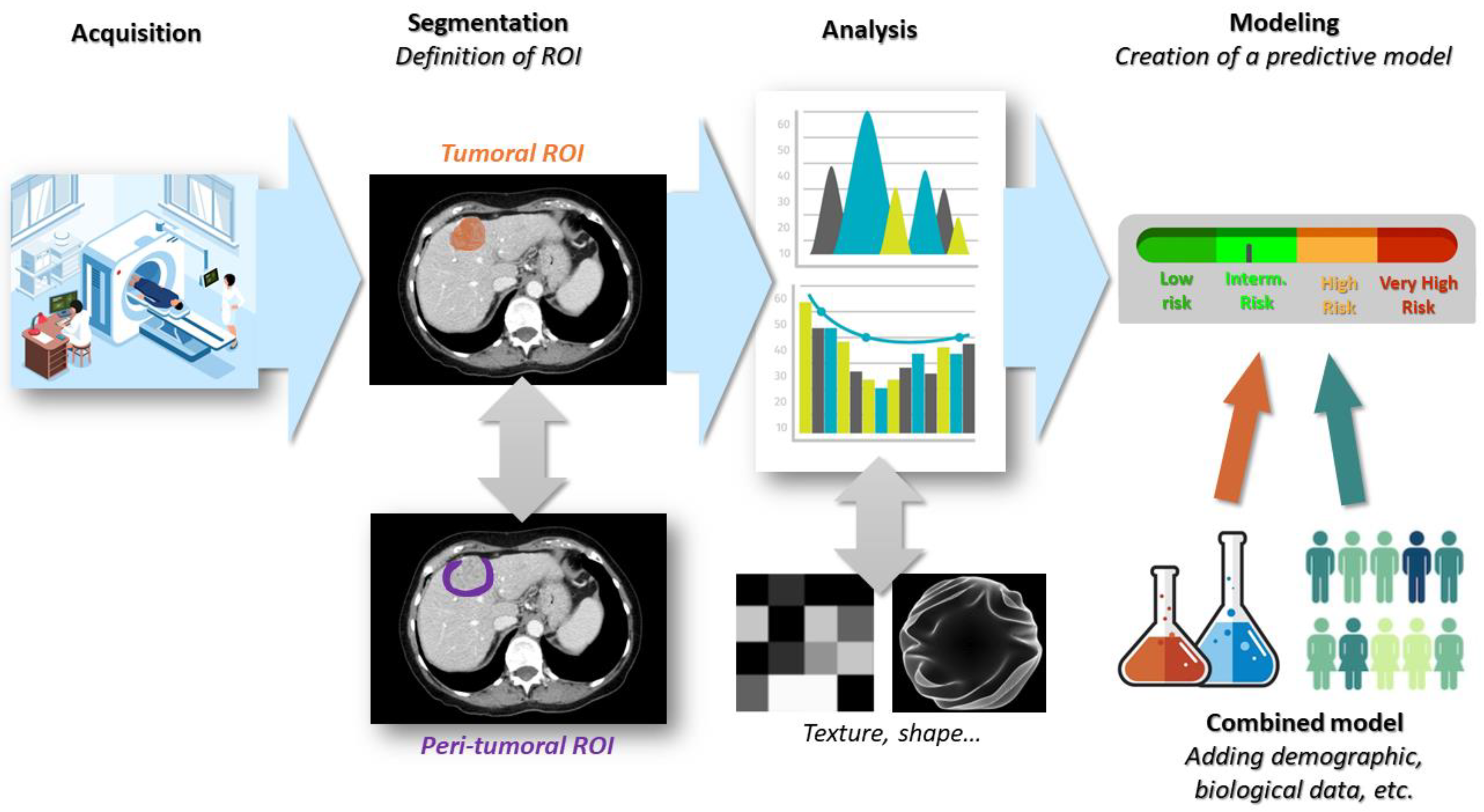
3. Conducting a Radiomics Study
4. Limitations
5. Radiomics in Practice: Example of Hepatocellular Carcinoma
5.1. Radiomics for Histological Diagnosis
5.2. Radiomics for Prediction of HCC Outcome
5.3. Radiomics for Prediction of HCC Response to Therapy
| Authors (Ref) | Objectives of the Study | Imaging Modality | Nmbr of Patients | Geographical Origin | Cause of Liver Disease | Performance of Radiomics Model Alone | Performance of Combined Model |
|---|---|---|---|---|---|---|---|
| Blanc-Durand et al. [13] | Predict survival after radio embolization | PET-CT | 47 | Europe | Alcool | NC | NC |
| Liang et al. [18] | Differentiate hypervascular liver tumors | CT MRI | CT: 170 MRI: 137 | China | NC | AUC (CT): 0.879 AUC (MRI): 0.925 | AUC (CT): 0.966 AUC (MRI): 0.971 |
| Zhong et al. [20] | Differentiate between HCC and benign liver nodules | MRI | 150 | China | HBV | AUC: 0.917 Sens: 93.8% Spe: 86.4% | AUC: 0.975 Sens: 97.3% Spe: 97.7% |
| Ji et al. [14] | Predict postoperative recurrence | CT | 470 | China | NC | NC | tdAUC: 0.803 |
| Feng et al. [25] | Predict microvascular invasion | MRI | 160 | China | HBV | AUC: 0.83 Sens: 90% Spe: 75% | Radiomics model only |
| Shi et al. [26] | Predicting microvascular invasion | MRI PET-CT | 97 | China | HBV | AUC (MVI+): 0.917 AUC (MVI−): 0.771 | AUC (MVI+): 0.996 AUC (MVI−): 0.953 |
| Sheen et al. [30] | Predict response to chemo-embolization | CT | 80 | Korea | HBV | NC | AUC: 0.95 Sens: 0.91 Spe: 0.75 |
| Chen et al. [33] | Predict response to immunotherapy | MRI | 207 | China | HBV | AUC: 0.640 Sens: 53.9% Spe: 61.4% | AUC: 0.934 Sens: 84.6% Spe: 84.1% |
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, P.-R.; Lu, L.; Zhang, J.-Y.; Huo, T.-T.; Liu, S.-X.; Ye, Z.-W. Application of Artificial Intelligence in Medicine: An Overview. Curr. Med Sci. 2021, 41, 1105–1115. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef] [Green Version]
- Guiot, J.; Vaidyanathan, A.; Deprez, L.; Zerka, F.; Danthine, D.; Frix, A.; Lambin, P.; Bottari, F.; Tsoutzidis, N.; Miraglio, B.; et al. A review in radiomics: Making personalized medicine a reality via routine imaging. Med. Res. Rev. 2022, 42, 426–440. [Google Scholar] [CrossRef] [PubMed]
- Bhinder, B.; Gilvary, C.; Madhukar, N.S.; Elemento, O. Artificial Intelligence in Cancer Research and Precision Medicine. Cancer Discov. 2021, 11, 900–915. [Google Scholar] [CrossRef]
- Burrell, R.A.; McGranahan, N.; Bartek, J.; Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013, 501, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Andor, N.; Graham, T.A.; Jansen, M.; Xia, L.C.; Aktipis, C.A.; Petritsch, C.; Ji, H.P.; Maley, C.C. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med. 2016, 22, 105–113. [Google Scholar] [CrossRef] [Green Version]
- Friemel, J.; Rechsteiner, M.; Frick, L.; Böhm, F.; Struckmann, K.; Egger, M.; Moch, H.; Heikenwalder, M.; Weber, A. Intratumor heterogeneity in hepatocellular carcinoma. Clin. Cancer Res. 2015, 21, 1951–1961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [Green Version]
- RRoy, S.; Whitehead, T.D.; Quirk, J.D.; Salter, A.; Ademuyiwa, F.O.; Li, S.; An, H.; Shoghi, K.I. Optimal co-clinical radiomics: Sensitivity of radiomic features to tumour volume, image noise and resolution in co-clinical T1-weighted and T2-weighted magnetic resonance imaging. EBioMedicine 2020, 59, 102963. [Google Scholar] [CrossRef]
- Sulaiman, A.; Wang, L. Bridging the divide: Preclinical research discrepancies between triple-negative breast cancer cell lines and patient tumors. Oncotarget 2017, 8, 113269–113281. [Google Scholar] [CrossRef] [Green Version]
- Roy, S.; Whitehead, T.D.; Li, S.; Ademuyiwa, F.O.; Wahl, R.L.; Dehdashti, F.; Shoghi, K.I. Co-clinical FDG-PET radiomic signature in predicting response to neoadjuvant chemotherapy in triple-negative breast cancer. Eur. J. Nucl. Med. 2021, 49, 550–562. [Google Scholar] [CrossRef] [PubMed]
- Limkin, E.J.; Sun, R.; Dercle, L.; Zacharaki, E.I.; Robert, C.; Reuzé, S.; Schernberg, A.; Paragios, N.; Deutsch, E.; Ferté, C. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann. Oncol. 2017, 28, 1191–1206. [Google Scholar] [CrossRef]
- Blanc-Durand, P.; Van Der Gucht, A.; Jreige, M.; Nicod-Lalonde, M.; Silva-Monteiro, M.; Prior, J.O.; Denys, A.; Depeursinge, A.; Schaefer, N. Signature of survival: A 18F-FDG PET based whole-liver radiomic analysis predicts survival after 90Y-TARE for hepatocellular carcinoma. Oncotarget 2017, 9, 4549–4558. [Google Scholar] [CrossRef] [Green Version]
- Ji, G.-W.; Zhu, F.-P.; Xu, Q.; Wang, K.; Wu, M.-Y.; Tang, W.-W.; Li, X.-C.; Wang, X.-H. Machine-learning analysis of contrast-enhanced CT radiomics predicts recurrence of hepatocellular carcinoma after resection: A multi-institutional study. EBioMedicine 2019, 50, 156–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef] [Green Version]
- Marrero, J.A.; Kulik, L.M.; Sirlin, C.B.; Zhu, A.X.; Finn, R.S.; Abecassis, M.M.; Roberts, L.R.; Heimbach, J.K. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2018, 68, 723–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef]
- Liang, W.; Shao, J.; Liu, W.; Ruan, S.; Tian, W.; Zhang, X.; Wan, D.; Huang, Q.; Ding, Y.; Xiao, W. Differentiating Hepatic Epithelioid Angiomyolipoma from Hepatocellular Carcinoma and Focal Nodular Hyperplasia via Radiomics Models. Front. Oncol. 2020, 10, 564307. [Google Scholar] [CrossRef]
- Elsayes, K.M.; Hooker, J.C.; Agrons, M.M.; Kielar, A.Z.; Tang, A.; Fowler, K.; Chernyak, V.; Bashir, M.R.; Kono, Y.; Do, R.K.; et al. 2017 Version of LI-RADS for CT and MR Imaging: An Update. RadioGraphics 2017, 37, 1994–2017. [Google Scholar] [CrossRef]
- Zhong, X.; Guan, T.; Tang, D.; Li, J.; Lu, B.; Cui, S.; Tang, H. Differentiation of small (≤3 cm) hepatocellular carcinomas from benign nodules in cirrhotic liver: The added additive value of MRI-based radiomics analysis to LI-RADS version 2018 algorithm. BMC Gastroenterol. 2021, 21, 155. [Google Scholar] [CrossRef]
- Chan, A.W.; Zhong, J.; Berhane, S.; Toyoda, H.; Cucchetti, A.; Shi, K.; Tada, T.; Chong, C.C.; Xiang, B.-D.; Li, L.-Q.; et al. Development of pre and post-operative models to predict early recurrence of hepatocellular carcinoma after surgical resection. J. Hepatol. 2018, 69, 1284–1293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, K.-C.; Chow, P.K.-H.; Allen, J.C.; Chia, G.-S.; Lim, M.; Cheow, P.-C.; Chung, A.Y.; Ooi, L.L.; Tan, S.-B. Microvascular invasion is a better predictor of tumor recurrence and overall survival following surgical resection for hepatocellular carcinoma compared to the Milan criteria. Ann. Surg. 2011, 254, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Gundlach, J.-P.; Schmidt, S.; Bernsmeier, A.; Günther, R.; Kataev, V.; Trentmann, J.; Schäfer, J.; Röcken, C.; Becker, T.; Braun, F. Indication of Liver Transplantation for Hepatocellular Carcinoma Should Be Reconsidered in Case of Microvascular Invasion and Multilocular Tumor Occurrence. J. Clin. Med. 2021, 10, 1155. [Google Scholar] [CrossRef]
- Lee, S.; Kim, S.H.; Lee, J.E.; Sinn, D.H.; Park, C.K. Preoperative gadoxetic acid-enhanced MRI for predicting microvascular invasion in patients with single hepatocellular carcinoma. J. Hepatol. 2017, 67, 526–534. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.-T.; Jia, Y.; Liao, B.; Huang, B.; Zhou, Q.; Li, X.; Wei, K.; Chen, L.; Li, B.; Wang, W.; et al. Preoperative prediction of microvascular invasion in hepatocellular cancer: A radiomics model using Gd-EOB-DTPA-enhanced MRI. Eur. Radiol. 2019, 29, 4648–4659. [Google Scholar] [CrossRef]
- Shi, H.; Duan, Y.; Shi, J.; Zhang, W.; Liu, W.; Shen, B.; Liu, F.; Mei, X.; Li, X.; Yuan, Z. Role of preoperative prediction of microvascular invasion in hepatocellular carcinoma based on the texture of FDG PET image: A comparison of quantitative metabolic parameters and MRI. Front. Physiol. 2022, 13, 928969. [Google Scholar] [CrossRef]
- Reig, M.; Forner, A.; Rimola, J.; Ferrer-Fàbrega, J.; Burrel, M.; Garcia-Criado, Á.; Kelley, R.K.; Galle, P.R.; Mazzaferro, V.; Salem, R.; et al. BCLC strategy for prognosis prediction and treatment recommendation: The 2022 update. J. Hepatol. 2022, 76, 681–693. [Google Scholar] [CrossRef]
- Burroughs, A.; Dufour, J.-F.; Galle, P.R.; Mazzaferro, V.; Piscaglia, F.; Raoul, J.L.; Sangro, B.; Bolondi, L. Heterogeneity of patients with intermediate (BCLC B) Hepatocellular Carcinoma: Proposal for a subclassification to facilitate treatment decisions. Semin. Liver Dis. 2012, 32, 348–359. [Google Scholar] [CrossRef]
- Kadalayil, L.; Benini, R.; Pallan, L.; O’Beirne, J.; Marelli, L.; Yu, D.; Hackshaw, A.; Fox, R.; Johnson, P.; Burroughs, A.; et al. A simple prognostic scoring system for patients receiving transarterial embolisation for hepatocellular cancer. Ann. Oncol. 2013, 24, 2565–2570. [Google Scholar] [CrossRef]
- Sheen, H.; Kim, J.S.; Lee, J.K.; Choi, S.Y.; Baek, S.Y.; Kim, J.Y. A radiomics nomogram for predicting transcatheter arterial chemoembolization refractoriness of hepatocellular carcinoma without extrahepatic metastasis or macrovascular invasion. Abdom. Radiol. 2021, 46, 2839–2849. [Google Scholar] [CrossRef]
- Kothari, G. Role of radiomics in predicting immunotherapy response. J. Med Imaging Radiat. Oncol. 2022, 66, 575–591. [Google Scholar] [CrossRef] [PubMed]
- Bruni, D.; Angell, H.K.; Galon, J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat. Rev. Cancer 2020, 20, 662–680. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Feng, S.; Wei, J.; Liu, F.; Li, B.; Li, X.; Hou, Y.; Gu, D.; Tang, M.; Xiao, H.; et al. Pretreatment prediction of immunoscore in hepatocellular cancer: A radiomics-based clinical model based on Gd-EOB-DTPA-enhanced MRI imaging. Eur. Radiol. 2019, 29, 4177–4187. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.K.; Ng, T.-T.; Loh, T.P. Machine learning assistive rapid, label-free molecular phenotyping of blood with two-dimensional NMR correlational spectroscopy. Commun. Biol. 2020, 3, 535. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lévi-Strauss, T.; Tortorici, B.; Lopez, O.; Viau, P.; Ouizeman, D.J.; Schall, B.; Adhoute, X.; Humbert, O.; Chevallier, P.; Gual, P.; et al. Radiomics, a Promising New Discipline: Example of Hepatocellular Carcinoma. Diagnostics 2023, 13, 1303. https://doi.org/10.3390/diagnostics13071303
Lévi-Strauss T, Tortorici B, Lopez O, Viau P, Ouizeman DJ, Schall B, Adhoute X, Humbert O, Chevallier P, Gual P, et al. Radiomics, a Promising New Discipline: Example of Hepatocellular Carcinoma. Diagnostics. 2023; 13(7):1303. https://doi.org/10.3390/diagnostics13071303
Chicago/Turabian StyleLévi-Strauss, Thomas, Bettina Tortorici, Olivier Lopez, Philippe Viau, Dann J. Ouizeman, Baptiste Schall, Xavier Adhoute, Olivier Humbert, Patrick Chevallier, Philippe Gual, and et al. 2023. "Radiomics, a Promising New Discipline: Example of Hepatocellular Carcinoma" Diagnostics 13, no. 7: 1303. https://doi.org/10.3390/diagnostics13071303
APA StyleLévi-Strauss, T., Tortorici, B., Lopez, O., Viau, P., Ouizeman, D. J., Schall, B., Adhoute, X., Humbert, O., Chevallier, P., Gual, P., Fillatre, L., & Anty, R. (2023). Radiomics, a Promising New Discipline: Example of Hepatocellular Carcinoma. Diagnostics, 13(7), 1303. https://doi.org/10.3390/diagnostics13071303






