Gastro-BaseNet: A Specialized Pre-Trained Model for Enhanced Gastroscopic Data Classification and Diagnosis of Gastric Cancer and Ulcer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Acquation
2.2. Study Environment
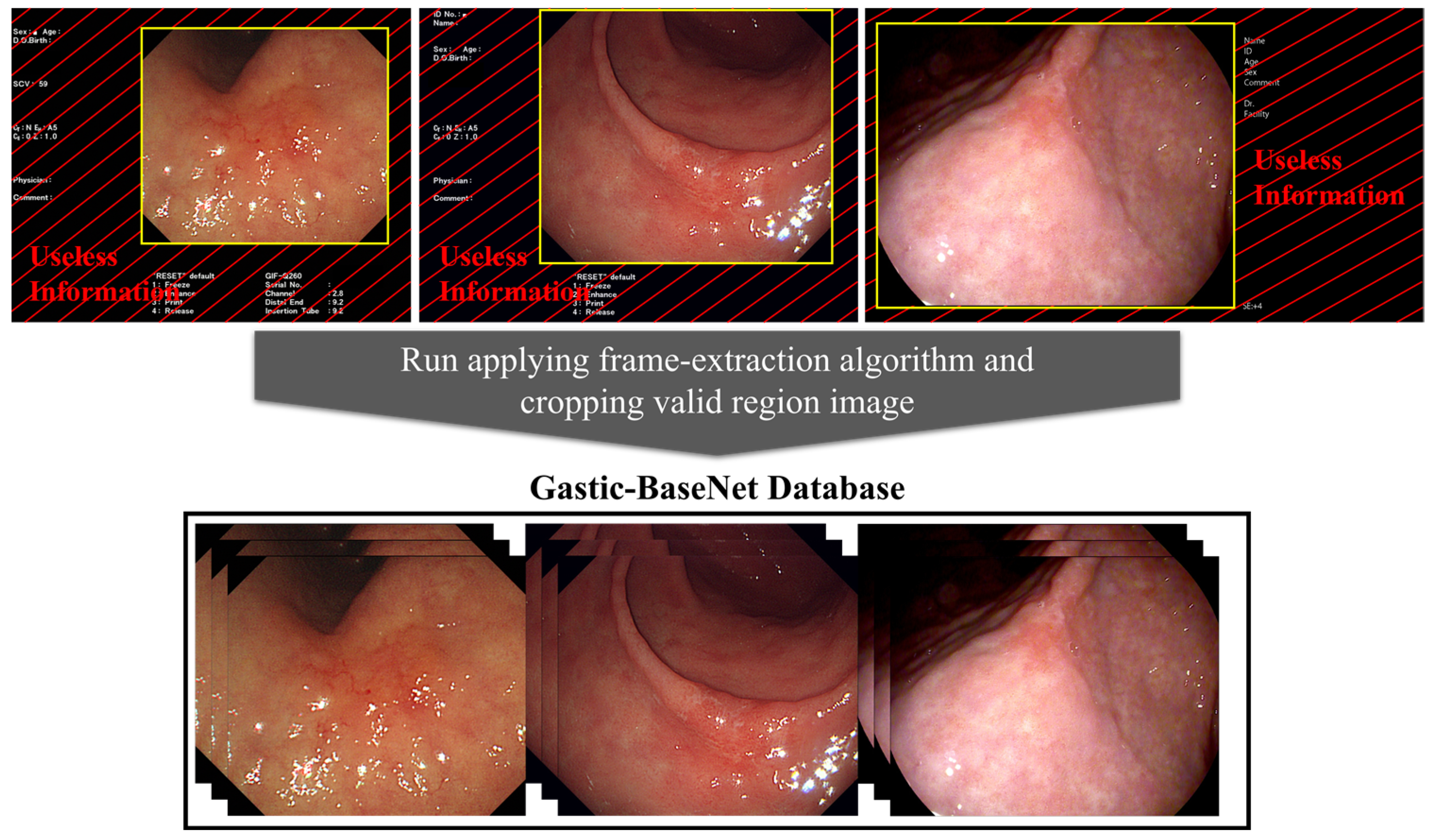
2.3. Data Preprocess
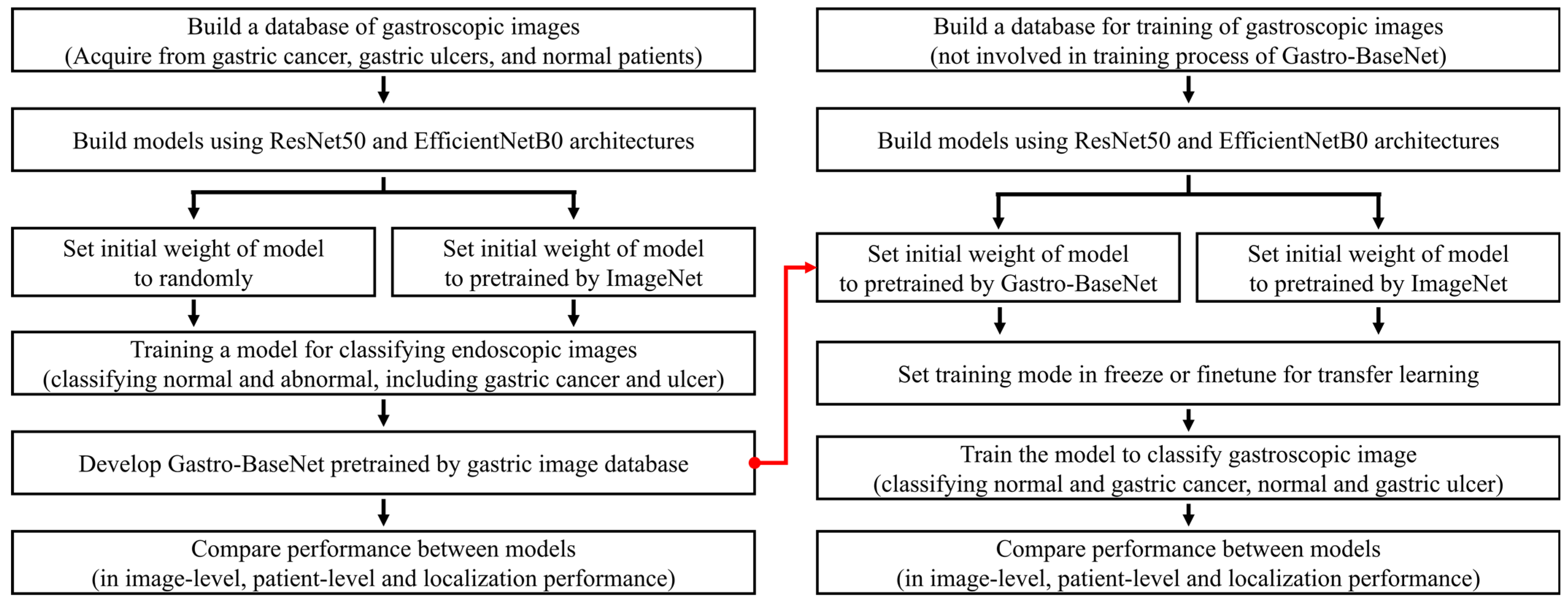
2.4. Transfer Learning
2.5. Setting Model Training Hyperparameters
2.6. Statistical Analysis
2.6.1. Image-Level Performance
2.6.2. Patient-Level Performance
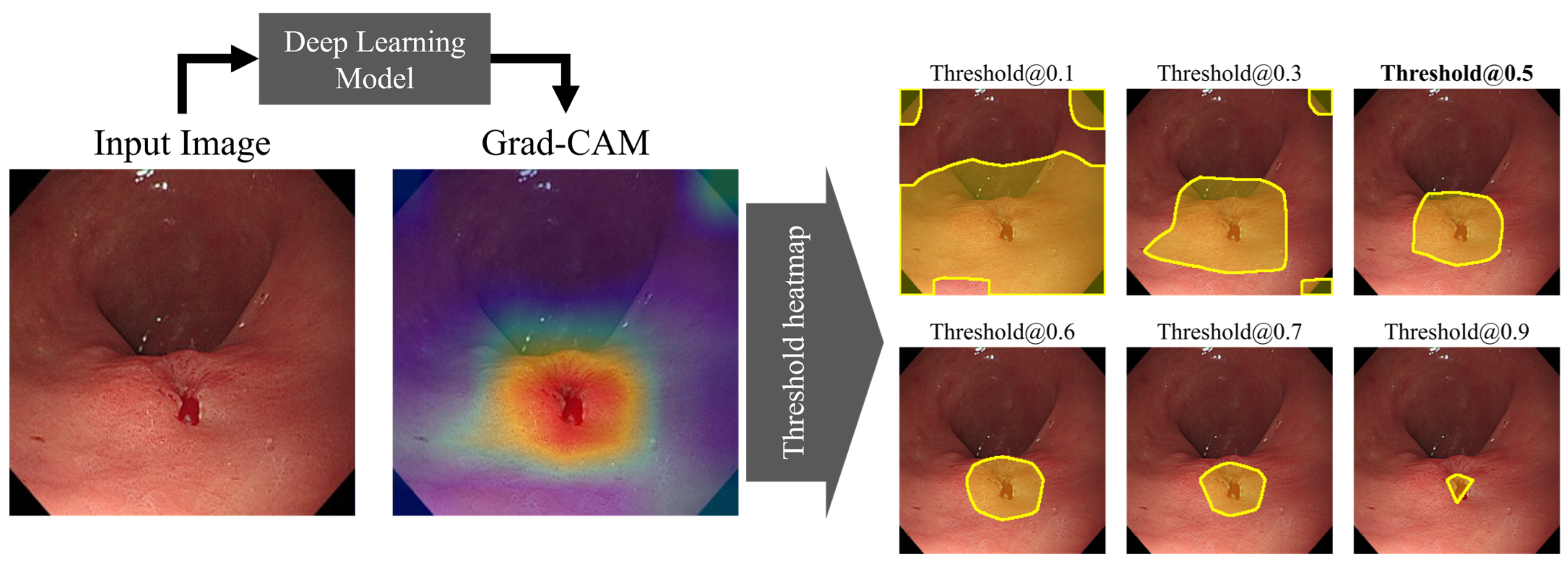
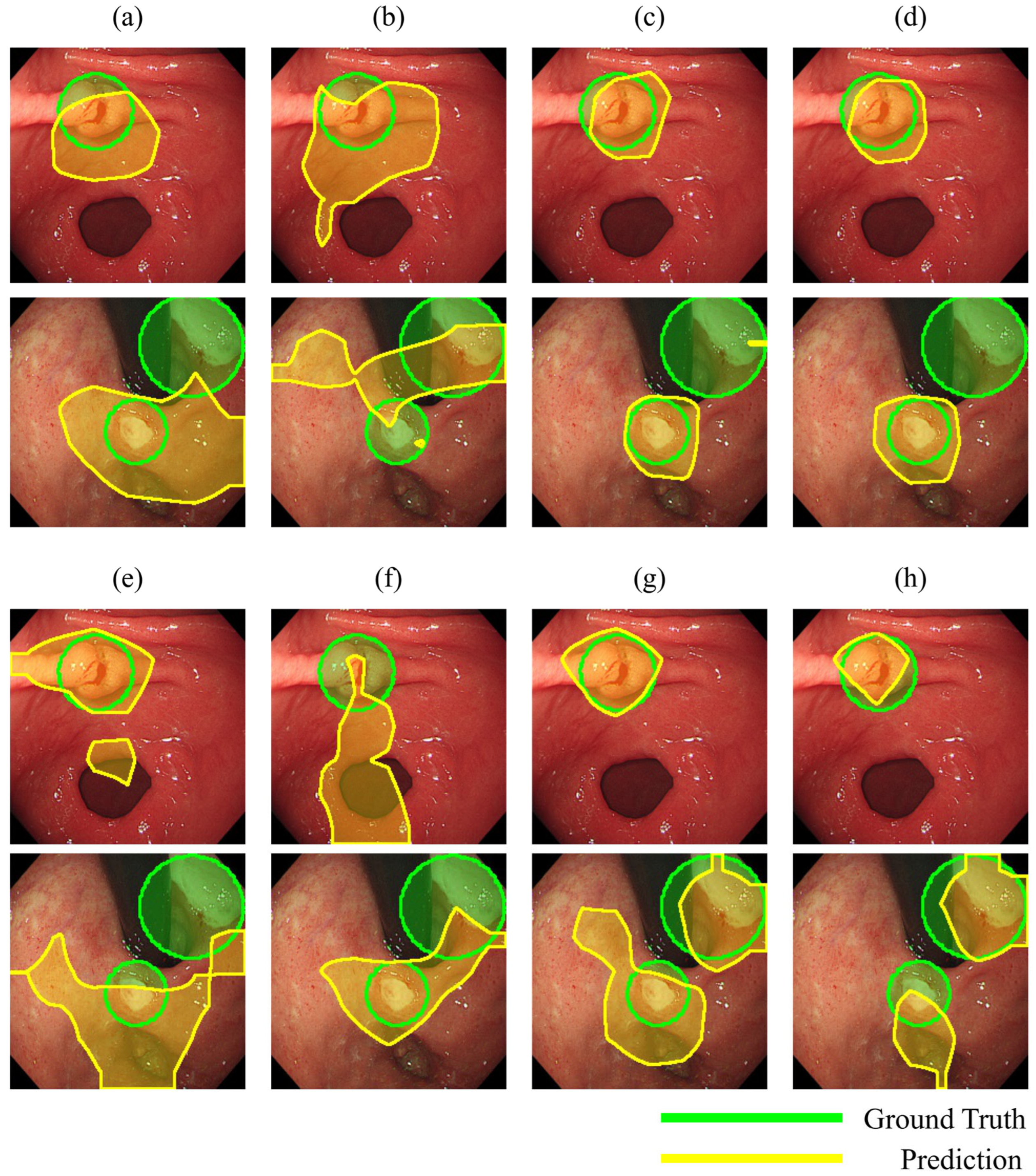
2.6.3. Localization Performance
3. Results
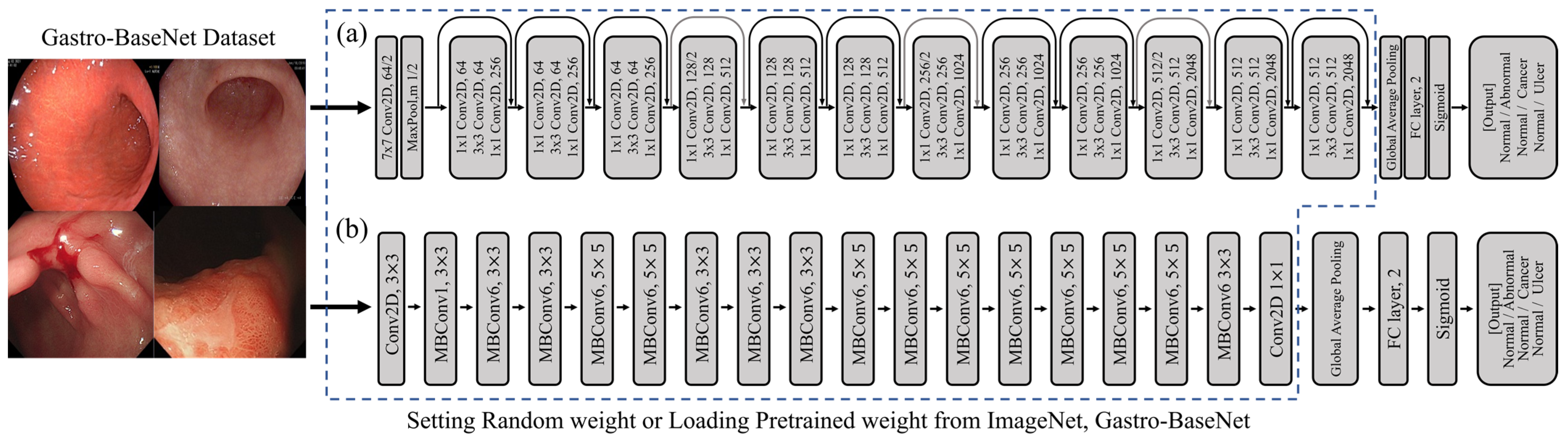
3.1. Gastro-BaseNet
3.2. Models Trained on Transfer Learning Based on Gastro-BaseNet
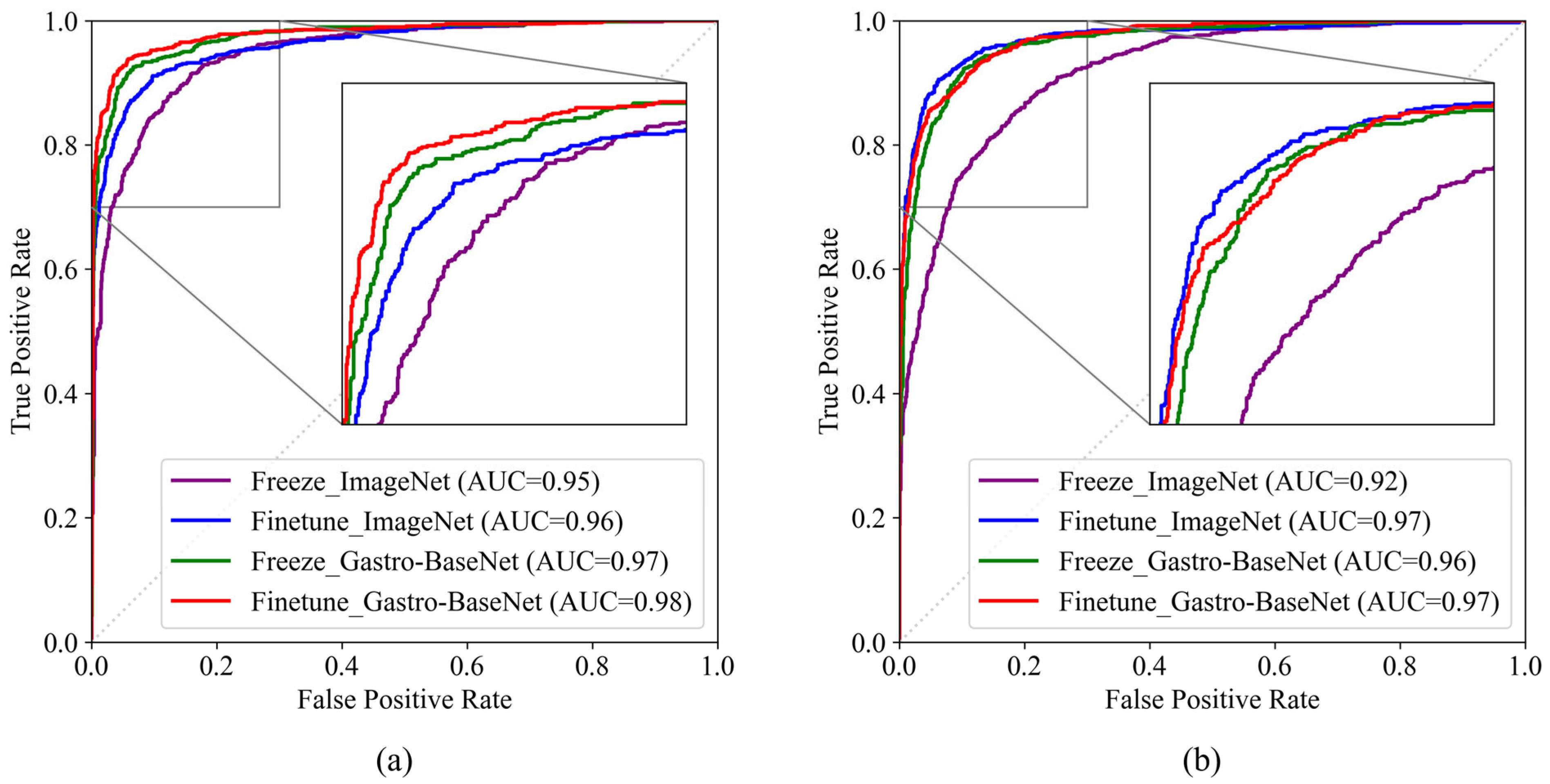
3.2.1. Gastric Cancer Classification Trained by Gastro-BaseNet
3.2.2. Gastric Ulcer Classification Trained by Gastro-BaseNet
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Smyth, E.C.; Nilsson, M.; Grabsch, H.I.; van Grieken, N.C.T.; Lordick, F. Gastric cancer. Lancet 2020, 396, 635–648. [Google Scholar] [CrossRef]
- Hernanz, N.; Rodríguez de Santiago, E.; Marcos Prieto, H.M.; Jorge Turrión, M.Á.; Barreiro Alonso, E.; Rodríguez Escaja, C.; Jiménez Jurado, A.; Sierra, M.; Pérez Valle, I.; Volpato, N.; et al. Characteristics and consequences of missed gastric cancer: A multicentric cohort study. Dig. Liver Dis. 2019, 51, 894–900. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, Y.J.; Kim, Y.W.; Park, S.; Choi, Y.-i.; Kim, Y.J.; Park, D.K.; Kim, K.G.; Chung, J.-W. Spotting malignancies from gastric endoscopic images using deep learning. Surg. Endosc. 2019, 33, 3790–3797. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Su, X.; Ma, L.; Gao, X.; Sun, M. Deep learning for classification and localization of early gastric cancer in endoscopic images. Biomed. Signal Process. Control 2023, 79, 104200. [Google Scholar] [CrossRef]
- Wei, W.; Wan, X.; Jun, Z.; Lei, S.; Shan, H.; Qianshan, D.; Ganggang, M.; Anning, Y.; Xu, H.; Jun, L.; et al. A deep neural network improves endoscopic detection of early gastric cancer without blind spots. Endoscopy 2019, 51, 522–531. [Google Scholar] [CrossRef]
- Teramoto, A.; Shibata, T.; Yamada, H.; Hirooka, Y.; Saito, K.; Fujita, H. Detection and characterization of gastric cancer using cascade deep learning model in endoscopic images. Diagnostics 2022, 12, 1996. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.-L.; Zhou, Y.; Liu, W.; Luo, Q.; Zeng, X.-H.; Yi, Z.; Hu, B. Artificial intelligence for diagnosing gastric lesions under white-light endoscopy. Surg. Endosc. 2022, 36, 9444–9453. [Google Scholar] [CrossRef] [PubMed]
- Cho, B.-J.; Bang, C.S.; Park, S.W.; Yang, Y.J.; Seo, S.I.; Lim, H.; Shin, W.G.; Hong, J.T.; Yoo, Y.T.; Hong, S.H. Automated classification of gastric neoplasms in endoscopic images using a convolutional neural network. Endoscopy 2019, 51, 1121–1129. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Kora, P.; Ooi, C.P.; Faust, O.; Raghavendra, U.; Gudigar, A.; Chan, W.Y.; Meenakshi, K.; Swaraja, K.; Plawiak, P.; Rajendra Acharya, U. Transfer learning techniques for medical image analysis: A review. Biocybern. Biomed. Eng. 2022, 42, 79–107. [Google Scholar] [CrossRef]
- Rawat, W.; Wang, Z. Deep Convolutional Neural Networks for Image Classification: A Comprehensive Review. Neural Comput. 2017, 29, 2352–2449. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.J.; Yang, Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Kim, H.E.; Cosa-Linan, A.; Santhanam, N.; Jannesari, M.; Maros, M.E.; Ganslandt, T. Transfer learning for medical image classification: A literature review. BMC Med. Imaging 2022, 22, 69. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Kumar, R.; Gupta, M. Review on Transfer Learning for Convolutional Neural Network. In Proceedings of the 2021 3rd International Conference on Advances in Computing, Communication Control and Networking (ICAC3N), Greater Noida, India, 17–18 December 2021; pp. 922–926. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Hussain, M.; Bird, J.J.; Faria, D.R. A Study on CNN Transfer Learning for Image Classification; Springer: Cham, Switzerland, 2019; pp. 191–202. [Google Scholar]
- Morid, M.A.; Borjali, A.; Del Fiol, G. A scoping review of transfer learning research on medical image analysis using ImageNet. Comput. Biol. Med. 2021, 128, 104115. [Google Scholar] [CrossRef] [PubMed]
- Raghu, M.; Zhang, C.; Kleinberg, J.; Bengio, S. Transfusion: Understanding transfer learning for medical imaging. Adv. Neural Inf. Process. Syst. 2019, 32, 1–22. [Google Scholar]
- He, K.; Girshick, R.; Dollár, P. Rethinking imagenet pre-training. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 4918–4927. [Google Scholar]
- Reverberi, C.; Rigon, T.; Solari, A.; Hassan, C.; Cherubini, P.; Antonelli, G.; Awadie, H.; Bernhofer, S.; Carballal, S.; Dinis-Ribeiro, M.; et al. Experimental evidence of effective human–AI collaboration in medical decision-making. Sci. Rep. 2022, 12, 14952. [Google Scholar] [CrossRef]
- Park, Y.R.; Kim, Y.J.; Chung, J.-W.; Kim, K.G. Convolution Neural Network Based Auto Classification Model Using Endoscopic Images of Gastric Cancer and Gastric Ulcer. J. Biomed. Eng. Res. 2020, 41, 101–106. [Google Scholar]
- Kim, Y.-j.; Cho, H.C.; Cho, H.-c. Deep learning-based computer-aided diagnosis system for gastroscopy image classification using synthetic data. Appl. Sci. 2021, 11, 760. [Google Scholar] [CrossRef]
- Garcea, F.; Serra, A.; Lamberti, F.; Morra, L. Data augmentation for medical imaging: A systematic literature review. Comput. Biol. Med. 2023, 152, 106391. [Google Scholar] [CrossRef]
- Jin, T.; Jiang, Y.; Mao, B.; Wang, X.; Lu, B.; Qian, J.; Zhou, H.; Ma, T.; Zhang, Y.; Li, S.; et al. Multi-center verification of the influence of data ratio of training sets on test results of an AI system for detecting early gastric cancer based on the YOLO-v4 algorithm. Front. Oncol. 2022, 12, 953090. [Google Scholar] [CrossRef]
- Ikenoyama, Y.; Hirasawa, T.; Ishioka, M.; Namikawa, K.; Yoshimizu, S.; Horiuchi, Y.; Ishiyama, A.; Yoshio, T.; Tsuchida, T.; Takeuchi, Y.; et al. Detecting early gastric cancer: Comparison between the diagnostic ability of convolutional neural networks and endoscopists. Dig. Endosc. 2021, 33, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Hirasawa, T.; Aoyama, K.; Tanimoto, T.; Ishihara, S.; Shichijo, S.; Ozawa, T.; Ohnishi, T.; Fujishiro, M.; Matsuo, K.; Fujisaki, J. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer 2018, 21, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Yacob, Y.M.; Alquran, H.; Mustafa, W.A.; Alsalatie, M.; Sakim, H.A.M.; Lola, M.S. H. pylori Related Atrophic Gastritis Detection Using Enhanced Convolution Neural Network (CNN) Learner. Diagnostics 2023, 13, 336. [Google Scholar] [CrossRef] [PubMed]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S. An image is worth 16 × 16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Touvron, H.; Cord, M.; Douze, M.; Massa, F.; Sablayrolles, A.; Jégou, H. Training Data-Efficient Image Transformers & Distillation through Attention. In Proceedings of the International Conference on Machine Learning, Virtual, 18–24 July 2021; pp. 10347–10357. [Google Scholar]
- Zhuang, F.; Qi, Z.; Duan, K.; Xi, D.; Zhu, Y.; Zhu, H.; Xiong, H.; He, Q. A Comprehensive Survey on Transfer Learning. Proc. IEEE 2021, 109, 43–76. [Google Scholar] [CrossRef]


| Normal | Abnormal | Total | ||
|---|---|---|---|---|
| Gastric Cancer | Gastric Ulcer | |||
| Train | 7297 | 4476 | 2409 | 14,182 |
| Validation | 738 | 493 | 263 | 1494 |
| Test | 2022 | 1195 | 625 | 3842 |
| Total (Number of patients) | 10,057 (300) | 6164 (1070) | 3297 (532) | 19,518 (1902) |
| Study 1: Gastric Cancer | Study 2: Gastric Ulcer | |||||
|---|---|---|---|---|---|---|
| Normal | Gastric Cancer | Total | Normal | Gastric Ulcer | Total | |
| Train | 3662 | 2671 | 6333 | 3617 | 789 | 4406 |
| Validation | 416 | 273 | 689 | 461 | 92 | 553 |
| Test | 1075 | 780 | 1855 | 1075 | 243 | 1318 |
| Total (Number of patients) | 5153 (148) | 3724 (707) | 8877 (855) | 5153 (148) | 1124 (178) | 6277 (326) |
| Model Architecture | Hyperparameters | Image-Level Performance (%) | Patient-Level Performance (%) | Localization Performance (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Training Mode | Pretrained Weight | Accuracy | Sensitivity | Specificity | F1 Score | AUC | Accuracy | Sensitivity | Specificity | Sensitivity | |
| ResNet50 | fine-tune | Random | 83.91 | 87.42 | 80.76 | 84.09 | 89.57 | 88.38 | 90.06 | 79.31 | 41.23 |
| freeze | ImageNet | 88.50 | 87.09 | 89.76 | 89.15 | 92.39 | 92.68 | 91.61 | 98.31 | 58.11 | |
| fine-tune | ImageNet | 91.88 | 90.38 | 93.22 | 92.36 | 96.54 | 95.42 | 94.53 | 100.0 | 82.19 | |
| EfficientNetB0 | fine-tune | Random | 79.54 | 84.29 | 75.27 | 79.48 | 86.10 | 83.11 | 85.94 | 68.33 | 63.43 |
| freeze | ImageNet | 87.79 | 87.25 | 88.28 | 88.39 | 91.44 | 92.43 | 91.94 | 95.00 | 61.52 | |
| fine-tune | ImageNet | 91.93 | 91.26 | 92.53 | 92.35 | 96.95 | 95.92 | 95.13 | 100.0 | 70.74 | |
| Model Architecture | Hyperparameters | Image-Level Performance (%) | Patient-Level Performance (%) | Localization Performance (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Training Mode | Pretrained Weight | Accuracy | Sensitivity | Specificity | F1 Score | AUC | Accuracy | Sensitivity | Specificity | Sensitivity | |
| ResNet50 | freeze | ImageNet | 90.46 | 90.26 | 90.60 | 91.67 | 94.81 | 96.99 | 96.32 | 100.0 | 80.26 |
| fine-tune | ImageNet | 90.67 | 92.44 | 89.40 | 91.74 | 96.22 | 95.29 | 94.29 | 100.0 | 82.94 | |
| freeze | Gastro-BaseNet | 94.07 | 93.72 | 94.33 | 94.86 | 97.43 | 97.66 | 97.16 | 100.0 | 83.99 | |
| fine-tune | Gastro-BaseNet | 94.72 | 94.10 | 95.16 | 95.43 | 97.90 | 97.66 | 97.16 | 100.0 | 87.19 | |
| EfficientNetB0 | freeze | ImageNet | 88.79 | 87.82 | 89.49 | 90.24 | 91.73 | 92.26 | 91.37 | 96.55 | 67.15 |
| fine-tune | ImageNet | 94.02 | 93.33 | 94.51 | 94.82 | 97.05 | 97.66 | 97.16 | 100.0 | 68.54 | |
| freeze | Gastro-BaseNet | 85.71 | 98.21 | 76.65 | 86.15 | 97.01 | 95.32 | 99.29 | 7667 | 75.72 | |
| fine-tune | Gastro-BaseNet | 83.45 | 98.97 | 72.19 | 83.49 | 96.42 | 95.88 | 100.0 | 7667 | 78.50 | |
| Model Architecture | Variable 1 | Variable 2 | p-Value | ||
|---|---|---|---|---|---|
| Training Mode | Pretrained Weight | Training Mode | Pretrained Weight | ||
| ResNet50 | ImageNet | freeze | ImageNet | fine-tune | 0.0006 |
| ImageNet | freeze | Gastro-BaseNet | freeze | <0.0001 | |
| ImageNet | freeze | Gastro-BaseNet | fine-tune | <0.0001 | |
| ImageNet | fine-tune | Gastro-BaseNet | freeze | 0.0002 | |
| ImageNet | fine-tune | Gastro-BaseNet | fine-tune | <0.0001 | |
| Gastro-BaseNet | fine-tune | Gastro-BaseNet | freeze | <0.0001 | |
| EfficientNetB0 | ImageNet | freeze | ImageNet | fine-tune | <0.0001 |
| ImageNet | freeze | Gastro-BaseNet | freeze | <0.0001 | |
| ImageNet | freeze | Gastro-BaseNet | fine-tune | <0.0001 | |
| ImageNet | fine-tune | Gastro-BaseNet | freeze | 0.0981 | |
| ImageNet | fine-tune | Gastro-BaseNet | fine-tune | 0.8909 | |
| Gastro-BaseNet | fine-tune | Gastro-BaseNet | freeze | 0.0270 | |
| Model Architecture | Hyperparameters | Image-Level Performance (%) | Patient-Level Performance (%) | Localization Performance (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Training Mode | Pretrained Weight | Accuracy | Sensitivity | Specificity | F1 Score | AUC | Accuracy | Sensitivity | Specificity | Sensitivity | |
| ResNet50 | freeze | ImageNet | 88.24 | 71.19 | 92.09 | 92.74 | 88.95 | 87.50 | 76.47 | 100.0 | 68.21 |
| fine-tune | ImageNet | 88.54 | 63.79 | 94.14 | 93.06 | 88.90 | 87.50 | 76.47 | 100.0 | 64.52 | |
| freeze | Gastro-BaseNet | 92.03 | 76.54 | 95.53 | 95.14 | 92.76 | 92.31 | 85.71 | 100.0 | 80.11 | |
| fine-tune | Gastro-BaseNet | 92.72 | 71.60 | 97.49 | 95.62 | 93.82 | 88.89 | 78.79 | 100.0 | 90.80 | |
| EfficientNetB0 | freeze | ImageNet | 88.54 | 63.79 | 94.14 | 93.06 | 83.65 | 87.50 | 76.47 | 100.0 | 64.52 |
| fine-tune | ImageNet | 88.62 | 63.79 | 94.23 | 93.11 | 81.97 | 85.71 | 72.73 | 100.0 | 72.90 | |
| freeze | Gastro-BaseNet | 74.51 | 95.06 | 69.86 | 81.72 | 90.84 | 89.39 | 100.0 | 76.67 | 81.82 | |
| fine-tune | Gastro-BaseNet | 83.76 | 76.54 | 85.40 | 89.56 | 90.04 | 88.89 | 78.79 | 100.0 | 68.28 | |
| Model Architecture | Variable 1 | Variable 2 | p-Value | ||
|---|---|---|---|---|---|
| Training Mode | Pretrained Weight | Training Mode | Pretrained Weight | ||
| ResNet50 | ImageNet | freeze | ImageNet | fine-tune | 0.4423 |
| ImageNet | freeze | Gastro-BaseNet | freeze | 0.0005 | |
| ImageNet | freeze | Gastro-BaseNet | fine-tune | <0.0001 | |
| ImageNet | fine-tune | Gastro-BaseNet | freeze | 0.0165 | |
| ImageNet | fine-tune | Gastro-BaseNet | fine-tune | 0.0004 | |
| Gastro-BaseNet | fine-tune | Gastro-BaseNet | freeze | 0.0280 | |
| EfficientNetB0 | ImageNet | freeze | ImageNet | fine-tune | 0.3943 |
| ImageNet | freeze | Gastro-BaseNet | freeze | <0.0001 | |
| ImageNet | freeze | Gastro-BaseNet | fine-tune | <0.0001 | |
| ImageNet | fine-tune | Gastro-BaseNet | freeze | <0.0001 | |
| ImageNet | fine-tune | Gastro-BaseNet | fine-tune | <0.0001 | |
| Gastro-BaseNet | fine-tune | Gastro-BaseNet | freeze | 0.4122 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, G.P.; Kim, Y.J.; Park, D.K.; Kim, Y.J.; Han, S.K.; Kim, K.G. Gastro-BaseNet: A Specialized Pre-Trained Model for Enhanced Gastroscopic Data Classification and Diagnosis of Gastric Cancer and Ulcer. Diagnostics 2024, 14, 75. https://doi.org/10.3390/diagnostics14010075
Lee GP, Kim YJ, Park DK, Kim YJ, Han SK, Kim KG. Gastro-BaseNet: A Specialized Pre-Trained Model for Enhanced Gastroscopic Data Classification and Diagnosis of Gastric Cancer and Ulcer. Diagnostics. 2024; 14(1):75. https://doi.org/10.3390/diagnostics14010075
Chicago/Turabian StyleLee, Gi Pyo, Young Jae Kim, Dong Kyun Park, Yoon Jae Kim, Su Kyeong Han, and Kwang Gi Kim. 2024. "Gastro-BaseNet: A Specialized Pre-Trained Model for Enhanced Gastroscopic Data Classification and Diagnosis of Gastric Cancer and Ulcer" Diagnostics 14, no. 1: 75. https://doi.org/10.3390/diagnostics14010075
APA StyleLee, G. P., Kim, Y. J., Park, D. K., Kim, Y. J., Han, S. K., & Kim, K. G. (2024). Gastro-BaseNet: A Specialized Pre-Trained Model for Enhanced Gastroscopic Data Classification and Diagnosis of Gastric Cancer and Ulcer. Diagnostics, 14(1), 75. https://doi.org/10.3390/diagnostics14010075







