Automated Classification of Physiologic, Glaucomatous, and Glaucoma-Suspected Optic Discs Using Machine Learning
Abstract
1. Introduction
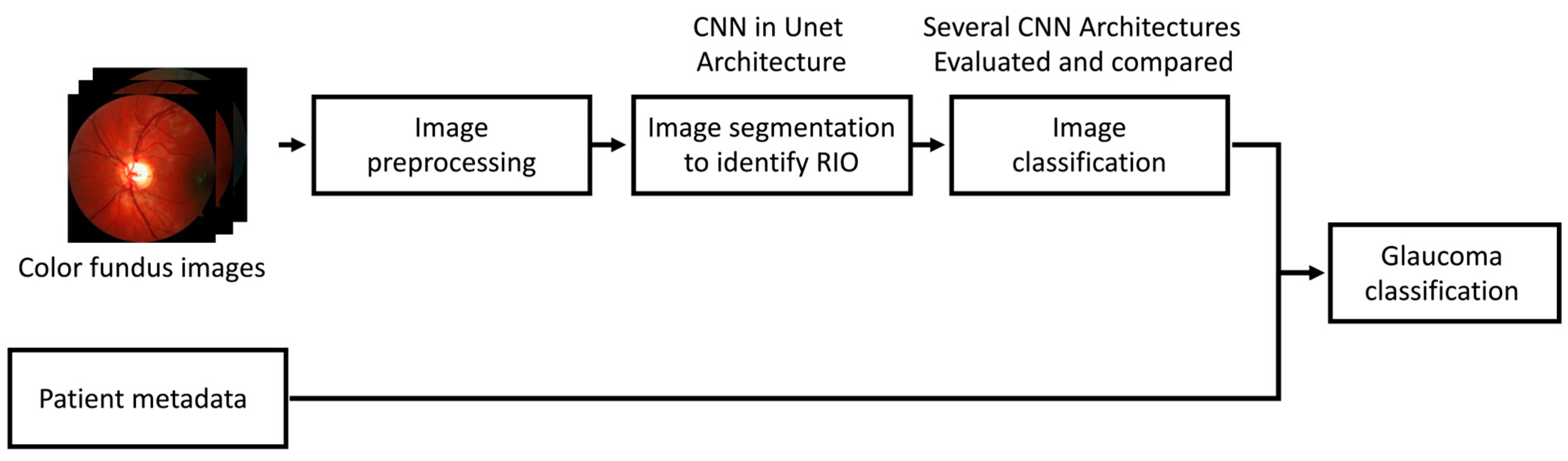
2. Materials and Methods
2.1. Data Acquisition and Ground Truth Labeling
2.2. Data Processing
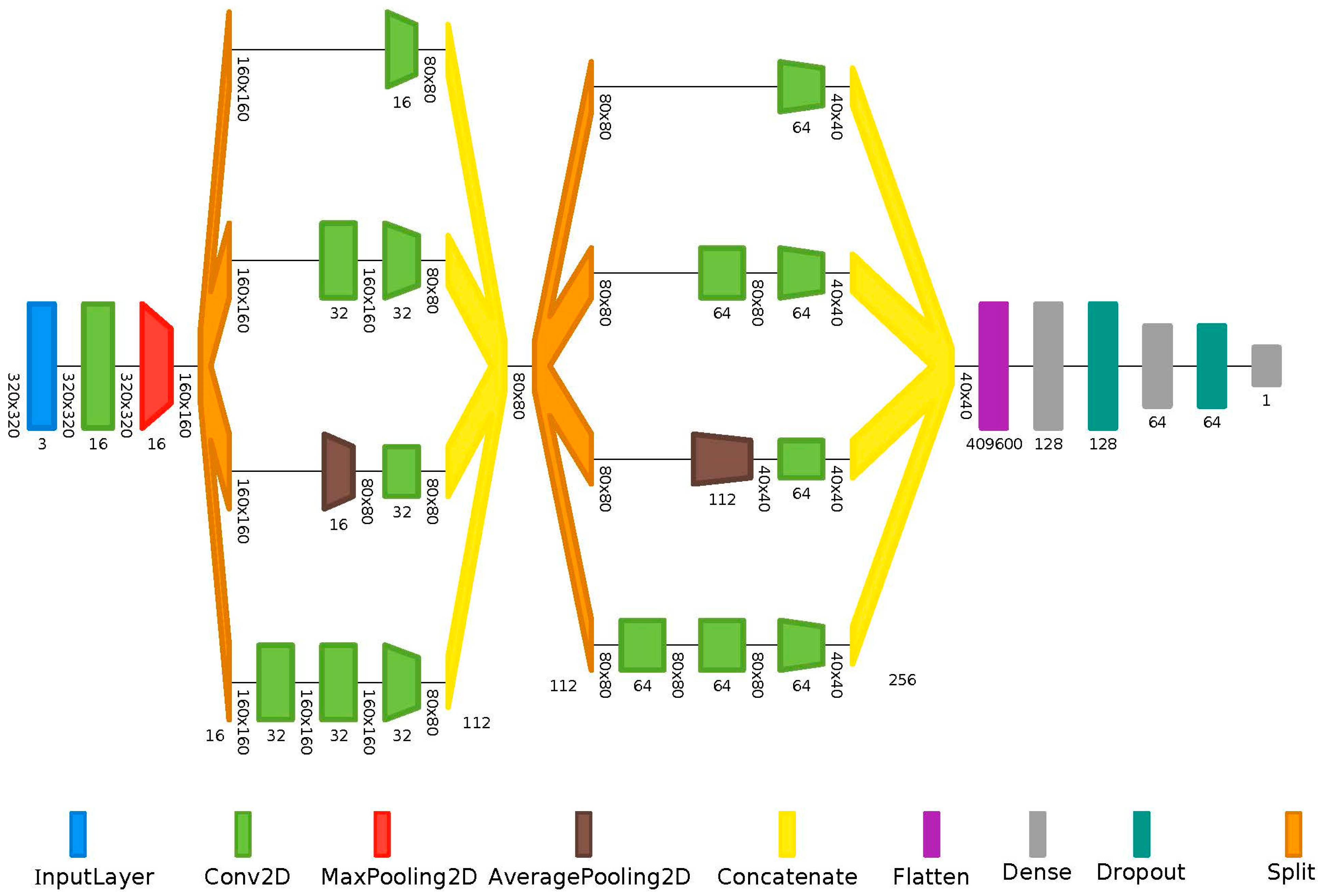
2.3. Machine Learning Workflow
2.4. Statistics
3. Results
3.1. Demographic Data
3.2. Performance of the Algorithm
4. Discussion
Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alencar, L.M.; Zangwill, L.M.; Weinreb, R.N.; Bowd, C.; Sample, P.A.; Girkin, C.A.; Liebmann, J.M.; Medeiros, F.A. A comparison of rates of change in neuroretinal rim area and retinal nerve fiber layer thickness in progressive glaucoma. Investig. Ophthalmol. Vis. Sci. 2010, 51, 3531–3539. [Google Scholar] [CrossRef]
- Medeiros, F.A.; Zangwill, L.M.; Bowd, C.; Mansouri, K.; Weinreb, R.N. The structure and function relationship in glaucoma: Implications for detection of progression and measurement of rates of change. Investig. Ophthalmol. Vis. Sci. 2012, 53, 6939–6946. [Google Scholar] [CrossRef] [PubMed]
- Kass, M.A.; Heuer, D.K.; Higginbotham, E.J.; Johnson, C.A.; Keltner, J.L.; Miller, J.P.; Parrish, R.K., 2nd; Wilson, M.R.; Gordon, M.O. The Ocular Hypertension Treatment Study: A randomized trial determines that topical ocular hypotensive medication delays or prevents the onset of primary open-angle glaucoma. Arch. Ophthalmol. 2002, 120, 701–713; discussion 829–730. [Google Scholar] [CrossRef]
- Anderson, D.R. Collaborative normal tension glaucoma study. Curr. Opin. Ophthalmol. 2003, 14, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Leske, M.C.; Heijl, A.; Hussein, M.; Bengtsson, B.; Hyman, L.; Komaroff, E. Factors for glaucoma progression and the effect of treatment: The early manifest glaucoma trial. Arch. Ophthalmol. 2003, 121, 48–56. [Google Scholar] [CrossRef]
- Stein, J.D.; Khawaja, A.P.; Weizer, J.S. Glaucoma in Adults-Screening, Diagnosis, and Management: A Review. JAMA 2021, 325, 164–174. [Google Scholar] [CrossRef]
- Hoffmann, E.M.; Zangwill, L.M.; Crowston, J.G.; Weinreb, R.N. Optic disk size and glaucoma. Surv. Ophthalmol. 2007, 52, 32–49. [Google Scholar] [CrossRef]
- Jonas, J.B.; Schmidt, A.M.; Müller-Bergh, J.A.; Schlötzer-Schrehardt, U.M.; Naumann, G.O. Human optic nerve fiber count and optic disc size. Investig. Ophthalmol. Vis. Sci. 1992, 33, 2012–2018. [Google Scholar]
- Okimoto, S.; Yamashita, K.; Shibata, T.; Kiuchi, Y. Morphological features and important parameters of large optic discs for diagnosing glaucoma. PLoS ONE 2015, 10, e0118920. [Google Scholar] [CrossRef]
- Senthil, S.; Nakka, M.; Sachdeva, V.; Goyal, S.; Sahoo, N.; Choudhari, N. Glaucoma Mimickers: A major review of causes, diagnostic evaluation, and recommendations. Semin. Ophthalmol. 2021, 36, 692–712. [Google Scholar] [CrossRef]
- Leske, M.C.; Heijl, A.; Hyman, L.; Bengtsson, B.; Dong, L.; Yang, Z. Predictors of long-term progression in the early manifest glaucoma trial. Ophthalmology 2007, 114, 1965–1972. [Google Scholar] [CrossRef] [PubMed]
- Zedan, M.J.M.; Zulkifley, M.A.; Ibrahim, A.A.; Moubark, A.M.; Kamari, N.A.M.; Abdani, S.R. Automated Glaucoma Screening and Diagnosis Based on Retinal Fundus Images Using Deep Learning Approaches: A Comprehensive Review. Diagnostics 2023, 13, 2180. [Google Scholar] [CrossRef] [PubMed]
- Singh, L.K.; Khanna, M.; Garg, H.; Singh, R. Emperor penguin optimization algorithm-and bacterial foraging optimization algorithm-based novel feature selection approach for glaucoma classification from fundus images. Soft Comput. 2024, 28, 2431–2467. [Google Scholar] [CrossRef]
- Singh, L.K.; Garg, H.; Pooja. Automated Glaucoma Type Identification Using Machine Learning or Deep Learning Techniques. In Advancement of Machine Intelligence in Interactive Medical Image Analysis; Verma, O.P., Roy, S., Pandey, S.C., Mittal, M., Eds.; Springer: Singapore, 2020; pp. 241–263. [Google Scholar]
- Lee, E.B.; Wang, S.Y.; Chang, R.T. Interpreting Deep Learning Studies in Glaucoma: Unresolved Challenges. Asia-Pac. J. Ophthalmol. 2021, 10, 261–267. [Google Scholar] [CrossRef]
- Gedde, S.J.; Lind, J.T.; Wright, M.M.; Chen, P.P.; Muir, K.W.; Vinod, K.; Li, T.; Mansberger, S.L. Primary open-angle glaucoma suspect preferred practice pattern®. Ophthalmology 2021, 128, P151–P192. [Google Scholar] [CrossRef] [PubMed]
- Issac, A.; Sarathi, M.P.; Dutta, M.K. An adaptive threshold based image processing technique for improved glaucoma detection and classification. Comput. Methods Programs Biomed. 2015, 122, 229–244. [Google Scholar] [CrossRef] [PubMed]
- Salam, A.A.; Khalil, T.; Akram, M.U.; Jameel, A.; Basit, I. Automated detection of glaucoma using structural and non structural features. Springerplus 2016, 5, 1519. [Google Scholar] [CrossRef]
- Tielsch, J.M.; Katz, J.; Singh, K.; Quigley, H.A.; Gottsch, J.D.; Javitt, J.; Sommer, A. A population-based evaluation of glaucoma screening: The Baltimore Eye Survey. Am. J. Epidemiol. 1991, 134, 1102–1110. [Google Scholar] [CrossRef] [PubMed]
- Muramatsu, C.; Hayashi, Y.; Sawada, A.; Hatanaka, Y.; Hara, T.; Yamamoto, T.; Fujita, H. Detection of retinal nerve fiber layer defects on retinal fundus images for early diagnosis of glaucoma. J. Biomed. Opt. 2010, 15, 016021. [Google Scholar] [CrossRef]
- Wang, F.; Casalino, L.P.; Khullar, D. Deep learning in medicine—Promise, progress, and challenges. JAMA Intern. Med. 2019, 179, 293–294. [Google Scholar] [CrossRef]
- Bengtsson, B.; Andersson, S.; Heijl, A. Performance of time-domain and spectral-domain optical coherence tomography for glaucoma screening. Acta Ophthalmol. 2012, 90, 310–315. [Google Scholar] [CrossRef]
- Mutasa, S.; Sun, S.; Ha, R. Understanding artificial intelligence based radiology studies: What is overfitting? Clin. Imaging 2020, 65, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Yoo, T.K.; Hong, S. Artificial Neural Network Approach for Differentiating Open-Angle Glaucoma From Glaucoma Suspect Without a Visual Field Test. Investig. Ophthalmol. Vis. Sci. 2015, 56, 3957–3966. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.B.; Cho, H.K. Deep learning classification of early normal-tension glaucoma and glaucoma suspects using Bruch’s membrane opening-minimum rim width and RNFL. Sci. Rep. 2020, 10, 19042. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, H.; Fuchs, T.J.; De Cuir, N.; De Moraes, C.G.; Blumberg, D.M.; Liebmann, J.M.; Ritch, R.; Hood, D.C. Hybrid Deep Learning on Single Wide-field Optical Coherence tomography Scans Accurately Classifies Glaucoma Suspects. J. Glaucoma 2017, 26, 1086–1094. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015, Proceedings, Part III 18; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Li, Z.; He, Y.; Keel, S.; Meng, W.; Chang, R.T.; He, M. Efficacy of a Deep Learning System for Detecting Glaucomatous Optic Neuropathy Based on Color Fundus Photographs. Ophthalmology 2018, 125, 1199–1206. [Google Scholar] [CrossRef]
- Liu, H.; Li, L.; Wormstone, I.M.; Qiao, C.; Zhang, C.; Liu, P.; Li, S.; Wang, H.; Mou, D.; Pang, R.; et al. Development and Validation of a Deep Learning System to Detect Glaucomatous Optic Neuropathy Using Fundus Photographs. JAMA Ophthalmol. 2019, 137, 1353–1360. [Google Scholar] [CrossRef]
- Varma, R.; Steinmann, W.C.; Scott, I.U. Expert agreement in evaluating the optic disc for glaucoma. Ophthalmology 1992, 99, 215–221. [Google Scholar] [CrossRef]
- Abrams, L.S.; Scott, I.U.; Spaeth, G.L.; Quigley, H.A.; Varma, R. Agreement among optometrists, ophthalmologists, and residents in evaluating the optic disc for glaucoma. Ophthalmology 1994, 101, 1662–1667. [Google Scholar] [CrossRef]
- Jampel, H.D.; Friedman, D.; Quigley, H.; Vitale, S.; Miller, R.; Knezevich, F.; Ding, Y. Agreement among glaucoma specialists in assessing progressive disc changes from photographs in open-angle glaucoma patients. Am. J. Ophthalmol. 2009, 147, 39–44.e31. [Google Scholar] [CrossRef]
- Chan, H.H.; Ong, D.N.; Kong, Y.X.; O’Neill, E.C.; Pandav, S.S.; Coote, M.A.; Crowston, J.G. Glaucomatous optic neuropathy evaluation (GONE) project: The effect of monoscopic versus stereoscopic viewing conditions on optic nerve evaluation. Am. J. Ophthalmol. 2014, 157, 936–944. [Google Scholar] [CrossRef] [PubMed]
- Trobe, J.D.; Glaser, J.S.; Cassady, J.; Herschler, J.; Anderson, D.R. Nonglaucomatous excavation of the optic disc. Arch. Ophthalmol. 1980, 98, 1046–1050. [Google Scholar] [CrossRef]
- Kupersmith, M.J.; Krohn, D. Cupping of the optic disc with compressive lesions of the anterior visual pathway. Ann. Ophthalmol. 1984, 16, 948–953. [Google Scholar] [PubMed]
- Mursch-Edlmayr, A.S.; Ng, W.S.; Diniz-Filho, A.; Sousa, D.C.; Arnold, L.; Schlenker, M.B.; Duenas-Angeles, K.; Keane, P.A.; Crowston, J.G.; Jayaram, H. Artificial Intelligence Algorithms to Diagnose Glaucoma and Detect Glaucoma Progression: Translation to Clinical Practice. Transl. Vis. Sci. Technol. 2020, 9, 55. [Google Scholar] [CrossRef]
- Ipp, E.; Liljenquist, D.; Bode, B.; Shah, V.N.; Silverstein, S.; Regillo, C.D.; Lim, J.I.; Sadda, S.; Domalpally, A.; Gray, G.; et al. Pivotal Evaluation of an Artificial Intelligence System for Autonomous Detection of Referrable and Vision-Threatening Diabetic Retinopathy. JAMA Netw. Open 2021, 4, e2134254. [Google Scholar] [CrossRef]
- Noury, E.; Mannil, S.S.; Chang, R.T.; Ran, A.R.; Cheung, C.Y.; Thapa, S.S.; Rao, H.L.; Dasari, S.; Riyazuddin, M.; Chang, D.; et al. Deep Learning for Glaucoma Detection and Identification of Novel Diagnostic Areas in Diverse Real-World Datasets. Transl. Vis. Sci. Technol. 2022, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Medeiros, F.A.; Jammal, A.A.; Mariottoni, E.B. Detection of Progressive Glaucomatous Optic Nerve Damage on Fundus Photographs with Deep Learning. Ophthalmology 2021, 128, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Yan, L.; Wang, Y.; Shi, J.; Chen, H.; Zhang, X.; Jiang, M.; Wu, Z.; Zhou, K. Deep learning-based automated detection of glaucomatous optic neuropathy on color fundus photographs. Graefe’s Arch. Clin. Exp. Ophthalmol. 2020, 258, 851–867. [Google Scholar] [CrossRef] [PubMed]
- Bhuiyan, A.; Govindaiah, A.; Smith, R.T. An Artificial-Intelligence- and Telemedicine-Based Screening Tool to Identify Glaucoma Suspects from Color Fundus Imaging. J. Ophthalmol. 2021, 2021, 6694784. [Google Scholar] [CrossRef]
- Atalay, E.; Özalp, O.; Devecioğlu, Ö.C.; Erdoğan, H.; İnce, T.; Yıldırım, N. Investigation of the Role of Convolutional Neural Network Architectures in the Diagnosis of Glaucoma using Color Fundus Photography. Turk. J. Ophthalmol. 2022, 52, 193–200. [Google Scholar] [CrossRef]
- Li, F.; Pan, J.; Yang, D.; Wu, J.; Ou, Y.; Li, H.; Huang, J.; Xie, H.; Ou, D.; Wu, X.; et al. A Multicenter Clinical Study of the Automated Fundus Screening Algorithm. Transl. Vis. Sci. Technol. 2022, 11, 22. [Google Scholar] [CrossRef] [PubMed]
- Grewal, D.S.; Jain, R.; Grewal, S.P.; Rihani, V. Artificial neural network-based glaucoma diagnosis using retinal nerve fiber layer analysis. Eur. J. Ophthalmol. 2008, 18, 915–921. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.H.; Maetschke, S.R.; Antony, B.J.; Ishikawa, H.; Wollstein, G.; Schuman, J.S.; Garnavi, R. Estimating Global Visual Field Indices in Glaucoma by Combining Macula and Optic Disc OCT Scans Using 3-Dimensional Convolutional Neural Networks. Ophthalmol. Glaucoma 2021, 4, 102–112. [Google Scholar] [CrossRef]
- Seo, S.B.; Cho, H.K. Deep learning classification of early normal-tension glaucoma and glaucoma suspect eyes using Bruch’s membrane opening-based disc photography. Front. Med. 2022, 9, 1037647. [Google Scholar] [CrossRef] [PubMed]
| Group | 1 = GOD | 2 = GSOD | 3 = POD |
|---|---|---|---|
| n | 336 | 310 | 321 |
| Group | GOD (1) | GSOD (2) | POD (3) | 1 vs. 2 p | 1 vs. 3 p | 2 vs. 3 p |
|---|---|---|---|---|---|---|
| Value | M ± SD | M ± SD | M ± SD | |||
| n | 336 | 310 | 321 | |||
| Age (years) * | 67 ± 13 69 (62; 78) | 35 ± 20 28 (18; 53) | 48 ± 19 48 (29; 59) | <0.001 | <0.001 | >0.05 |
| Visual acuity (decimal) | 0.68 ± 0.30 0.70 (0.50; 0.80) | 0.90 ± 0.20 1.00 (0.80; 1.00) | 0.81 ± 0.27 0.80 (0.63; 1.00) | >0.05 | >0.05 | >0.05 |
| Spherical equivalent (dpt.) | −0.81 ± 2.00 −0.38 (−2.00; 0.50) | −0.61 ± 2.48 0.00 (−1.60; 0.60) | −0.44 ± 2.38 −0.13 (−1.00; 0.75) | >0.05 | >0.05 | >0.05 |
| MD (dB) | −7.96 ± 9.82 −4.62 (−12.52; −1.28) | −1.01 ± 2.15 −0.77 (−1.99; 0.39) | −3.34 ± 6.41 −1.41 (−3.52; 0.24) | <0.001 | <0.001 | >0.05 |
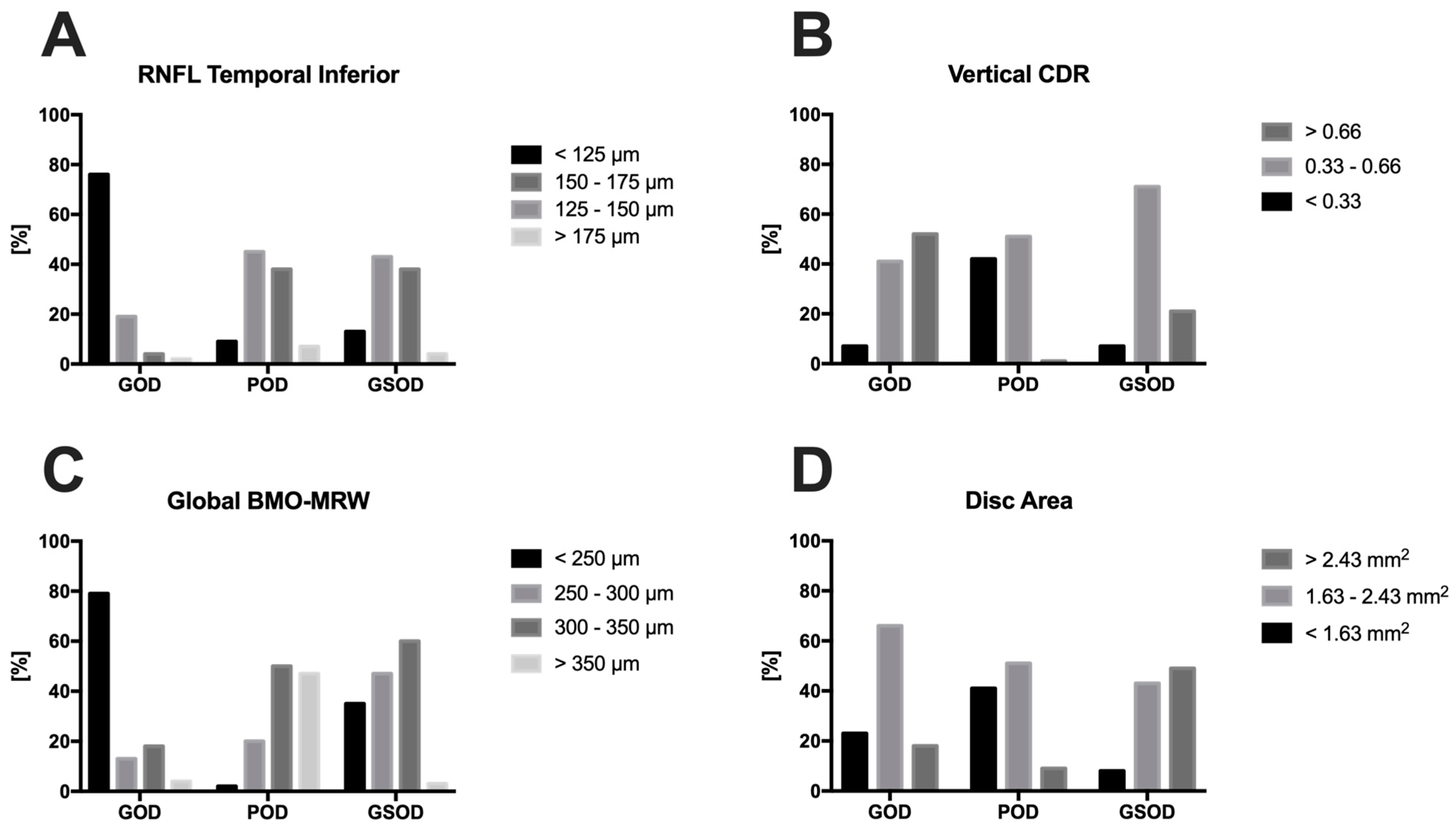
| CDR horizontal | 0.66 ± 0.20 0.67 (0.55; 0.80) | 0.56 ± 0.16 0.59 (0.50; 0.66) | 0.33 ± 0.20 0.36 (0.20; 0.45) | >0.05 | 0.01 | >0.05 |
| CDR vertical | 0.68 ± 0.20 0.69 (0.55; 0.85) | 0.53 ± 0.24 0.55 (0.50; 0.61) | 0.32 ± 0.20 0.33 (0.20; 0.43) | 0.03 | 0.002 | >0.05 |
| BMO area (mm2) | 1.97 ± 0.43 1.94 (1.68; 2.27) | 2.57 ± 0.57 2.52 (2.21; 2.90) | 1.93 ± 0.43 1.88 (1.68; 2.16) | <0.001 | >0.05 | <0.001 |
| Global BMO-MRW (µm) | 197 ± 77 192 (141; 239) | 264 ± 40 261 (239; 288) | 352 ± 68 347 (305; 389) | <0.001 | <0.001 | <0.001 |
| Global RNFL (µm) | 69 ± 19 69 (54; 82) | 96 ± 11 98 (90; 104) | 99 ± 10 100 (92; 105) | <0.001 | <0.001 | >0.05 |
| NS (µm) | 78 ± 28 77 (57; 98) | 108 ± 23 109 (93; 125) | 113 ± 22 113 (98; 127) | <0.001 | <0.001 | >0.05 |
| N (µm) | 59 ± 19 60 (47; 72) | 80 ± 13 80 (72; 89) | 81 ± 14 81 (71; 91) | <0.001 | <0.001 | >0.05 |
| NI (µm) | 78 ± 27 76 (57; 96) | 110 ± 26 112 (94; 127) | 110 ± 24 109 (94; 126) | <0.001 | <0.001 | >0.05 |
| TI (µm) | 91 ± 40 87 (58; 123) | 143 ± 23 146 (133; 158) | 151 ± 56 147 (137; 160) | <0.001 | <0.001 | >0.05 |
| T (µm) | 54 ± 17 53 (42; 66) | 68 ± 10 69 (61; 75) | 70 ± 12 70 (62; 76) | <0.001 | <0.001 | >0.05 |
| TS (µm) | 88 ± 34 87 (58; 113) | 127 ± 23 129 (114; 144) | 134 ± 20 135 (122; 147) | <0.001 | <0.001 | >0.05 |
| Disc area (mm2) | 1.89 ± 0.42 1.83 (1.63; 2.14) | 2.37 ± 0.80 2.43 (2.07; 2.73) | 1.80 ± 0.49 1.79 (1.48; 2.13) | <0.001 | <0.05 | <0.001 |
| ML Performance | TP | TN | FP | FN | Sn | Sp | Acc. | Pr. | F1 | AUC |
|---|---|---|---|---|---|---|---|---|---|---|
| GOD vs. POD | 59 | 47 | 17 | 5 | 92% | 73% | 83% | 78% | 84% | 0.88 |
| GOD vs. GSOD | 48 | 47 | 15 | 14 | 77% | 76% | 77% | 76% | 77% | 0.84 |
| ML Performance | TP | TN | FP | FN | Sn | Sp | Acc. | Pr. | F1 | AUC |
|---|---|---|---|---|---|---|---|---|---|---|
| GOD vs. POD | 60 | 60 | 4 | 4 | 94% | 93% | 94% | 94% | 94% | 0.99 |
| GOD vs. GSOD | 52 | 56 | 6 | 10 | 84% | 90% | 87% | 89% | 86% | 0.92 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diener, R.; Renz, A.W.; Eckhard, F.; Segbert, H.; Eter, N.; Malcherek, A.; Biermann, J. Automated Classification of Physiologic, Glaucomatous, and Glaucoma-Suspected Optic Discs Using Machine Learning. Diagnostics 2024, 14, 1073. https://doi.org/10.3390/diagnostics14111073
Diener R, Renz AW, Eckhard F, Segbert H, Eter N, Malcherek A, Biermann J. Automated Classification of Physiologic, Glaucomatous, and Glaucoma-Suspected Optic Discs Using Machine Learning. Diagnostics. 2024; 14(11):1073. https://doi.org/10.3390/diagnostics14111073
Chicago/Turabian StyleDiener, Raphael, Alexander W. Renz, Florian Eckhard, Helmar Segbert, Nicole Eter, Arnim Malcherek, and Julia Biermann. 2024. "Automated Classification of Physiologic, Glaucomatous, and Glaucoma-Suspected Optic Discs Using Machine Learning" Diagnostics 14, no. 11: 1073. https://doi.org/10.3390/diagnostics14111073
APA StyleDiener, R., Renz, A. W., Eckhard, F., Segbert, H., Eter, N., Malcherek, A., & Biermann, J. (2024). Automated Classification of Physiologic, Glaucomatous, and Glaucoma-Suspected Optic Discs Using Machine Learning. Diagnostics, 14(11), 1073. https://doi.org/10.3390/diagnostics14111073





