Artificial Intelligence in Point-of-Care Biosensing: Challenges and Opportunities
Abstract
1. Introduction
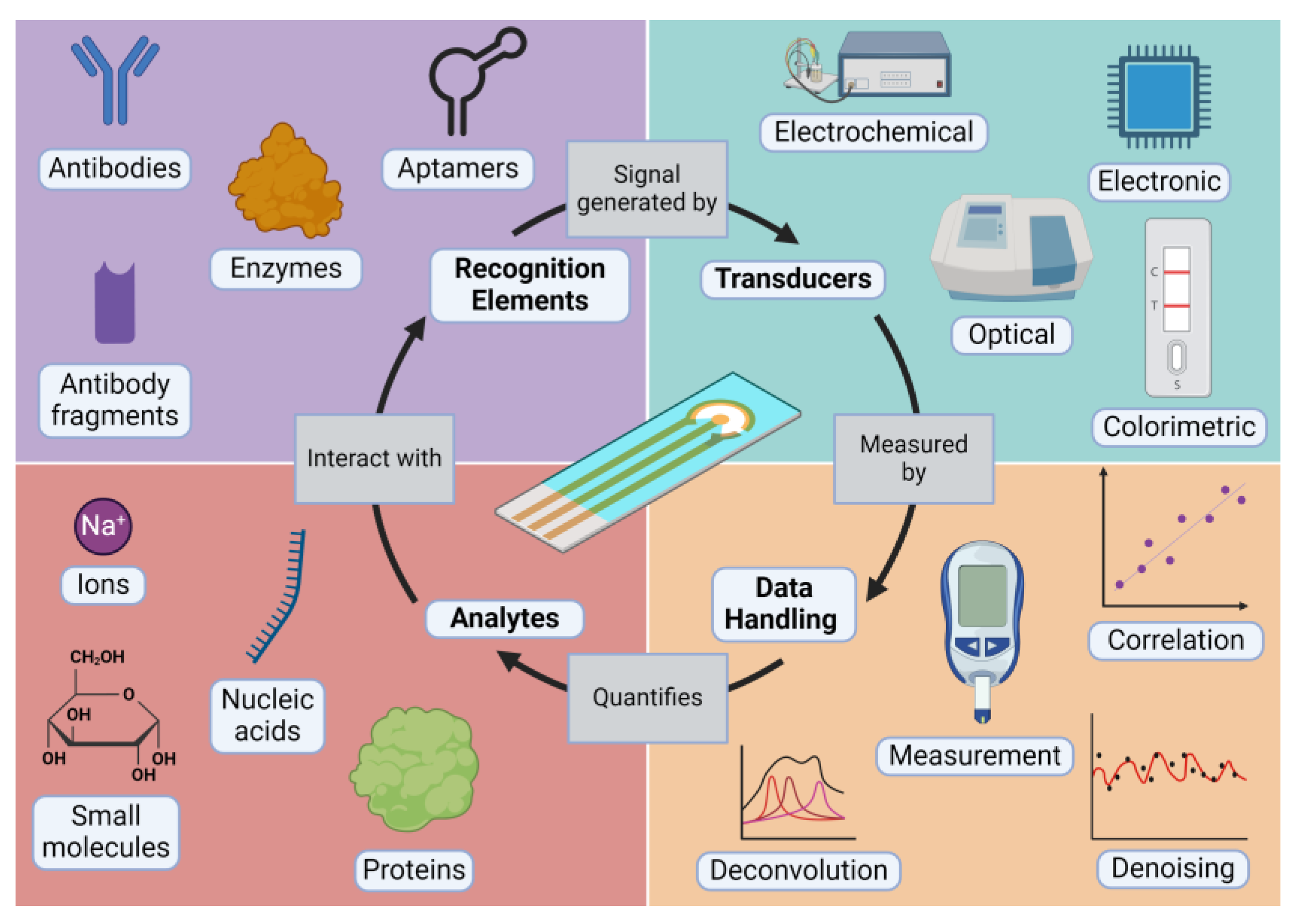
2. What Is a Biosensor?
2.1. Analytes
2.2. Recognition Elements
2.3. Transducers
2.4. Data Handling
3. Sensing at the Point-of-Care
3.1. Sensing at the Bedside
3.2. Sensing at Home
3.3. Sensing in the Field
3.4. Challenges in POC Sensing
4. Artificial Intelligence: A Brief Overview
5. Artificial Intelligence in Biosensing
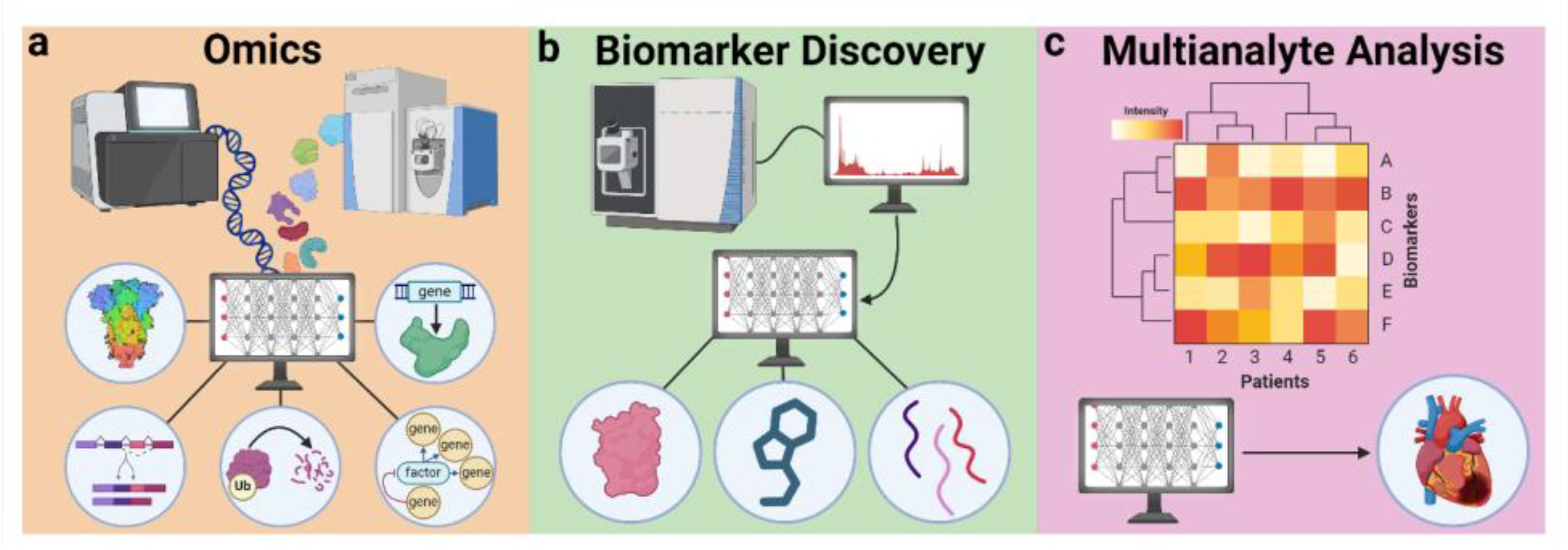
5.1. AI in Analyte Selection
5.1.1. Omics
5.1.2. Biomarker Discovery
5.1.3. Multianalyte Analysis
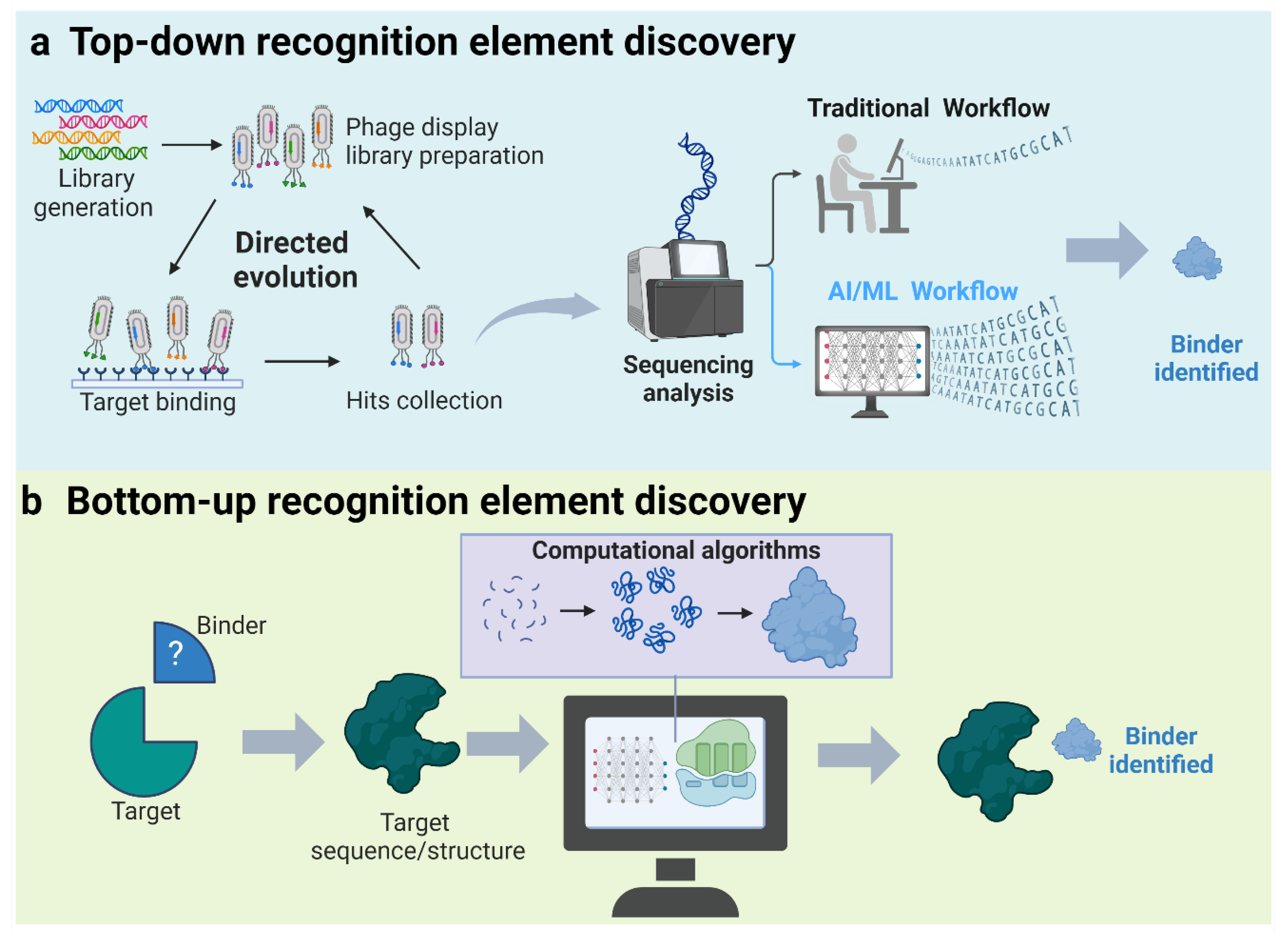
5.2. AI in Recognition Element Selection
5.2.1. Top-Down Discovery of Recognition Elements
5.2.2. Bottom-Up Discovery of Recognition Elements
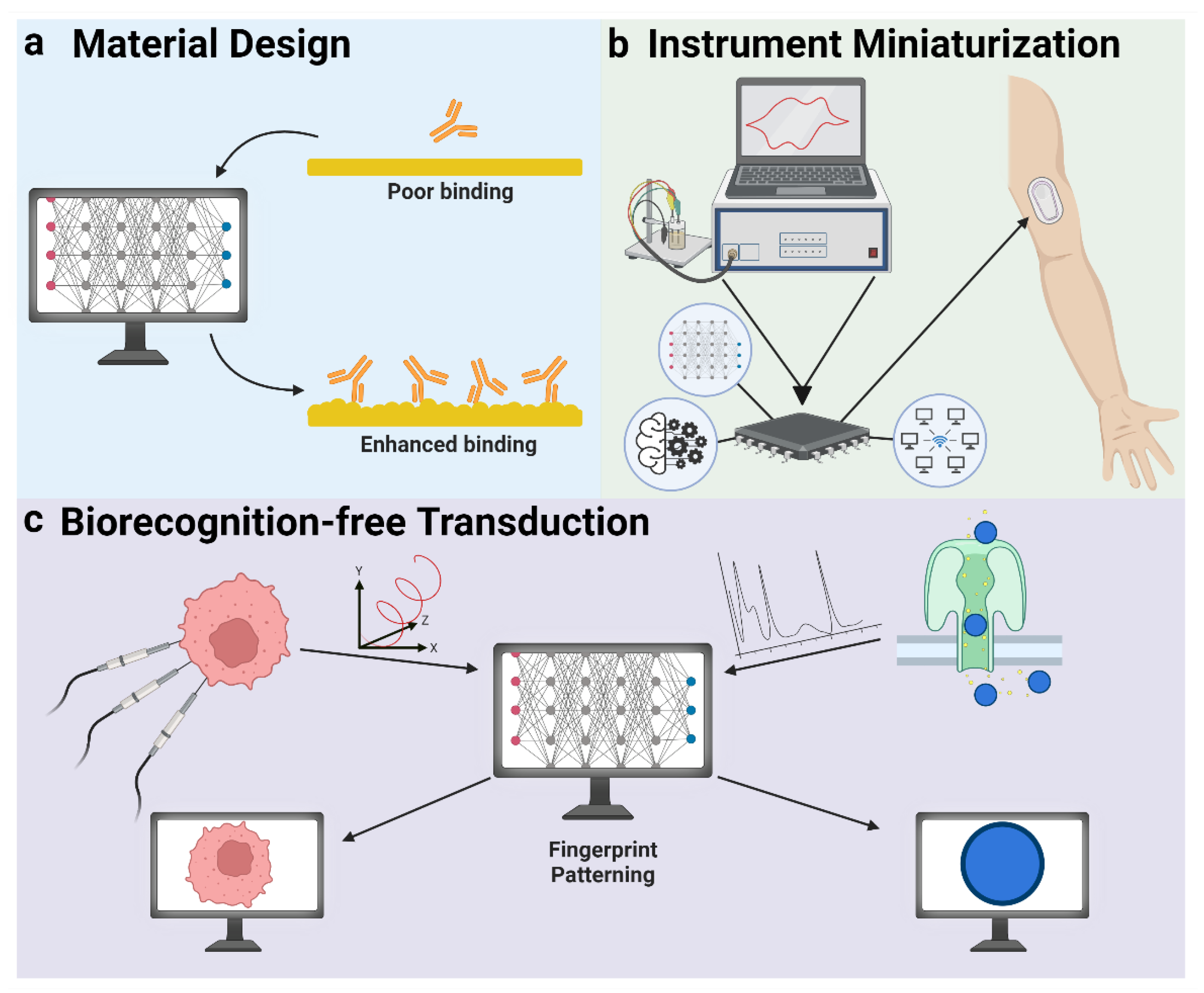
5.3. AI in Transduction
5.3.1. Material Design
5.3.2. Instrument Miniaturization
5.3.3. Biorecognition-Free Transduction
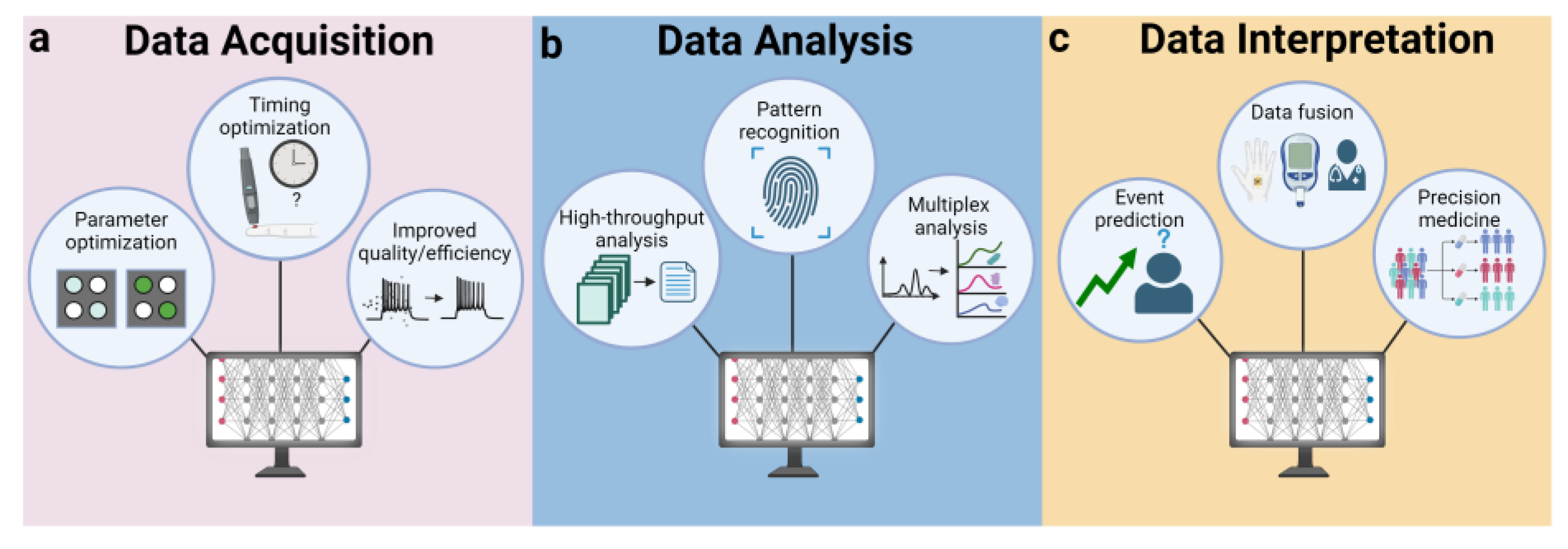
5.4. AI in Data Handling
5.4.1. Data Acquisition
5.4.2. Data Analysis
5.4.3. Data Interpretation
5.5. Artificial Intelligence Workflow
6. Challenges in AI POC Biosensing
7. Future Outlooks for AI-Assisted Biosensing
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gubala, V.; Harris, L.F.; Ricco, A.J.; Tan, M.X.; Williams, D.E. Point of Care Diagnostics: Status and Future. Anal. Chem. 2012, 84, 487–515. [Google Scholar] [CrossRef] [PubMed]
- Flynn, C.D.; Chang, D.; Mahmud, A.; Yousefi, H.; Das, J.; Riordan, K.T.; Sargent, E.H.; Kelley, S.O. Biomolecular Sensors for Advanced Physiological Monitoring. Nat. Rev. Bioeng. 2023, 1, 560–575. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zeng, J.; Wang, C.; Feng, L.; Song, Z.; Zhao, W.; Wang, Q.; Liu, C. The Application of Wearable Glucose Sensors in Point-of-Care Testing. Front. Bioeng. Biotechnol. 2021, 9, 774210. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Tian, F.; Liu, X.; Zhou, Q.; Pan, J.; Luo, Z.; Yang, M.; Yi, C. Virus Detection: From State-of-the-art Laboratories to Smartphone-based Point-of-care Testing. Adv. Sci. 2022, 9, 2105904. [Google Scholar] [CrossRef] [PubMed]
- Collinson, P. Cardiac Biomarker Measurement by Point of Care Testing—Development, Rationale, Current State and Future Developments. Clin. Chim. Acta 2020, 508, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Kline, R.R. Cybernetics, Automata Studies, and the Dartmouth Conference on Artificial Intelligence. IEEE Ann. Hist. Comput. 2011, 33, 5–16. [Google Scholar] [CrossRef]
- Roumeliotis, K.I.; Tselikas, N.D. ChatGPT and Open-AI Models: A Preliminary Review. Future Internet 2023, 15, 192. [Google Scholar] [CrossRef]
- Li, L.; Wen, Y.; Xu, L.; Xu, Q.; Song, S.; Zuo, X.; Yan, J.; Zhang, W.; Liu, G. Development of Mercury (II) Ion Biosensors Based on Mercury-Specific Oligonucleotide Probes. Biosens. Bioelectron. 2016, 75, 433–445. [Google Scholar] [CrossRef]
- Li, J.; Si, Y.; Park, Y.E.; Choi, J.-S.; Jung, S.M.; Lee, J.E.; Lee, H.J. A Serotonin Voltammetric Biosensor Composed of Carbon Nanocomposites and DNA Aptamer. Microchim. Acta 2021, 188, 146. [Google Scholar] [CrossRef]
- Ren, Y.; Deng, H.; Shen, W.; Gao, Z. A Highly Sensitive and Selective Electrochemical Biosensor for Direct Detection of MicroRNAs in Serum. Anal. Chem. 2013, 85, 4784–4789. [Google Scholar] [CrossRef]
- Mahmud, A.; Chang, D.; Das, J.; Gomis, S.; Foroutan, F.; Chen, J.B.; Pandey, L.; Flynn, C.D.; Yousefi, H.; Geraili, A.; et al. Monitoring Cardiac Biomarkers with Aptamer-based Molecular Pendulum Sensors. Angew. Chem. 2023, 135, e202213567. [Google Scholar] [CrossRef]
- Yousefi, H.; Mahmud, A.; Chang, D.; Das, J.; Gomis, S.; Chen, J.B.; Wang, H.; Been, T.; Yip, L.; Coomes, E.; et al. Detection of SARS-CoV-2 Viral Particles Using Direct, Reagent-Free Electrochemical Sensing. J. Am. Chem. Soc. 2021, 143, 1722–1727. [Google Scholar] [CrossRef] [PubMed]
- Flynn, C.D.; Sandomierski, M.; Kim, K.; Lewis, J.; Lloyd, V.; Ignaszak, A. Electrochemical Detection of Borrelia Burgdorferi Using a Biomimetic Flow Cell System. ACS Meas. Sci. Au 2023, 3, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.N.; Retterer, S.; Zieziulewicz, T.J.; Isaacson, M.; Szarowski, D.; Mousseau, D.E.; Lawrence, D.A.; Turner, J.N. On-Chip Micro-Biosensor for the Detection of Human CD4+ Cells Based on AC Impedance and Optical Analysis. Biosens. Bioelectron. 2005, 21, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Griesche, C.; Baeumner, A.J. Biosensors to Support Sustainable Agriculture and Food Safety. TrAC Trends Anal. Chem. 2020, 128, 115906. [Google Scholar] [CrossRef]
- Bruen, D.; Delaney, C.; Florea, L.; Diamond, D. Glucose Sensing for Diabetes Monitoring: Recent Developments. Sensors 2017, 17, 1866. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Byrne, H.; O’Kennedy, R.J. Antibodies and Antibody-Derived Analytical Biosensors. Essays Biochem. 2016, 60, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Weisser, N.E.; Hall, J.C. Applications of Single-Chain Variable Fragment Antibodies in Therapeutics and Diagnostics. Biotechnol. Adv. 2009, 27, 502–520. [Google Scholar] [CrossRef]
- Hosseindokht, M.; Bakherad, H.; Zare, H. Nanobodies: A Tool to Open New Horizons in Diagnosis and Treatment of Prostate Cancer. Cancer Cell Int. 2021, 21, 580. [Google Scholar] [CrossRef]
- Qian, S.; Chang, D.; He, S.; Li, Y. Aptamers from Random Sequence Space: Accomplishments, Gaps and Future Considerations. Anal. Chim. Acta 2022, 1196, 339511. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- McConnell, E.M.; Cozma, I.; Mou, Q.; Brennan, J.D.; Lu, Y.; Li, Y. Biosensing with DNAzymes. Chem. Soc. Rev. 2021, 50, 8954–8994. [Google Scholar] [CrossRef]
- Lohse, P.A.; Szostak, J.W. Ribozyme-Catalysed Amino-Acid Transfer Reactions. Nature 1996, 381, 442–444. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wei, H.; Zhang, Z.; Wang, E.; Dong, S. Nanozyme: An Emerging Alternative to Natural Enzyme for Biosensing and Immunoassay. TrAC Trends Anal. Chem. 2018, 105, 218–224. [Google Scholar] [CrossRef]
- Cieplak, M.; Kutner, W. Artificial Biosensors: How Can Molecular Imprinting Mimic Biorecognition? Trends Biotechnol. 2016, 34, 922–941. [Google Scholar] [CrossRef] [PubMed]
- Kirste, R.; Rohrbaugh, N.; Bryan, I.; Bryan, Z.; Collazo, R.; Ivanisevic, A. Electronic Biosensors Based on III-Nitride Semiconductors. Annu. Rev. Anal. Chem. 2015, 8, 149–169. [Google Scholar] [CrossRef]
- Aldewachi, H.; Chalati, T.; Woodroofe, M.N.; Bricklebank, N.; Sharrack, B.; Gardiner, P. Gold Nanoparticle-Based Colorimetric Biosensors. Nanoscale 2017, 10, 18–33. [Google Scholar] [CrossRef]
- Chang, D.; Li, J.; Liu, R.; Liu, M.; Tram, K.; Schmitt, N.; Li, Y. A Colorimetric Biosensing Platform with Aptamers, Rolling Circle Amplification and Urease-mediated Litmus Test. Angew. Chem. 2023, 135, e202315185. [Google Scholar] [CrossRef]
- Chen, C.; Wang, J. Optical Biosensors: An Exhaustive and Comprehensive Review. Analyst 2020, 145, 1605–1628. [Google Scholar] [CrossRef]
- Fogel, R.; Limson, J.; Seshia, A.A. Acoustic Biosensors. Essays Biochem. 2016, 60, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, K.; Danielsson, B. Principles and Applications of Thermal Biosensors. Biosens. Bioelectron. 2001, 16, 417–423. [Google Scholar] [CrossRef]
- Skládal, P. Piezoelectric Biosensors. TrAC Trends Anal. Chem. 2016, 79, 127–133. [Google Scholar] [CrossRef]
- Zargartalebi, H.; Yousefi, H.; Flynn, C.D.; Gomis, S.; Das, J.; Young, T.L.; Chien, E.; Mubareka, S.; McGeer, A.; Wang, H.; et al. Capillary-Assisted Molecular Pendulum Bioanalysis. J. Am. Chem. Soc. 2022, 144, 18338–18349. [Google Scholar] [CrossRef]
- Downs, A.M.; Gerson, J.; Hossain, M.N.; Ploense, K.; Pham, M.; Kraatz, H.-B.; Kippin, T.; Plaxco, K.W. Nanoporous Gold for the Miniaturization of in Vivo Electrochemical Aptamer-Based Sensors. ACS Sens. 2021, 6, 2299–2306. [Google Scholar] [CrossRef]
- Idili, A.; Parolo, C.; Alvarez-Diduk, R.; Merkoçi, A. Rapid and Efficient Detection of the SARS-CoV-2 Spike Protein Using an Electrochemical Aptamer-Based Sensor. ACS Sens. 2021, 6, 3093–3101. [Google Scholar] [CrossRef] [PubMed]
- Jolly, P.; Formisano, N.; Tkáč, J.; Kasák, P.; Frost, C.G.; Estrela, P. Label-Free Impedimetric Aptasensor with Antifouling Surface Chemistry: A Prostate Specific Antigen Case Study. Sens. Actuators B Chem. 2015, 209, 306–312. [Google Scholar] [CrossRef]
- Vu, C.-A.; Chen, W.-Y. Field-Effect Transistor Biosensors for Biomedical Applications: Recent Advances and Future Prospects. Sensors 2019, 19, 4214. [Google Scholar] [CrossRef] [PubMed]
- Ehrenkranz, J.R.L. Home and Point-of-Care Pregnancy Tests: A Review of the Technology. Epidemiology 2002, 13, S15–S18. [Google Scholar] [CrossRef]
- Arshavsky-Graham, S.; Segal, E. Lab-on-a-Chip Devices for Point-of-Care Medical Diagnostics. In Microfluidics in Biotechnology; Advances in Biochemical Engineering/Biotechnology; Springer: Berlin/Heidelberg, Germany, 2020; Volume 179, pp. 247–265. ISBN 9783031041877. [Google Scholar]
- Akceoglu, G.A.; Saylan, Y.; Inci, F. A Snapshot of Microfluidics in Point-of-care Diagnostics: Multifaceted Integrity with Materials and Sensors. Adv. Mater. Technol. 2021, 6, 2100049. [Google Scholar] [CrossRef]
- Ozer, T.; McMahon, C.; Henry, C.S. Advances in Paper-Based Analytical Devices. Annu. Rev. Anal. Chem. 2020, 13, 85–109. [Google Scholar] [CrossRef]
- Yang, Y.; Gao, W. Wearable and Flexible Electronics for Continuous Molecular Monitoring. Chem. Soc. Rev. 2018, 48, 1465–1491. [Google Scholar] [CrossRef]
- van Dongen, D.N.; Fokkert, M.J.; Tolsma, R.T.; van der Sluis, A.; Slingerland, R.J.; Badings, E.A.; van ’t Hof, A.W.J.; Ottervanger, J.P. Accuracy of Pre-Hospital HEART Score Risk Classification Using Point of Care versus High Sensitive Troponin in Suspected NSTE-ACS. Am. J. Emerg. Med. 2020, 38, 1616–1620. [Google Scholar] [CrossRef]
- Müller, N.; Schneider, L.; Breuer, J.; Freudenthal, N.J. Evaluation of the Roche Point of Care System for Determination of NT-ProBNP in Urine Samples. Clin. Chim. Acta 2022, 537, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, N.; van Pelt, J. Validation and Evaluation of Eight Commercially Available Point of Care CRP Methods. Clin. Chim. Acta 2015, 439, 195–201. [Google Scholar] [CrossRef]
- Harris, L.F.; Castro-López, V.; Killard, A.J. Coagulation Monitoring Devices: Past, Present, and Future at the Point of Care. TrAC Trends Anal. Chem. 2013, 50, 85–95. [Google Scholar] [CrossRef]
- Kucherenko, I.S.; Topolnikova, Y.V.; Soldatkin, O.O. Advances in the Biosensors for Lactate and Pyruvate Detection for Medical Applications: A Review. TrAC Trends Anal. Chem. 2019, 110, 160–172. [Google Scholar] [CrossRef]
- Harnan, S.E.; Tappenden, P.; Essat, M.; Gomersall, T.; Minton, J.; Wong, R.; Pavord, I.; Everard, M.; Lawson, R. Measurement of Exhaled Nitric Oxide Concentration in Asthma: A Systematic Review and Economic Evaluation of NIOX MINO, NIOX VERO and Nobreath. Health Technol. Assess. 2015, 19, 1–330. [Google Scholar] [CrossRef] [PubMed]
- Dalcin, D.; Bogoch, I.I. Point-of-Care Testing for HIV. CMAJ 2018, 190, E569. [Google Scholar] [CrossRef]
- Song, Q.; Sun, X.; Dai, Z.; Gao, Y.; Gong, X.; Zhou, B.; Wu, J.; Wen, W. Point-of-Care Testing Detection Methods for COVID-19. Lab A Chip 2021, 21, 1634–1660. [Google Scholar] [CrossRef]
- Hong, J.M.; Lee, H.; Menon, N.V.; Lim, C.T.; Lee, L.P.; Ong, C.W.M. Point-of-Care Diagnostic Tests for Tuberculosis Disease. Sci. Transl. Med. 2022, 14, eabj4124. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.; Zakaria, S.; Samani, S.E.; Chang, Y.; Filipe, C.D.M.; Soleymani, L.; Brennan, J.D.; Liu, M.; Li, Y. Functional Nucleic Acids for Pathogenic Bacteria Detection. Acc. Chem. Res. 2021, 54, 3540–3549. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, M.; Wang, Z.; Li, S.; Deng, Y.; He, N. Point-of-Care Diagnostics for Infectious Diseases: From Methods to Devices. Nano Today 2021, 37, 101092. [Google Scholar] [CrossRef]
- Pandey, R.; Chang, D.; Smieja, M.; Hoare, T.; Li, Y.; Soleymani, L. Integrating Programmable DNAzymes with Electrical Readout for Rapid and Culture-Free Bacterial Detection Using a Handheld Platform. Nat. Chem. 2021, 13, 895–901. [Google Scholar] [CrossRef] [PubMed]
- Bahadır, E.B.; Sezgintürk, M.K. Applications of Commercial Biosensors in Clinical, Food, Environmental, and Biothreat/Biowarfare Analyses. Anal. Biochem. 2015, 478, 107–120. [Google Scholar] [CrossRef]
- Clarke, S.F.; Foster, J.R. A History of Blood Glucose Meters and Their Role in Self-Monitoring of Diabetes Mellitus. Br. J. Biomed. Sci. 2018, 69, 83–93. [Google Scholar] [CrossRef]
- Drain, P.K. Rapid Diagnostic Testing for SARS-CoV-2. N. Engl. J. Med. 2022, 386, 264–272. [Google Scholar] [CrossRef]
- Dashtian, K.; Amourizi, F.; Shahbazi, N.; Mousavi, A.; Saboorizadeh, B.; Astaraei, S.S.; Zare-Dorabei, R. Biosensors for Drug of Abuse Detection. In Advanced Sensor Technology; Elsevier: Amsterdam, The Netherlands, 2023; pp. 125–172. ISBN 9780323902229. [Google Scholar]
- Barnett, B.S.; Chai, P.R.; Suzuki, J. Scaling up Point-of-Care Fentanyl Testing—A Step Forward. N. Engl. J. Med. 2023, 389, 1643–1645. [Google Scholar] [CrossRef] [PubMed]
- Kim, U.; Ghanbari, S.; Ravikumar, A.; Seubert, J.; Figueira, S. Rapid, Affordable, and Point-of-Care Water Monitoring via a Microfluidic DNA Sensor and a Mobile Interface for Global Health. IEEE J. Transl. Eng. Health Med. 2013, 1, 3700207. [Google Scholar] [CrossRef]
- Maity, A.; Pu, H.; Sui, X.; Chang, J.; Bottum, K.J.; Jin, B.; Zhou, G.; Wang, Y.; Lu, G.; Chen, J. Scalable Graphene Sensor Array for Real-Time Toxins Monitoring in Flowing Water. Nat. Commun. 2023, 14, 4184. [Google Scholar] [CrossRef] [PubMed]
- Samani, S.E.; Chang, D.; McConnell, E.M.; Rothenbroker, M.; Filipe, C.D.M.; Li, Y. Highly Sensitive RNA-cleaving DNAzyme Sensors from Surface-to-surface Product Enrichment. ChemBioChem 2020, 21, 632–637. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.Z.H.; Hasan, M.R.; Hossain, S.I.; Ahommed, M.S.; Daizy, M. Ultrasensitive Detection of Pathogenic Viruses with Electrochemical Biosensor: State of the Art. Biosens. Bioelectron. 2020, 166, 112431. [Google Scholar] [CrossRef] [PubMed]
- Briganti, G.; Moine, O.L. Artificial Intelligence in Medicine: Today and Tomorrow. Front. Med. 2020, 7, 27. [Google Scholar] [CrossRef]
- Wang, H.; Fu, T.; Du, Y.; Gao, W.; Huang, K.; Liu, Z.; Chandak, P.; Liu, S.; Katwyk, P.V.; Deac, A.; et al. Scientific Discovery in the Age of Artificial Intelligence. Nature 2023, 620, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Gradus, J.L.; Rosellini, A.J. Supervised Machine Learning: A Brief Primer. Behav. Ther. 2020, 51, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Eckhardt, C.M.; Madjarova, S.J.; Williams, R.J.; Ollivier, M.; Karlsson, J.; Pareek, A.; Nwachukwu, B.U. Unsupervised Machine Learning Methods and Emerging Applications in Healthcare. Knee Surg. Sports Traumatol. Arthrosc. 2023, 31, 376–381. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Morgan, D.; Jacobs, R. Opportunities and Challenges for Machine Learning in Materials Science. Annu. Rev. Mater. Res. 2020, 50, 71–103. [Google Scholar] [CrossRef]
- L’Heureux, A.; Grolinger, K.; Elyamany, H.F.; Capretz, M.A.M. Machine Learning with Big Data: Challenges and Approaches. IEEE Access 2017, 5, 7776–7797. [Google Scholar] [CrossRef]
- Biswas, N.; Chakrabarti, S. Artificial Intelligence (AI)-Based Systems Biology Approaches in Multi-Omics Data Analysis of Cancer. Front. Oncol. 2020, 10, 588221. [Google Scholar] [CrossRef]
- Sancesario, G.M.; Bernardini, S. Alzheimer’s Disease in the Omics Era. Clin. Biochem. 2018, 59, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, L.; Xu, Y.; Yang, J. Machine Learning Meets Omics: Applications and Perspectives. Brief. Bioinform. 2021, 23, bbab460. [Google Scholar] [CrossRef] [PubMed]
- Scherer, M.; Schmidt, F.; Lazareva, O.; Walter, J.; Baumbach, J.; Schulz, M.H.; List, M. Machine Learning for Deciphering Cell Heterogeneity and Gene Regulation. Nat. Comput. Sci. 2021, 1, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Leman, R.; Parfait, B.; Vidaud, D.; Girodon, E.; Pacot, L.; Gac, G.L.; Ka, C.; Ferec, C.; Fichou, Y.; Quesnelle, C.; et al. SPiP: Splicing Prediction Pipeline, a Machine Learning Tool for Massive Detection of Exonic and Intronic Variant Effects on MRNA Splicing. Hum. Mutat. 2022, 43, 2308–2323. [Google Scholar] [CrossRef] [PubMed]
- AlQuraishi, M. Machine Learning in Protein Structure Prediction. Curr. Opin. Chem. Biol. 2021, 65, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Burman, S.S.R.; Chen, J.; Donovan, K.A.; Cao, Y.; Shu, C.; Zhang, B.; Zeng, Z.; Gu, S.; Zhang, Y.; et al. Machine Learning Modeling of Protein-Intrinsic Features Predicts Tractability of Targeted Protein Degradation. Genom. Proteom. Bioinform. 2022, 20, 882–898. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.-W.; Xu, J.-Y.; Zhang, T. DGMP: Identifying Cancer Driver Genes by Jointing DGCN and MLP from Multi-Omics Genomic Data. Genom. Proteom. Bioinform. 2022, 20, 928–938. [Google Scholar] [CrossRef] [PubMed]
- Quinodoz, M.; Royer-Bertrand, B.; Cisarova, K.; Gioia, S.A.D.; Superti-Furga, A.; Rivolta, C. DOMINO: Using Machine Learning to Predict Genes Associated with Dominant Disorders. Am. J. Hum. Genet. 2017, 101, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Albaradei, S.; Thafar, M.; Alsaedi, A.; Neste, C.V.; Gojobori, T.; Essack, M.; Gao, X. Machine Learning and Deep Learning Methods That Use Omics Data for Metastasis Prediction. Comput. Struct. Biotechnol. J. 2021, 19, 5008–5018. [Google Scholar] [CrossRef]
- Siristatidis, C.; Stavros, S.; Drakeley, A.; Bettocchi, S.; Pouliakis, A.; Drakakis, P.; Papapanou, M.; Vlahos, N. Omics and Artificial Intelligence to Improve in Vitro Fertilization (IVF) Success: A Proposed Protocol. Diagnostics 2021, 11, 743. [Google Scholar] [CrossRef]
- Yu, H.; Samuels, D.C.; Zhao, Y.; Guo, Y. Architectures and Accuracy of Artificial Neural Network for Disease Classification from Omics Data. BMC Genom. 2019, 20, 167. [Google Scholar] [CrossRef]
- Sharma, A.; Lysenko, A.; Boroevich, K.A.; Tsunoda, T. DeepInsight-3D Architecture for Anti-Cancer Drug Response Prediction with Deep-Learning on Multi-Omics. Sci. Rep. 2023, 13, 2483. [Google Scholar] [CrossRef]
- Henriksen, K.; O’Bryant, S.E.; Hampel, H.; Trojanowski, J.Q.; Montine, T.J.; Jeromin, A.; Blennow, K.; Lönneborg, A.; Wyss-Coray, T.; Soares, H.; et al. The Future of Blood-Based Biomarkers for Alzheimer’s Disease. Alzheimer’s Dement. 2014, 10, 115–131. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.; Jo, Y.; Ryu, D.; Jeong, C.; Choe, S.; Lee, J. Artificial-intelligence-driven Discovery of Prognostic Biomarker for Sarcopenia. J. Cachexia Sarcopenia Muscle 2021, 12, 2220–2230. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, Z.-P. Robust Biomarker Discovery for Hepatocellular Carcinoma from High-Throughput Data by Multiple Feature Selection Methods. BMC Med. Genom. 2021, 14, 112. [Google Scholar] [CrossRef]
- Xie, Y.; Meng, W.-Y.; Li, R.-Z.; Wang, Y.-W.; Qian, X.; Chan, C.; Yu, Z.-F.; Fan, X.-X.; Pan, H.-D.; Xie, C.; et al. Early Lung Cancer Diagnostic Biomarker Discovery by Machine Learning Methods. Transl. Oncol. 2021, 14, 100907. [Google Scholar] [CrossRef]
- Zafeiris, D.; Rutella, S.; Ball, G.R. An Artificial Neural Network Integrated Pipeline for Biomarker Discovery Using Alzheimer’s Disease as a Case Study. Comput. Struct. Biotechnol. J. 2018, 16, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Seth, S.; Mallik, S.; Bhadra, T.; Zhao, Z. Dimensionality Reduction and Louvain Agglomerative Hierarchical Clustering for Cluster-Specified Frequent Biomarker Discovery in Single-Cell Sequencing Data. Front. Genet. 2022, 13, 828479. [Google Scholar] [CrossRef]
- Yang, P.; Zhang, Z.; Zhou, B.B.; Zomaya, A.Y. A Clustering Based Hybrid System for Biomarker Selection and Sample Classification of Mass Spectrometry Data. Neurocomputing 2010, 73, 2317–2331. [Google Scholar] [CrossRef]
- Wang, M.; Chen, J.Y. A GMM-IG Framework for Selecting Genes as Expression Panel Biomarkers. Artif. Intell. Med. 2010, 48, 75–82. [Google Scholar] [CrossRef]
- Pawar, S.; Liew, T.O.; Stanam, A.; Lahiri, C. Common Cancer Biomarkers of Breast and Ovarian Types Identified through Artificial Intelligence. Chem. Biol. Drug Des. 2020, 96, 995–1004. [Google Scholar] [CrossRef] [PubMed]
- Yagin, F.H.; Yasar, S.; Gormez, Y.; Yagin, B.; Pinar, A.; Alkhateeb, A.; Ardigò, L.P. Explainable Artificial Intelligence Paves the Way in Precision Diagnostics and Biomarker Discovery for the Subclass of Diabetic Retinopathy in Type 2 Diabetics. Metabolites 2023, 13, 1204. [Google Scholar] [CrossRef] [PubMed]
- Yagin, F.H.; Cicek, İ.B.; Alkhateeb, A.; Yagin, B.; Colak, C.; Azzeh, M.; Akbulut, S. Explainable Artificial Intelligence Model for Identifying COVID-19 Gene Biomarkers. Comput. Biol. Med. 2023, 154, 106619. [Google Scholar] [CrossRef] [PubMed]
- Sempionatto, J.R.; Lasalde-Ramírez, J.A.; Mahato, K.; Wang, J.; Gao, W. Wearable Chemical Sensors for Biomarker Discovery in the Omics Era. Nat. Rev. Chem. 2022, 6, 899–915. [Google Scholar] [CrossRef] [PubMed]
- Mann, M.; Kumar, C.; Zeng, W.-F.; Strauss, M.T. Artificial Intelligence for Proteomics and Biomarker Discovery. Cell Syst. 2021, 12, 759–770. [Google Scholar] [CrossRef] [PubMed]
- Zupančič, U.; Rainbow, J.; Flynn, C.; Aidoo-Brown, J.; Estrela, P.; Moschou, D. Strategies for Multiplexed Electrochemical Sensor Development. In Modern Techniques in Biosensors, Detection Methods and Commercial Aspects; Studies in Systems, Decision and Control; Springer: Berlin/Heidelberg, Germany, 2021; pp. 63–93. ISBN 9789811596117. [Google Scholar]
- Malekzadeh, A.; Twaalfhoven, H.; Wijnstok, N.J.; Killestein, J.; Blankenstein, M.A.; Teunissen, C.E. Comparison of Multiplex Platforms for Cytokine Assessments and Their Potential Use for Biomarker Profiling in Multiple Sclerosis. Cytokine 2017, 91, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Marty, P.K.; Pathakumari, B.; Cox, T.M.; Keulen, V.P.V.; Erskine, C.L.; Shah, M.; Vadiyala, M.; Arias-Sanchez, P.; Karnakoti, S.; Pennington, K.M.; et al. Multiparameter Immunoprofiling for the Diagnosis and Differentiation of Progressive versus Nonprogressive Nontuberculous Mycobacterial Lung Disease—A Pilot Study. PLoS ONE 2024, 19, e0301659. [Google Scholar] [CrossRef] [PubMed]
- Bethmann, D.; Feng, Z.; Fox, B.A. Immunoprofiling as a Predictor of Patient’s Response to Cancer Therapy—Promises and Challenges. Curr. Opin. Immunol. 2017, 45, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Yang, Y.; Wu, Y.; Liu, S. Review: Multiplexed Profiling of Biomarkers in Extracellular Vesicles for Cancer Diagnosis and Therapy Monitoring. Anal. Chim. Acta 2021, 1175, 338633. [Google Scholar] [CrossRef]
- Song, S.; Lee, J.U.; Jeon, M.J.; Kim, S.; Lee, C.-N.; Sim, S.J. Precise Profiling of Exosomal Biomarkers via Programmable Curved Plasmonic Nanoarchitecture-Based Biosensor for Clinical Diagnosis of Alzheimer’s Disease. Biosens. Bioelectron. 2023, 230, 115269. [Google Scholar] [CrossRef]
- Ballard, Z.; Brown, C.; Madni, A.M.; Ozcan, A. Machine Learning and Computation-Enabled Intelligent Sensor Design. Nat. Mach. Intell. 2021, 3, 556–565. [Google Scholar] [CrossRef]
- Joung, H.-A.; Ballard, Z.S.; Wu, J.; Tseng, D.K.; Teshome, H.; Zhang, L.; Horn, E.J.; Arnaboldi, P.M.; Dattwyler, R.J.; Garner, O.B.; et al. Point-of-Care Serodiagnostic Test for Early-Stage Lyme Disease Using a Multiplexed Paper-Based Immunoassay and Machine Learning. ACS Nano 2020, 14, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Ballard, Z.S.; Joung, H.-A.; Goncharov, A.; Liang, J.; Nugroho, K.; Carlo, D.D.; Garner, O.B.; Ozcan, A. Deep Learning-Enabled Point-of-Care Sensing Using Multiplexed Paper-Based Sensors. npj Digit. Med. 2020, 3, 66. [Google Scholar] [CrossRef]
- Flynn, C.; Ignaszak, A. Lyme Disease Biosensors: A Potential Solution to a Diagnostic Dilemma. Biosensors 2020, 10, 137. [Google Scholar] [CrossRef] [PubMed]
- Bennett, N.R.; Coventry, B.; Goreshnik, I.; Huang, B.; Allen, A.; Vafeados, D.; Peng, Y.P.; Dauparas, J.; Baek, M.; Stewart, L.; et al. Improving de Novo Protein Binder Design with Deep Learning. Nat. Commun. 2023, 14, 2625. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.; Wang, Z.; Flynn, C.D.; Mahmud, A.; Labib, M.; Wang, H.; Geraili, A.; Li, X.; Zhang, J.; Sargent, E.H.; et al. A High-Dimensional Microfluidic Approach for Selection of Aptamers with Programmable Binding Affinities. Nat. Chem. 2023, 15, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Song, H.; Chen, Q.; Yu, J.; Xian, M.; Nian, R.; Feng, D. Recent Advances in the Selection and Identification of Antigen-Specific Nanobodies. Mol. Immunol. 2018, 96, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Parola, C.; Neumeier, D.; Reddy, S.T. Integrating High-throughput Screening and Sequencing for Monoclonal Antibody Discovery and Engineering. Immunology 2018, 153, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Sun, M.; Zhang, J.; Lin, X.; Zhang, Y.; Lin, F.; Zhang, P.; Yang, C.; Song, J. Computational Tools for Aptamer Identification and Optimization. TrAC Trends Anal. Chem. 2022, 157, 116767. [Google Scholar] [CrossRef]
- Hoinka, J.; Berezhnoy, A.; Sauna, Z.E.; Gilboa, E.; Przytycka, T.M. AptaCluster—A Method to Cluster HT-SELEX Aptamer Pools and Lessons from Its Application. Lect. Notes Comput. Sci. 2014, 8394, 115–128. [Google Scholar] [CrossRef]
- Alam, K.K.; Chang, J.L.; Burke, D.H. FASTAptamer: A Bioinformatic Toolkit for High-Throughput Sequence Analysis of Combinatorial Selections. Mol. Ther. Nucleic Acids 2015, 4, e230. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Zheng, Y.; Huang, M.; Wu, L.; Wang, W.; Zhu, Z.; Song, Y.; Yang, C. A Sequential Multidimensional Analysis Algorithm for Aptamer Identification Based on Structure Analysis and Machine Learning. Anal. Chem. 2020, 92, 3307–3314. [Google Scholar] [CrossRef]
- Tobia, J.P.; Huang, P.-J.J.; Ding, Y.; Narayan, R.S.; Narayan, A.; Liu, J. Machine Learning Directed Aptamer Search from Conserved Primary Sequences and Secondary Structures. ACS Synth. Biol. 2023, 12, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Bashir, A.; Yang, Q.; Wang, J.; Hoyer, S.; Chou, W.; McLean, C.; Davis, G.; Gong, Q.; Armstrong, Z.; Jang, J.; et al. Machine Learning Guided Aptamer Refinement and Discovery. Nat. Commun. 2021, 12, 2366. [Google Scholar] [CrossRef]
- Iwano, N.; Adachi, T.; Aoki, K.; Nakamura, Y.; Hamada, M. Generative Aptamer Discovery Using RaptGen. Nat. Comput. Sci. 2022, 2, 378–386. [Google Scholar] [CrossRef]
- Rube, H.T.; Rastogi, C.; Feng, S.; Kribelbauer, J.F.; Li, A.; Becerra, B.; Melo, L.A.N.; Do, B.V.; Li, X.; Adam, H.H.; et al. Prediction of Protein–Ligand Binding Affinity from Sequencing Data with Interpretable Machine Learning. Nat. Biotechnol. 2022, 40, 1520–1527. [Google Scholar] [CrossRef]
- Li, L.; Gupta, E.; Spaeth, J.; Shing, L.; Jaimes, R.; Engelhart, E.; Lopez, R.; Caceres, R.S.; Bepler, T.; Walsh, M.E. Machine Learning Optimization of Candidate Antibody Yields Highly Diverse Sub-Nanomolar Affinity Antibody Libraries. Nat. Commun. 2023, 14, 3454. [Google Scholar] [CrossRef] [PubMed]
- Porebski, B.T.; Balmforth, M.; Browne, G.; Riley, A.; Jamali, K.; Fürst, M.J.L.J.; Velic, M.; Buchanan, A.; Minter, R.; Vaughan, T.; et al. Rapid Discovery of High-Affinity Antibodies via Massively Parallel Sequencing, Ribosome Display and Affinity Screening. Nat. Biomed. Eng. 2024, 8, 214–232. [Google Scholar] [CrossRef]
- Irvine, E.B.; Reddy, S.T. Advancing Antibody Engineering through Synthetic Evolution and Machine Learning. J. Immunol. 2024, 212, 235–243. [Google Scholar] [CrossRef]
- Yu, T.; Boob, A.G.; Singh, N.; Su, Y.; Zhao, H. In Vitro Continuous Protein Evolution Empowered by Machine Learning and Automation. Cell Syst. 2023, 14, 633–644. [Google Scholar] [CrossRef]
- Makowski, E.K.; Chen, H.-T.; Tessier, P.M. Simplifying Complex Antibody Engineering Using Machine Learning. Cell Syst. 2023, 14, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 2021, 50, D439–D444. [Google Scholar] [CrossRef]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate Prediction of Protein Structures and Interactions Using a Three-Track Neural Network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lisanza, S.; Juergens, D.; Tischer, D.; Watson, J.L.; Castro, K.M.; Ragotte, R.; Saragovi, A.; Milles, L.F.; Baek, M.; et al. Scaffolding Protein Functional Sites Using Deep Learning. Science 2022, 377, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Xu, X.; Chen, S.-J. Predicting RNA Structure with Vfold. Methods Mol. Biol. 2017, 1654, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Krokhotin, A.; Houlihan, K.; Dokholyan, N.V. IFoldRNA v2: Folding RNA with Constraints. Bioinformatics 2015, 31, 2891–2893. [Google Scholar] [CrossRef] [PubMed]
- Watkins, A.M.; Rangan, R.; Das, R. FARFAR2: Improved de Novo Rosetta Prediction of Complex Global RNA Folds. Structure 2020, 28, 963–976.e6. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.; McHugh, R.; Anishchenko, I.; Jiang, H.; Baker, D.; DiMaio, F. Accurate Prediction of Protein–Nucleic Acid Complexes Using RoseTTAFoldNA. Nat. Methods 2024, 21, 117–121. [Google Scholar] [CrossRef]
- Krishna, R.; Wang, J.; Ahern, W.; Sturmfels, P.; Venkatesh, P.; Kalvet, I.; Lee, G.R.; Morey-Burrows, F.S.; Anishchenko, I.; Humphreys, I.R.; et al. Generalized Biomolecular Modeling and Design with RoseTTAFold All-Atom. Science 2024, 384, eadl2528. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Su, Y.; Peng, X.; Wang, S.; Peng, J.; Ma, J. Antigen-Specific Antibody Design and Optimization with Diffusion-Based Generative Models for Protein Structures. bioRxiv 2022, 35, 9754–9767. [Google Scholar] [CrossRef]
- Anand, N.; Achim, T. Protein Structure and Sequence Generation with Equivariant Denoising Diffusion Probabilistic Models. arXiv 2022. [Google Scholar] [CrossRef]
- Anishchenko, I.; Pellock, S.J.; Chidyausiku, T.M.; Ramelot, T.A.; Ovchinnikov, S.; Hao, J.; Bafna, K.; Norn, C.; Kang, A.; Bera, A.K.; et al. De Novo Protein Design by Deep Network Hallucination. Nature 2021, 600, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Dauparas, J.; Anishchenko, I.; Bennett, N.; Bai, H.; Ragotte, R.J.; Milles, L.F.; Wicky, B.I.M.; Courbet, A.; de Haas, R.J.; Bethel, N.; et al. Robust Deep Learning–Based Protein Sequence Design Using ProteinMPNN. Science 2022, 378, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Ferruz, N.; Schmidt, S.; Höcker, B. ProtGPT2 Is a Deep Unsupervised Language Model for Protein Design. Nat. Commun. 2022, 13, 4348. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.L.; Juergens, D.; Bennett, N.R.; Trippe, B.L.; Yim, J.; Eisenach, H.E.; Ahern, W.; Borst, A.J.; Ragotte, R.J.; Milles, L.F.; et al. De Novo Design of Protein Structure and Function with RFdiffusion. Nature 2023, 620, 1089–1100. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Coventry, B.; Goreshnik, I.; Huang, B.; Sheffler, W.; Park, J.S.; Jude, K.M.; Marković, I.; Kadam, R.U.; Verschueren, K.H.G.; et al. Design of Protein-Binding Proteins from the Target Structure Alone. Nature 2022, 605, 551–560. [Google Scholar] [CrossRef]
- Torres, S.V.; Leung, P.J.Y.; Venkatesh, P.; Lutz, I.D.; Hink, F.; Huynh, H.-H.; Becker, J.; Yeh, A.H.-W.; Juergens, D.; Bennett, N.R.; et al. De Novo Design of High-Affinity Binders of Bioactive Helical Peptides. Nature 2024, 626, 435–442. [Google Scholar] [CrossRef]
- Sahtoe, D.D.; Coscia, A.; Mustafaoglu, N.; Miller, L.M.; Olal, D.; Vulovic, I.; Yu, T.-Y.; Goreshnik, I.; Lin, Y.-R.; Clark, L.; et al. Transferrin Receptor Targeting by de Novo Sheet Extension. Proc. Natl. Acad. Sci. USA 2021, 118, e2021569118. [Google Scholar] [CrossRef]
- Sahtoe, D.D.; Andrzejewska, E.A.; Han, H.L.; Rennella, E.; Schneider, M.M.; Meisl, G.; Ahlrichs, M.; Decarreau, J.; Nguyen, H.; Kang, A.; et al. Design of Amyloidogenic Peptide Traps. Nat. Chem. Biol. 2024, 1–10. [Google Scholar] [CrossRef]
- Cao, L.; Goreshnik, I.; Coventry, B.; Case, J.B.; Miller, L.; Kozodoy, L.; Chen, R.E.; Carter, L.; Walls, A.C.; Park, Y.-J.; et al. De Novo Design of Picomolar SARS-CoV-2 Miniprotein Inhibitors. Science 2020, 370, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Lamoureux, P.S.; Winther, K.T.; Torres, J.A.G.; Streibel, V.; Zhao, M.; Bajdich, M.; Abild-Pedersen, F.; Bligaard, T. Machine Learning for Computational Heterogeneous Catalysis. ChemCatChem 2019, 11, 3581–3601. [Google Scholar] [CrossRef]
- Zhuang, J.; Midgley, A.C.; Wei, Y.; Liu, Q.; Kong, D.; Huang, X. Machine-learning-assisted Nanozyme Design: Lessons from Materials and Engineered Enzymes. Adv. Mater. 2024, 36, e2210848. [Google Scholar] [CrossRef]
- Yeh, A.H.-W.; Norn, C.; Kipnis, Y.; Tischer, D.; Pellock, S.J.; Evans, D.; Ma, P.; Lee, G.R.; Zhang, J.Z.; Anishchenko, I.; et al. De Novo Design of Luciferases Using Deep Learning. Nature 2023, 614, 774–780. [Google Scholar] [CrossRef] [PubMed]
- Margraf, J.T.; Jung, H.; Scheurer, C.; Reuter, K. Exploring Catalytic Reaction Networks with Machine Learning. Nat. Catal. 2023, 6, 112–121. [Google Scholar] [CrossRef]
- Musa, E.; Doherty, F.; Goldsmith, B.R. Accelerating the Structure Search of Catalysts with Machine Learning. Curr. Opin. Chem. Eng. 2022, 35, 100771. [Google Scholar] [CrossRef]
- Mai, H.; Le, T.C.; Chen, D.; Winkler, D.A.; Caruso, R.A. Machine Learning for Electrocatalyst and Photocatalyst Design and Discovery. Chem. Rev. 2022, 122, 13478–13515. [Google Scholar] [CrossRef]
- Zheludev, N.I.; Kivshar, Y.S. From Metamaterials to Metadevices. Nat. Mater. 2012, 11, 917–924. [Google Scholar] [CrossRef]
- Song, J.; Lee, J.; Kim, N.; Min, K. Artificial Intelligence in the Design of Innovative Metamaterials: A Comprehensive Review. Int. J. Precis. Eng. Manuf. 2024, 25, 225–244. [Google Scholar] [CrossRef]
- Potyrailo, R.A.; Brewer, J.; Cheng, B.; Carpenter, M.A.; Houlihan, N.; Kolmakov, A. Bio-Inspired Gas Sensing: Boosting Performance with Sensor Optimization Guided by “Machine Learning”. Faraday Discuss. 2020, 223, 161–182. [Google Scholar] [CrossRef] [PubMed]
- Malkiel, I.; Mrejen, M.; Nagler, A.; Arieli, U.; Wolf, L.; Suchowski, H. Plasmonic Nanostructure Design and Characterization via Deep Learning. Light Sci. Appl. 2018, 7, 60. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, G.J.C.; Ayres, L.B.; Costa, J.N.Y.; Paschoalino, W.J.; Whitehead, K.; Kubota, L.T.; Piazzetta, M.H.d.O.; Gobbi, A.L.; Shimizu, F.M.; Garcia, C.D.; et al. Ultradense Electrochemical Chips with Arrays of Nanostructured Microelectrodes to Enable Sensitive Diffusion-Limited Bioassays. ACS Appl. Mater. Interfaces 2024. [Google Scholar] [CrossRef] [PubMed]
- McGlennen, R.C. Miniaturization Technologies for Molecular Diagnostics. Clin. Chem. 2020, 47, 393–402. [Google Scholar] [CrossRef]
- Yang, Z.; Albrow-Owen, T.; Cai, W.; Hasan, T. Miniaturization of Optical Spectrometers. Science 2021, 371, eabe0722. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.H.; Fernandez, H.A.; Nigmatulin, F.; Cai, W.; Yang, Z.; Cui, H.; Ahmed, F.; Cui, X.; Uddin, M.G.; Minot, E.D.; et al. Miniaturized Spectrometers with a Tunable van Der Waals Junction. Science 2022, 378, 296–299. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Abid, M.; Zerara, M.; Cho, J.; Choi, M.; Coileáin, C.Ó.; Hung, K.-M.; Chang, C.-R.; Shvets, I.V.; Wu, H.-C. Miniaturized Spectrometer with Intrinsic Long-Term Image Memory. Nat. Commun. 2024, 15, 676. [Google Scholar] [CrossRef] [PubMed]
- Schackart, K.E.; Yoon, J.-Y. Machine Learning Enhances the Performance of Bioreceptor-Free Biosensors. Sensors 2021, 21, 5519. [Google Scholar] [CrossRef]
- Nicoliche, C.Y.N.; de Oliveira, R.A.G.; da Silva, G.S.; Ferreira, L.F.; Rodrigues, I.L.; Faria, R.C.; Fazzio, A.; Carrilho, E.; de Pontes, L.G.; Schleder, G.R.; et al. Converging Multidimensional Sensor and Machine Learning toward High-Throughput and Biorecognition Element-Free Multidetermination of Extracellular Vesicle Biomarkers. ACS Sens. 2020, 5, 1864–1871. [Google Scholar] [CrossRef]
- Dutt, S.; Shao, H.; Karawdeniya, B.; Bandara, Y.M.N.D.Y.; Daskalaki, E.; Suominen, H.; Kluth, P. High Accuracy Protein Identification: Fusion of Solid-state Nanopore Sensing and Machine Learning. Small Methods 2023, 7, e2300676. [Google Scholar] [CrossRef]
- Taniguchi, M.; Takei, H.; Tomiyasu, K.; Sakamoto, O.; Naono, N. Sensing the Performance of Artificially Intelligent Nanopores Developed by Integrating Solid-State Nanopores with Machine Learning Methods. J. Phys. Chem. C 2022, 126, 12197–12209. [Google Scholar] [CrossRef]
- Taniguchi, M.; Minami, S.; Ono, C.; Hamajima, R.; Morimura, A.; Hamaguchi, S.; Akeda, Y.; Kanai, Y.; Kobayashi, T.; Kamitani, W.; et al. Combining Machine Learning and Nanopore Construction Creates an Artificial Intelligence Nanopore for Coronavirus Detection. Nat. Commun. 2021, 12, 3726. [Google Scholar] [CrossRef]
- Tian, Y.; Chao, M.A.; Kulkarni, C.; Goebel, K.; Fink, O. Real-Time Model Calibration with Deep Reinforcement Learning. Mech. Syst. Signal Process. 2022, 165, 108284. [Google Scholar] [CrossRef]
- Durand, A.; Wiesner, T.; Gardner, M.-A.; Robitaille, L.-É.; Bilodeau, A.; Gagné, C.; Koninck, P.D.; Lavoie-Cardinal, F. A Machine Learning Approach for Online Automated Optimization of Super-Resolution Optical Microscopy. Nat. Commun. 2018, 9, 5247. [Google Scholar] [CrossRef] [PubMed]
- Ismaiel, E.; Zátonyi, A.; Fekete, Z. Dimensionality Reduction and Prediction of Impedance Data of Biointerface. Sensors 2022, 22, 4191. [Google Scholar] [CrossRef] [PubMed]
- Porr, B.; Daryanavard, S.; Bohollo, L.M.; Cowan, H.; Dahiya, R. Real-Time Noise Cancellation with Deep Learning. PLoS ONE 2022, 17, e0277974. [Google Scholar] [CrossRef] [PubMed]
- Thirukovalluru, R.; Dixit, S.; Sevakula, R.K.; Verma, N.K.; Salour, A. Generating Feature Sets for Fault Diagnosis Using Denoising Stacked Auto-Encoder. In Proceedings of the 2016 IEEE International Conference on Prognostics and Health Management (ICPHM), Ottawa, ON, Canada, 20–22 June 2016; pp. 1–7. [Google Scholar] [CrossRef]
- Ha, N.; Xu, K.; Ren, G.; Mitchell, A.; Ou, J.Z. Machine Learning-enabled Smart Sensor Systems. Adv. Intell. Syst. 2020, 2, 2000063. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, J.; Liu, T.; Luo, Y.; Loh, X.J.; Chen, X. Machine Learning-reinforced Noninvasive Biosensors for Healthcare. Adv. Healthc. Mater. 2021, 10, e2100734. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Yue, Y.; Zhang, Y.; Zhang, Z.; Zhou, H.S. Advancing Biosensors with Machine Learning. ACS Sens. 2020, 5, 3346–3364. [Google Scholar] [CrossRef]
- Alafeef, M.; Srivastava, I.; Pan, D. Machine Learning for Precision Breast Cancer Diagnosis and Prediction of the Nanoparticle Cellular Internalization. ACS Sens. 2020, 5, 1689–1698. [Google Scholar] [CrossRef]
- Ganjalizadeh, V.; Meena, G.G.; Stott, M.A.; Hawkins, A.R.; Schmidt, H. Machine Learning at the Edge for AI-Enabled Multiplexed Pathogen Detection. Sci. Rep. 2023, 13, 4744. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Cai, A.; Xu, T.; Zhang, X. Artificial Intelligence Biosensors for Continuous Glucose Monitoring. Interdiscip. Mater. 2023, 2, 290–307. [Google Scholar] [CrossRef]
- Chen, X.; Xie, H.; Tao, X.; Wang, F.L.; Leng, M.; Lei, B. Artificial Intelligence and Multimodal Data Fusion for Smart Healthcare: Topic Modeling and Bibliometrics. Artif. Intell. Rev. 2024, 57, 91. [Google Scholar] [CrossRef]
- Zeng, Z.; Huang, Z.; Leng, K.; Han, W.; Niu, H.; Yu, Y.; Ling, Q.; Liu, J.; Wu, Z.; Zang, J. Nonintrusive Monitoring of Mental Fatigue Status Using Epidermal Electronic Systems and Machine-Learning Algorithms. ACS Sens. 2020, 5, 1305–1313. [Google Scholar] [CrossRef]
- Nguyen, D.C.; Pham, Q.-V.; Pathirana, P.N.; Ding, M.; Seneviratne, A.; Lin, Z.; Dobre, O.; Hwang, W.-J. Federated Learning for Smart Healthcare: A Survey. ACM Comput. Surv. CSUR 2022, 55, 1–37. [Google Scholar] [CrossRef]
- Tian, S.; Yang, W.; Grange, J.M.L.; Wang, P.; Huang, W.; Ye, Z. Smart Healthcare: Making Medical Care More Intelligent. Glob. Health J. 2019, 3, 62–65. [Google Scholar] [CrossRef]



| Biosensing Challenges | AI Biosensing Challenges | |||||||
|---|---|---|---|---|---|---|---|---|
| Analyte | ||||||||
| Lack of available biomarkers | Lack of high-quality training data | |||||||
| Lack of biomarker characterization data | Lack of open access omics data | |||||||
| Poor understanding of biomarker relationships | Limited disease-specific omics data | |||||||
| Many diseases without defined clinical tests | ||||||||
| Recognition Element | ||||||||
| Lack of available receptors | Biased screening data | |||||||
| Lack of available catalysts | Lack of high-quality training data | |||||||
| Unknown biomolecular structures | Lack of standardized characterization | |||||||
| Inability to replicate natural receptors | ||||||||
| Transduction | ||||||||
| Low material diversity for sensors | Hardware limitations | |||||||
| Need for biocompatible materials | Lack of sensor material data | |||||||
| Bulky instrumentation | ||||||||
| Biomolecule instability & degradability | ||||||||
| Data Analysis | ||||||||
| Excessive background noise | Lack of open access analysis programs | |||||||
| Inability to adjust to dynamic environments | Lack of generalizable models | |||||||
| Limited interpretative ability | Data privacy and security | |||||||
| Unable to consider outside factors | ||||||||
| Development | ||||
| Expensive and inaccessible datasets | ||||
| Expensive computational resources | ||||
| Biased data | ||||
| Lack of standardized characterization methods | ||||
| Integration | ||||
| Biased models | ||||
| Overfitting or underfitting of data | ||||
| Difficulty evaluating models for uncharted territories | ||||
| Deployment | ||||
| Narrow application scopes; lack of universality | ||||
| Abundance of repeated models | ||||
| Issues with user trust | ||||
| Lack of regulatory guidelines for evolving models | ||||
| Lack of guidelines for user data | ||||
| Lack of explainability | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Flynn, C.D.; Chang, D. Artificial Intelligence in Point-of-Care Biosensing: Challenges and Opportunities. Diagnostics 2024, 14, 1100. https://doi.org/10.3390/diagnostics14111100
Flynn CD, Chang D. Artificial Intelligence in Point-of-Care Biosensing: Challenges and Opportunities. Diagnostics. 2024; 14(11):1100. https://doi.org/10.3390/diagnostics14111100
Chicago/Turabian StyleFlynn, Connor D., and Dingran Chang. 2024. "Artificial Intelligence in Point-of-Care Biosensing: Challenges and Opportunities" Diagnostics 14, no. 11: 1100. https://doi.org/10.3390/diagnostics14111100
APA StyleFlynn, C. D., & Chang, D. (2024). Artificial Intelligence in Point-of-Care Biosensing: Challenges and Opportunities. Diagnostics, 14(11), 1100. https://doi.org/10.3390/diagnostics14111100







