Gastro-Esophageal Cancer: Can Radiomic Parameters from Baseline 18F-FDG-PET/CT Predict the Development of Distant Metastatic Disease?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Cohort
2.2. 18F-FDG-PET/CT Acquisition and Image Analysis
2.3. Statistical Analysis
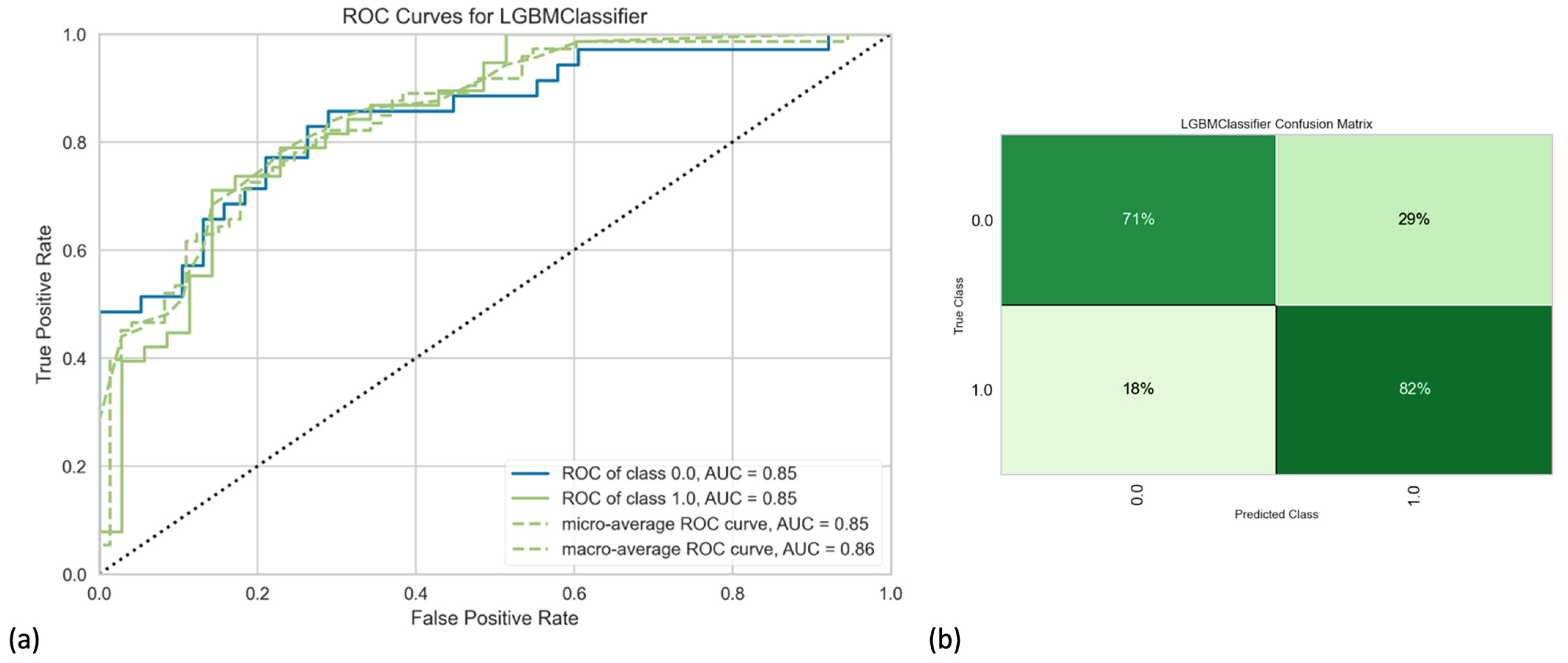
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cummings, D.; Wong, J.; Palm, R.; Hoffe, S.; Almhanna, K.; Vignesh, S. Epidemiology, diagnosis, staging and multimodal therapy of esophageal and gastric tumors. Cancers 2021, 13, 582. [Google Scholar] [CrossRef]
- Janmaat, V.T.; Steyerberg, E.W.; van der Gaast, A.; Mathijssen, R.H.; Bruno, M.J.; Peppelenbosch, M.P.; Kuipers, E.J.; Spaander, M.C. Palliative chemotherapy and targeted therapies for esophageal and gastroesophageal junction cancer. Cochrane Database Syst. Rev. 2017, 11, CD004063. [Google Scholar] [CrossRef]
- Hinzpeter, R.; Mirshahvalad, S.A.; Kulanthaivelu, R.; Ortega, C.; Metser, U.; Liu, Z.A.; Elimova, E.; Wong, R.K.S.; Yeung, J.; Jang, R.W.J.; et al. Prognostic Value of [18F]-FDG PET/CT Radiomics Combined with Sarcopenia Status among Patients with Advanced Gastroesophageal Cancer. Cancers 2022, 14, 5314. [Google Scholar] [CrossRef]
- Akin, E.A.; Qazi, Z.N.; Osman, M.; Zeman, R.K. Clinical impact of FDG PET/CT in alimentary tract malignancies: An updated review. Abdom. Radiol. 2020, 45, 1018–1035. [Google Scholar] [CrossRef]
- Mantziari, S.; Pomoni, A.; Prior, J.O.; Winiker, M.; Allemann, P.; Demartines, N.; Schäfer, M. 18F-FDG PET/CT-derived parameters predict clinical stage and prognosis of esophageal cancer. BMC Med. Imaging 2020, 20, 7. [Google Scholar] [CrossRef]
- Pan, L.; Gu, P.; Huang, G.; Xue, H.; Wu, S. Prognostic significance of SUV on PET/CT in patients with esophageal cancer: A systematic review and meta-analysis. Eur. J. Gastroenterol. Hepatol. 2009, 21, 1008–1015. [Google Scholar] [CrossRef]
- Amrane, K.; Thuillier, P.; Bourhis, D.; Le Meur, C.; Quere, C.; Leclere, J.C.; Ferec, M.; Jestin-Le Tallec, V.; Doucet, L.; Alemany, P.; et al. Prognostic value of pre-therapeutic FDG-PET radiomic analysis in gastro-esophageal junction cancer. Sci. Rep. 2023, 13, 5789. [Google Scholar] [CrossRef]
- Deantonio, L.; Garo, M.L.; Paone, G.; Valli, M.C.; Cappio, S.; La Regina, D.; Cefali, M.; Palmarocchi, M.C.; Vannelli, A.; De Dosso, S. 18F-FDG PET Radiomics as Predictor of Treatment Response in Oesophageal Cancer: A Systematic Review and Meta-Analysis. Front. Oncol. 2022, 12, 861638. [Google Scholar] [CrossRef]
- Jayaprakasam, V.S.; Gibbs, P.; Gangai, N.; Bajwa, R.; Sosa, R.E.; Yeh, R.; Greally, M.; Ku, G.Y.; Gollub, M.J.; Paroder, V. Can18F-FDG PET/CT Radiomics Features Predict Clinical Outcomes in Patients with Locally Advanced Esophageal Squamous Cell Carcinoma? Cancers 2022, 14, 3035. [Google Scholar] [CrossRef]
- Mirshahvalad, S.A.; Eisazadeh, R.; Shahbazi-Akbari, M.; Pirich, C.; Beheshti, M. Application of Artificial Intelligence in Oncologic Molecular PET-Imaging: A Narrative Review on Beyond [18F] F-FDG Tracers-Part I. PSMA, Choline, and DOTA Radiotracers. Semin. Nucl. Med. 2023, 54, 171–180. [Google Scholar] [CrossRef]
- Gupta, V.; Coburn, N.; Kidane, B.; Hess, K.R.; Compton, C.; Ringash, J.; Darling, G.; Mahar, A.L. Survival prediction tools for esophageal and gastroesophageal junction cancer: A systematic review. J. Thorac. Cardiovasc. Surg. 2018, 156, 847–856. [Google Scholar] [CrossRef]
- Deans, D.A.C.; Wigmore, S.J.; De Beaux, A.C.; Paterson-Brown, S.; Garden, O.J.; Fearon, K.C.H. Clinical prognostic scoring system to aid decision-making in gastro-oesophageal cancer. J. Br. Surg. 2007, 94, 1501–1508. [Google Scholar] [CrossRef]
- Zeng, Y.; Liang, W.; Liu, J.; He, J.; Ng, C.S.; Liu, C.-C.; Petersen, R.H.; Rocco, G.; D’Amico, T.; Brunelli, A.; et al. Esophageal cancer in elderly patients: A population-based study. J. Thorac. Dis. 2018, 10, 448. [Google Scholar] [CrossRef]
- Hinzpeter, R.; Mirshahvalad, S.A.; Kulanthaivelu, R.; Murad, V.; Ortega, C.; Metser, U.; Liu, Z.A.; Elimova, E.; Wong, R.K.S.; Yeung, J.; et al. Prognostic Value of Sarcopenia and Metabolic Parameters of 18F-FDG-PET/CT in Patients with Advanced Gastroesophageal Cancer. Diagnostics 2023, 13, 838. [Google Scholar] [CrossRef]
- Anconina, R.; Ortega, C.; Metser, U.; Liu, Z.A.; Suzuki, C.; McInnis, M.; Darling, G.E.; Wong, R.; Taylor, K.; Yeung, J.; et al. Influence of sarcopenia, clinical data, and 2-[18F] FDG PET/CT in outcome prediction of patients with early-stage adenocarcinoma esophageal cancer. Eur. J. Nucl. Med. Mol. Imaging 2021, 49, 1012–1020. [Google Scholar] [CrossRef]
- Nioche, C.; Orlhac, F.; Boughdad, S.; Reuzé, S.; Goya-Outi, J.; Robert, C.; Pellot-Barakat, C.; Soussan, M.; Frouin, F.; Buvat, I. LIFEx: A freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Res. 2018, 78, 4786–4789. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The image biomarker standardization initiative: Standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 2020, 295, 328. [Google Scholar] [CrossRef]
- Bu, S.; Pang, H.; Li, X.; Zhao, M.; Wang, J.; Liu, Y.; Yu, H. Multi-parametric radiomics of conventional T1 weighted and susceptibility-weighted imaging for differential diagnosis of idiopathic Parkinson’s disease and multiple system atrophy. BMC Med. Imaging 2023, 23, 204. [Google Scholar] [CrossRef]
- Lam, L.H.; Chu, N.T.; Tran, T.O.; Do, D.T.; Le, N.Q. A Radiomics-Based Machine Learning Model for Prediction of Tumor Mutational Burden in Lower-Grade Gliomas. Cancers 2022, 14, 3492. [Google Scholar] [CrossRef]
- Zhang, C.; Shi, Z.; Kalendralis, P.; Whybra, P.; Parkinson, C.; Berbee, M.; Spezi, E.; Roberts, A.; Christian, A.; Lewis, W.; et al. Prediction of lymph node metastases using pre-treatment PET radiomics of the primary tumour in esophageal adenocarcinoma: An external validation study. Br. J. Radiol. 2021, 94, 20201042. [Google Scholar] [CrossRef]
- Wu, L.; Wang, C.; Tan, X.; Cheng, Z.; Zhao, K.; Yan, L.; Liang, Y.; Liu, Z.; Liang, C. Radiomics approach for preoperative identification of stages I–II and III–IV of esophageal cancer. Chin. J. Cancer Res. 2018, 30, 396. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Mu, F.; Wang, S.; Qiu, Q.; Wang, S.; Wang, L. Prediction of distant metastasis in esophageal cancer using a radiomics–clinical model. Eur. J. Med. Res. 2022, 27, 272. [Google Scholar] [CrossRef] [PubMed]
- Foley, K.G.; Hills, R.K.; Berthon, B.; Marshall, C.; Parkinson, C.; Lewis, W.G.; Crosby, T.D.; Spezi, E.; Roberts, S.A. Development and validation of a prognostic model incorporating texture analysis derived from standardised segmentation of PET in patients with oesophageal cancer. Eur. Radiol. 2018, 28, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Mali, S.A.; Ibrahim, A.; Woodruff, H.C.; Andrearczyk, V.; Müller, H.; Primakov, S.; Salahuddin, Z.; Chatterjee, A.; Lambin, P. Making Radiomics More Reproducible across Scanner and Imaging Protocol Variations: A Review of Harmonization Methods. J. Pers. Med. 2021, 11, 842. [Google Scholar] [CrossRef] [PubMed]
- Nyflot, M.J.; Yang, F.; Byrd, D.; Bowen, S.R.; Sandison, G.A.; Kinahan, P.E. Quantitative radiomics: Impact of stochastic effects on textural feature analysis implies the need for standards. J. Med. Imaging 2015, 2, 041002. [Google Scholar] [CrossRef] [PubMed]
- Zwanenburg, A. Radiomics in nuclear medicine: Robustness, reproducibility, standardization, and how to avoid data analysis traps and replication crisis. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 2638–2655. [Google Scholar] [CrossRef]
- Yang, F.; Simpson, G.; Young, L.; Ford, J.; Dogan, N.; Wang, L. Impact of contouring variability on oncological PET radiomics features in the lung. Sci. Rep. 2020, 10, 369. [Google Scholar] [CrossRef]

| Non-Metastatic (n = 115) | Metastatic (n = 128) | p-Value | |
|---|---|---|---|
| Age | 64.8 ± 11.1 | 63.5 ± 11.7 | 0.399 |
| Age ≥ 70 (%) | 40 (35%) | 43 (34%) | 0.572 |
| Sex [male] (%) | 90 (78%) | 102 (80%) | 0.875 |
| Race [Asian] (%) | 4 (4%) | 11 (9%) | 0.115 |
| ECOG ≥ 2 (%) | 8 (7%) | 27 (21%) | 0.001 * |
| BMI | 28.1 ± 5.6 | 24.4 ± 4.9 | <0.001 * |
| Sarcopenia score | 48.4 ± 8.7 | 43.2 ± 9.7 | <0.001 * |
| Sarcopenic patients (%) | 29 (25%) | 60 (47%) | <0.001 * |
| Histology [adenocarcinoma] (%) | 112 (97%) | 82 (64%) | <0.001 * |
| - Grade (%): | - | ||
| - Gx | 21 (18%) | 30 (23%) | |
| - G1 | 6 (5%) | 5 (4%) | |
| - G2 | 39 (34%) | 42 (33%) | |
| - G3 | 49 (43%) | 51 (40%) | |
| Grade [G3] (%) | 49 (43%) | 51 (40%) | 0.696 |
| - T Stage (%): | - | ||
| - Tx | 29 (25%) | 83 (65%) | |
| - T0 | 0 (0%) | 1 (1%) | |
| - Tis | 1 (1%) | 0 (0%) | |
| - T1 | 6 (5%) | 1 (1%) | |
| - T2 | 13 (11%) | 2 (2%) | |
| - T3 | 65 (57%) | 33 (26%) | |
| - T4 | 1 (1%) | 8 (6%) | |
| T Stage ≥ T3 (%) | 66 (57%) | 41 (32%) | <0.001 * |
| Primary tumor SUVmax | 12.9 ± 7.8 | 15.4 ± 7.8 | 0.011 * |
| Primary tumor SUVmean | 8.5 ± 4.6 | 8.4 ± 3.8 | 0.816 |
| Primary tumor SUVpeak | 10.7 ± 6.9 | 12.9 ± 6.6 | 0.012 * |
| Primary tumor SULmax | 9.1 ± 5.6 | 11.5 ± 5.8 | 0.001 * |
| Primary tumor SULmean | 6.1 ± 3.3 | 6.3 ± 2.8 | 0.627 |
| Primary tumor SULpeak | 7.9 ± 5.0 | 9.7 ± 4.9 | 0.005 * |
| Accuracy (%) | AUC (%) | Recall (%) | Precision (%) | F1 Score (%) | |
|---|---|---|---|---|---|
| Baseline clinical model * | 64 | 70 | 60 | 68 | 63 |
| CT radiomics | 72 | 78 | 77 | 73 | 74 |
| PET radiomics | 51 | 54 | 57 | 54 | 55 |
| PET/CT radiomics | 67 | 78 | 71 | 69 | 34 |
| CT radiomics + clinical data | 76 | 84 | 78 | 77 | 77 |
| CT radiomics + sarcopenia score | 68 | 77 | 72 | 69 | 70 |
| CT radiomics + clinical + sarcopenia score | 77 | 83 | 79 | 79 | 78 |
| PET radiomics + clinical data | 69 | 77 | 67 | 72 | 68 |
| PET radiomics + sarcopenia score | 68 | 69 | 72 | 69 | 70 |
| PET radiomics + clinical + sarcopenia score | 74 | 80 | 74 | 76 | 75 |
| PET/CT radiomics + clinical data | 76 | 85 | 80 | 77 | 78 |
| PET/CT radiomics + sarcopenia score | 71 | 78 | 76 | 72 | 73 |
| PET/CT radiomics + clinical + sarcopenia ** | 79 | 85 | 80 | 80 | 80 |
| Finalized highly iterated cross-validated model | 80 | 88 | 84 | 80 | 82 |
| Modality | Parameter (Defined Cut-Off) | Hazard Ratio (95% CI) | p-Value |
|---|---|---|---|
| Clinical (n = 5) | Having advanced disease | 2.606 (1.921–3.535) | <0.001 |
| Age ≥ 70 years | 1.545 (1.136–2.102) | 0.006 | |
| ECOG ≥ 2 | 3.403 (2.286–5.066) | <0.001 | |
| Being Sarcopenic | 1.871 (1.385–2.526) | <0.001 | |
| Having SCC/undifferentiated pathology | 1.580 (1.102–2.265) | 0.013 | |
| CT (n = 4) | CT SHAPE volume (mL) (14.2) | 1.006 (1.002–1.010) | 0.002 |
| CT SHAPE sphericity (0.545) | 0.064 (0.015–0.270) | <0.001 | |
| CT NGLDM contrast (0.05) | 0.006 (0.000–0.389) | 0.016 | |
| CT GLZLM GLNU (67) | 1.001 (1.000–1.002) | 0.009 | |
| PET (whole tumor; n = 2) | SHAPE sphericity (0.743) | 0.089 (0.024–0.338) | <0.001 |
| GLZLM ZLNU (72) | 1.001 (1.001–1.002) | <0.001 | |
| PET (40% SUVmax; n = 2) | GLRLM SRLGE (0.0018) | 0.001 (0.001–0.082) | 0.044 |
| NGLDM coarseness (0.11) | 0.001 (0.001–0.041) | 0.015 | |
| PET (70% SUVmax; n = 1) | SHAPE volume (Voxel) (100) | 1.002 (1.001–1.003) | <0.001 |
| Modality | Parameter | Hazard Ratio (95% CI) | p-Value |
|---|---|---|---|
| Clinical (n = 3) | Having advanced disease (reference: no) | 2.566 (1.849–3.561) | <0.001 |
| Age ≥ 70 years (reference: no) | 1.538 (1.099–2.153) | 0.012 | |
| ECOG ≥ 2 (reference: no) | 2.799 (1.794–4.366) | <0.001 | |
| CT (n = 1) | CT SHAPE volume (mL) (reference: <14.2) | 1.043 (1.012–1.074) | 0.005 |
| PET (whole tumor; n = 1) | GLZLM ZLNU (reference: <72) | 1.001 (1.001–1.002) | 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hinzpeter, R.; Mirshahvalad, S.A.; Kulanthaivelu, R.; Kohan, A.; Ortega, C.; Metser, U.; Liu, A.; Farag, A.; Elimova, E.; Wong, R.K.S.; et al. Gastro-Esophageal Cancer: Can Radiomic Parameters from Baseline 18F-FDG-PET/CT Predict the Development of Distant Metastatic Disease? Diagnostics 2024, 14, 1205. https://doi.org/10.3390/diagnostics14111205
Hinzpeter R, Mirshahvalad SA, Kulanthaivelu R, Kohan A, Ortega C, Metser U, Liu A, Farag A, Elimova E, Wong RKS, et al. Gastro-Esophageal Cancer: Can Radiomic Parameters from Baseline 18F-FDG-PET/CT Predict the Development of Distant Metastatic Disease? Diagnostics. 2024; 14(11):1205. https://doi.org/10.3390/diagnostics14111205
Chicago/Turabian StyleHinzpeter, Ricarda, Seyed Ali Mirshahvalad, Roshini Kulanthaivelu, Andres Kohan, Claudia Ortega, Ur Metser, Amy Liu, Adam Farag, Elena Elimova, Rebecca K. S. Wong, and et al. 2024. "Gastro-Esophageal Cancer: Can Radiomic Parameters from Baseline 18F-FDG-PET/CT Predict the Development of Distant Metastatic Disease?" Diagnostics 14, no. 11: 1205. https://doi.org/10.3390/diagnostics14111205






