Performance Comparison of Convolutional Neural Network-Based Hearing Loss Classification Model Using Auditory Brainstem Response Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Auditory Brainstem Response Data
2.2. CNN Classification Model
2.2.1. VGG16 and VGG19
2.2.2. DenseNet121 and DenseNet201
2.2.3. AlexNet
2.2.4. InceptionV3
3. Results
Model Training and ABR Data Classification Results
4. Discussion
4.1. Classification Data Analysis
4.2. Analysis of ABR Data That Are Not Classified Correctly
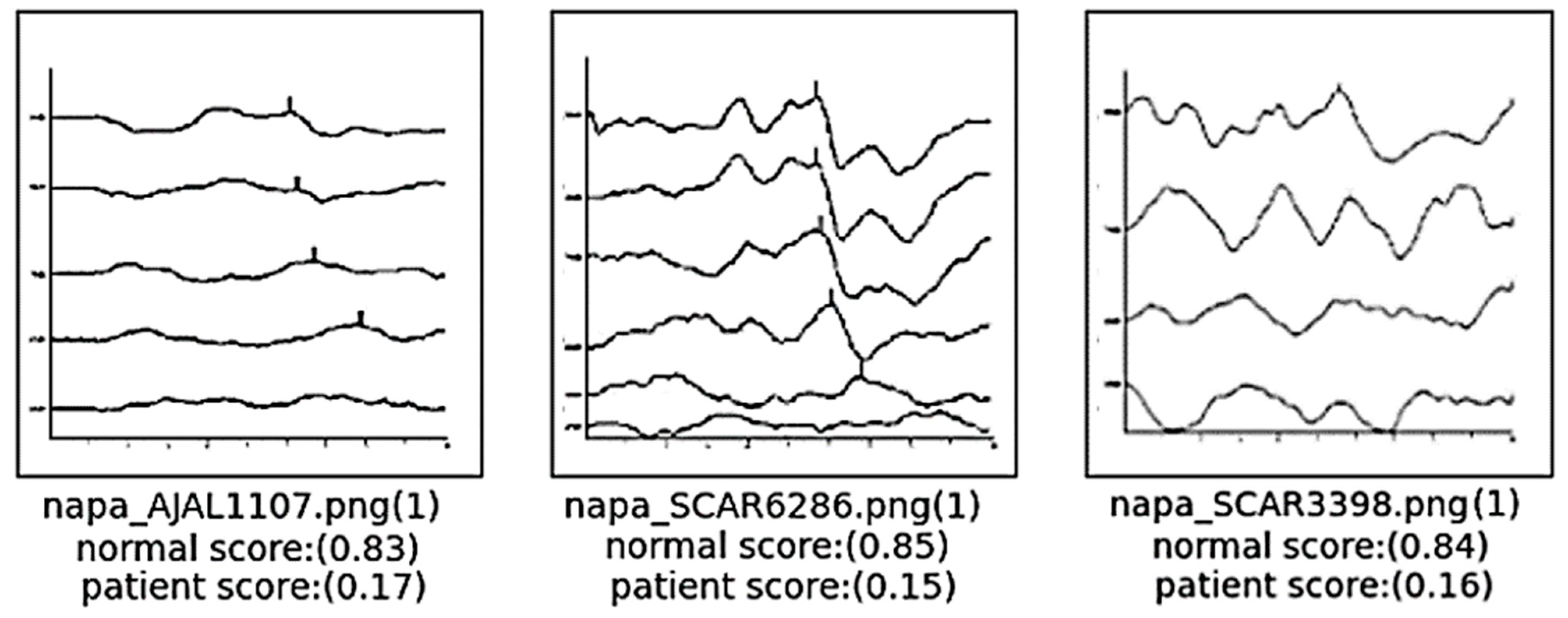
4.2.1. False Negative: In Case the Data Are actually Normal but Are Classified as a Patient with Hearing Loss
4.2.2. False Positive: In Case the Data Represent Patients with Actual Hearing Loss but Classified as Normal
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Q.; Cai, W.; Wang, X.; Zhou, Y.; Feng, D.D.; Chen, M. Medical image classification with convolutional neural network. In Proceedings of the 2014 13th International Conference on Control Automation Robotics & Vision (ICARCV), Singapore, 10–12 December 2014; pp. 844–848. [Google Scholar]
- Mu, R.; Zeng, X. A review of deep learning research. KSII Trans. Internet Inf. Syst. (TIIS) 2019, 13, 1738–1764. [Google Scholar]
- Yadav, S.S.; Jadhav, S.M. Deep convolutional neural network based medical image classification for disease diagnosis. J. Big Data 2019, 6, 1–18. [Google Scholar] [CrossRef]
- Eggermont, J.J. Auditory Brainstem Response. In Handbook of Clinical Neurology, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2019; Volume 160, pp. 451–464. [Google Scholar]
- Sun, J.; Liu, H.; Wang, X.; Li, Y.; Ni, X. Application of auditory brainstem response to different types of hearing loss in infants. J. Clin. Otorhinolaryngol. Head Neck Surg. 2022, 36, 120–125. [Google Scholar]
- Aldè, M.; Binda, S.; Primache, V.; Pellegrinelli, L.; Pariani, E.; Pregliasco, F.; Berardino, F.D.; Cantarella, G.; Ambrosetti, U. Congenital cytomegalovirus and hearing loss: The state of the art. J. Clin. Med. 2023, 12, 4465. [Google Scholar] [CrossRef]
- Elberling, C.; Parbo, J. Reference data for ABRs in retrocochlear diagnosis. Scand. Audiol. 1987, 16, 49–55. [Google Scholar] [CrossRef]
- Ma, J.; Seo, J.H.; Moon, I.J.; Park, M.K.; Lee, J.B.; Kim, H.; Ahn, J.H.; Jang, J.H.; Lee, J.D.; Choi, S.J.; et al. Auditory Brainstem Response Data Preprocessing Method for the Automatic Classification of Hearing Loss Patients. Diagnostics 2023, 13, 3538. [Google Scholar] [CrossRef] [PubMed]
- Hood, L.J. Principles and applications in auditory evoked potentials. Ear Hear. 1996, 17, 178. [Google Scholar] [CrossRef]
- Sininger, Y.S. Auditory brain stem response for objective measures of hearing. Ear Hear. 1993, 14, 23–30. [Google Scholar] [CrossRef]
- Sininger, Y.S.; Abdala, C.; Cone-Wesson, B. Auditory threshold sensitivity of the human neonate as measured by the auditory brainstem response. Hear. Res. 1997, 104, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Aiyer, R.G.; Parikh, B. Evaluation of auditory brainstem responses for hearing screening of high-risk infants. Indian J. Otolaryngol. Head Neck Surg. 2009, 61, 47–53. [Google Scholar] [CrossRef]
- Verhulst, S.; Jagadeesh, A.; Mauermann, M.; Ernst, F. Individual differences in auditory brainstem response wave characteristics: Relations to different aspects of peripheral hearing loss. Trends Hear. 2016, 20, 2331216516672186. [Google Scholar] [CrossRef] [PubMed]
- Galambos, R.; Despland, P.A. The auditory brainstem response (ABR) evaluates risk factors for hearing loss in the newborn. Pediatr. Res. 1980, 14, 159–163. [Google Scholar] [CrossRef] [PubMed]
- McCreery, R.W.; Kaminski, J.; Beauchaine, K.; Lenzen, N.; Simms, K.; Gorga, M.P. The impact of degree of hearing loss on auditory brainstem response predictions of behavioral thresholds. Ear Hear. 2015, 36, 309. [Google Scholar] [CrossRef] [PubMed]
- Stapells, D.R.; Gravel, J.S.; Martin, B.A. Thresholds for auditory brain stem responses to tones in notched noise from infants and young children with normal hearing or sensorineural hearing loss. Ear Hear. 1995, 16, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014. [Google Scholar] [CrossRef]
- Qassim, H.; Verma, A.; Feinzimer, D. Compressed residual-VGG16 CNN model for big data places image recognition. In Proceedings of the 2018 IEEE 8th Annual Computing and Communication Workshop and Conference (CCWC), Las Vegas, NV, USA, 8–10 January 2018; pp. 169–175. [Google Scholar]
- Mascarenhas, S.; Agarwal, M. A comparison between VGG16, VGG19 and ResNet50 architecture frameworks for Image Classification. In Proceedings of the 2021 International Conference on Disruptive Technologies for Multi-Disciplinary Research and Applications (CENTCON), Bengaluru, India, 19–21 November 2021; Volume 1, pp. 96–99. [Google Scholar]
- Carvalho, T.; De Rezende, E.R.; Alves, M.T.; Balieiro, F.K.; Sovat, R.B. Exposing computer generated images by eye’s region classification via transfer learning of VGG19 CNN. In Proceedings of the 2017 16th IEEE International Conference on Machine Learning and Applications (ICMLA), Cancun, Mexico, 18–21 December 2017; pp. 866–870. [Google Scholar]
- Yin, J.; Qu, J.; Huang, W.; Chen, Q. Road Damage Detection and Classification based on Multi-level Feature Pyramids. KSII Trans. Internet Inf. Syst. 2021, 15, 786. [Google Scholar]
- Dey, N.; Zhang, Y.D.; Rajinikanth, V.; Pugalenthi, R.; Raja, N.S.M. Customized VGG19 architecture for pneumonia detection in chest X-rays. Pattern Recognit. Lett. 2021, 143, 67–74. [Google Scholar] [CrossRef]
- Mateen, M.; Wen, J.; Nasrullah; Song, S.; Huang, Z. Fundus image classification using VGG-19 architecture with PCA and SVD. Symmetry 2018, 11, 1. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Singh, D.; Kumar, V.; Kaur, M. Densely connected convolutional networks-based COVID-19 screening model. Appl. Intell. 2021, 51, 3044–3051. [Google Scholar] [CrossRef]
- Jaiswal, A.; Gianchandani, N.; Singh, D.; Kumar, V.; Kaur, M. Classification of the COVID-19 infected patients using DenseNet201 based deep transfer learning. J. Biomol. Struct. Dyn. 2021, 39, 5682–5689. [Google Scholar] [CrossRef]
- Chhabra, M.; Kumar, R. A Smart Healthcare System Based on Classifier DenseNet 121 Model to Detect Multiple Diseases. In Mobile Radio Communications and 5G Networks, Proceedings of the Second MRCN 2021; Springer Nature: Singapore, 2022; pp. 297–312. [Google Scholar]
- Frimpong, E.A.; Qin, Z.; Turkson, R.E.; Cobbinah, B.M.; Baagyere, E.Y.; Tenagyei, E.K. Enhancing Alzheimer’s Disease Classification using 3D Convolutional Neural Network and Multilayer Perceptron Model with Attention Network. KSII Trans. Internet Inf. Syst. 2023, 17, 2924–2944. [Google Scholar]
- Chauhan, T.; Palivela, H.; Tiwari, S. Optimization and fine-tuning of DenseNet model for classification of COVID-19 cases in medical imaging. Int. J. Inf. Manag. Data Insights 2021, 1, 100020. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Yuan, Z.W.; Zhang, J. Feature extraction and image retrieval based on AlexNet. In Proceedings of the 8th International Conference on Digital Image Processing (ICDIP 2016), Chengdu, China, 20–22 May 2016; SPIE: Bellingham, WA, USA, 2016; Volume 10033, pp. 65–69. [Google Scholar]
- Alippi, C.; Disabato, S.; Roveri, M. Moving convolutional neural networks to embedded systems: The alexnet and VGG-16 case. In Proceedings of the 2018 17th ACM/IEEE International Conference on Information Processing in Sensor Networks (IPSN), Porto, Portugal, 11–13 April 2018; pp. 212–223. [Google Scholar]
- Abd Almisreb, A.; Jamil, N.; Din, N.M. Utilizing AlexNet deep transfer learning for ear recognition. In Proceedings of the 2018 Fourth International Conference on Information Retrieval and Knowledge Management (CAMP), Kota Kinabalu, Malaysia, 26–28 March 2018; pp. 1–5. [Google Scholar]
- Chen, J.; Wan, Z.; Zhang, J.; Li, W.; Chen, Y.; Li, Y.; Duan, Y. Medical image segmentation and reconstruction of prostate tumor based on 3D AlexNet. Comput. Methods Programs Biomed. 2021, 200, 105878. [Google Scholar] [CrossRef]
- Titoriya, A.; Sachdeva, S. Breast cancer histopathology image classification using AlexNet. In Proceedings of the 2019 4th International Conference on Information Systems and Computer Networks (ISCON), Mathura, India, 21–22 November 2019; pp. 708–712. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 2818–2826. [Google Scholar]
- Wang, C.; Chen, D.; Hao, L.; Liu, X.; Zeng, Y.; Chen, J.; Zhang, G. Pulmonary image classification based on inception-v3 transfer learning model. IEEE Access 2019, 7, 146533–146541. [Google Scholar] [CrossRef]








| Accuracy | TNR | TPR | FPR | FNR | Precision | F1 Score | |
|---|---|---|---|---|---|---|---|
| VGG16 | 92.37% | 93.67% | 90.99% | 6.33% | 9.01% | 93.15% | 0.9206 |
| VGG19 | 94.18% | 98.47% | 90.07% | 1.53% | 9.93% | 98.39% | 0.9405 |
| DenseNet121 | 92.62% | 93.69% | 91.61% | 6.31% | 8.39% | 93.89% | 0.9273 |
| DenseNet201 | 93.30% | 95.67% | 91.01% | 4.33% | 8.99% | 95.60% | 0.9325 |
| AlexNet | 95.93% | 93.62% | 98.25% | 6.38% | 1.75% | 93.90% | 0.9602 |
| InceptionV3 | 90.43% | 93.14% | 87.60% | 6.86% | 12.40% | 92.44% | 0.8995 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, J.; Choi, S.J.; Kim, S.; Hong, M. Performance Comparison of Convolutional Neural Network-Based Hearing Loss Classification Model Using Auditory Brainstem Response Data. Diagnostics 2024, 14, 1232. https://doi.org/10.3390/diagnostics14121232
Ma J, Choi SJ, Kim S, Hong M. Performance Comparison of Convolutional Neural Network-Based Hearing Loss Classification Model Using Auditory Brainstem Response Data. Diagnostics. 2024; 14(12):1232. https://doi.org/10.3390/diagnostics14121232
Chicago/Turabian StyleMa, Jun, Seong Jun Choi, Sungyeup Kim, and Min Hong. 2024. "Performance Comparison of Convolutional Neural Network-Based Hearing Loss Classification Model Using Auditory Brainstem Response Data" Diagnostics 14, no. 12: 1232. https://doi.org/10.3390/diagnostics14121232






