The Role of Biomarkers in HPV-Positive Head and Neck Squamous Cell Carcinoma: Towards Precision Medicine
Abstract
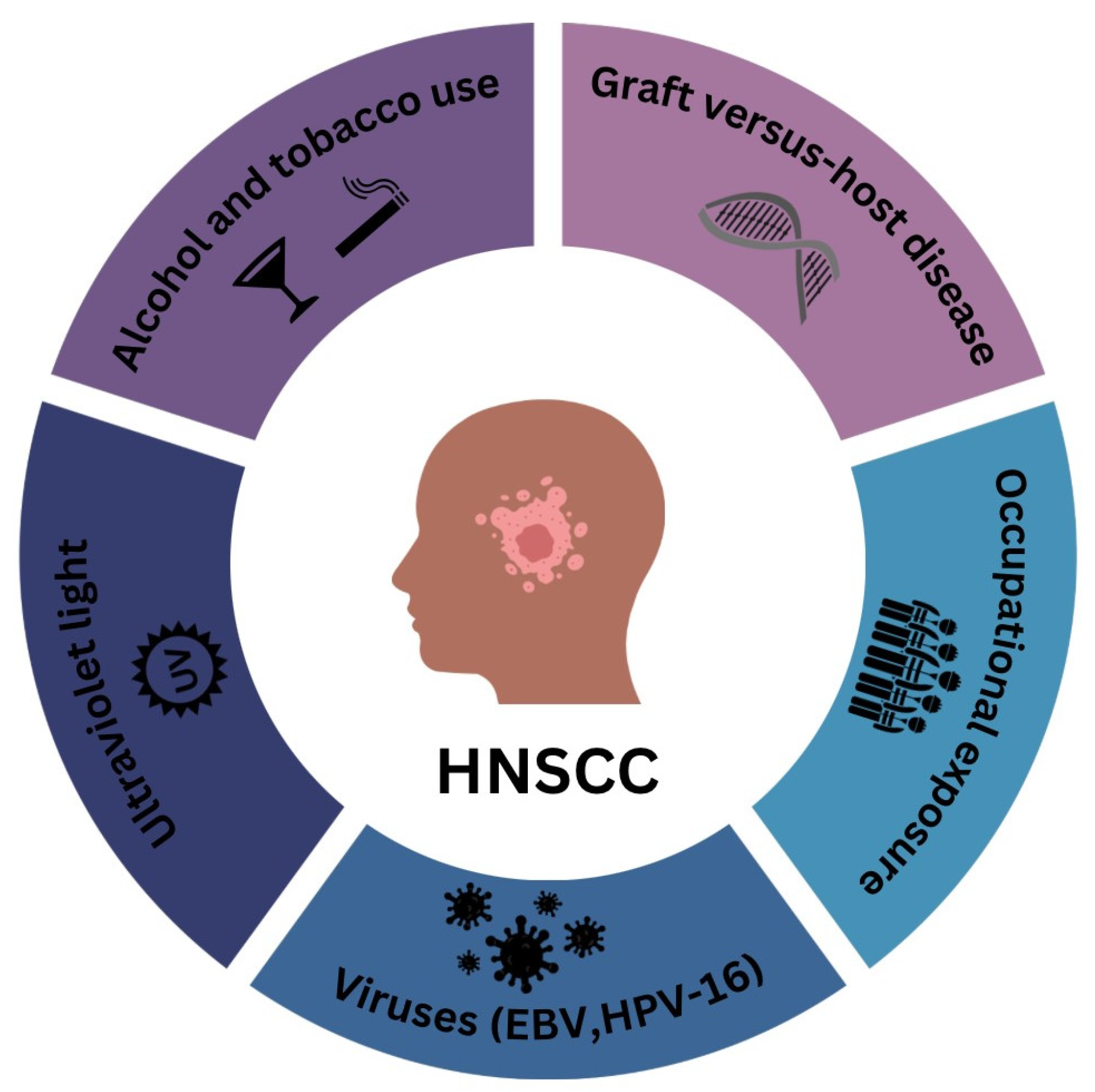
1. Introduction
2. Molecular Biomarkers in HPV-Positive Head and Neck Cancer: A Focus on DNA, RNA, and Protein Indicators
2.1. DNA Biomarkers
2.2. RNA Biomarkers
2.2.1. MicroRNA
2.2.2. Mechanism of Action of MicroRNA
2.2.3. Long Noncoding RNA
2.3. Protein Biomarkers
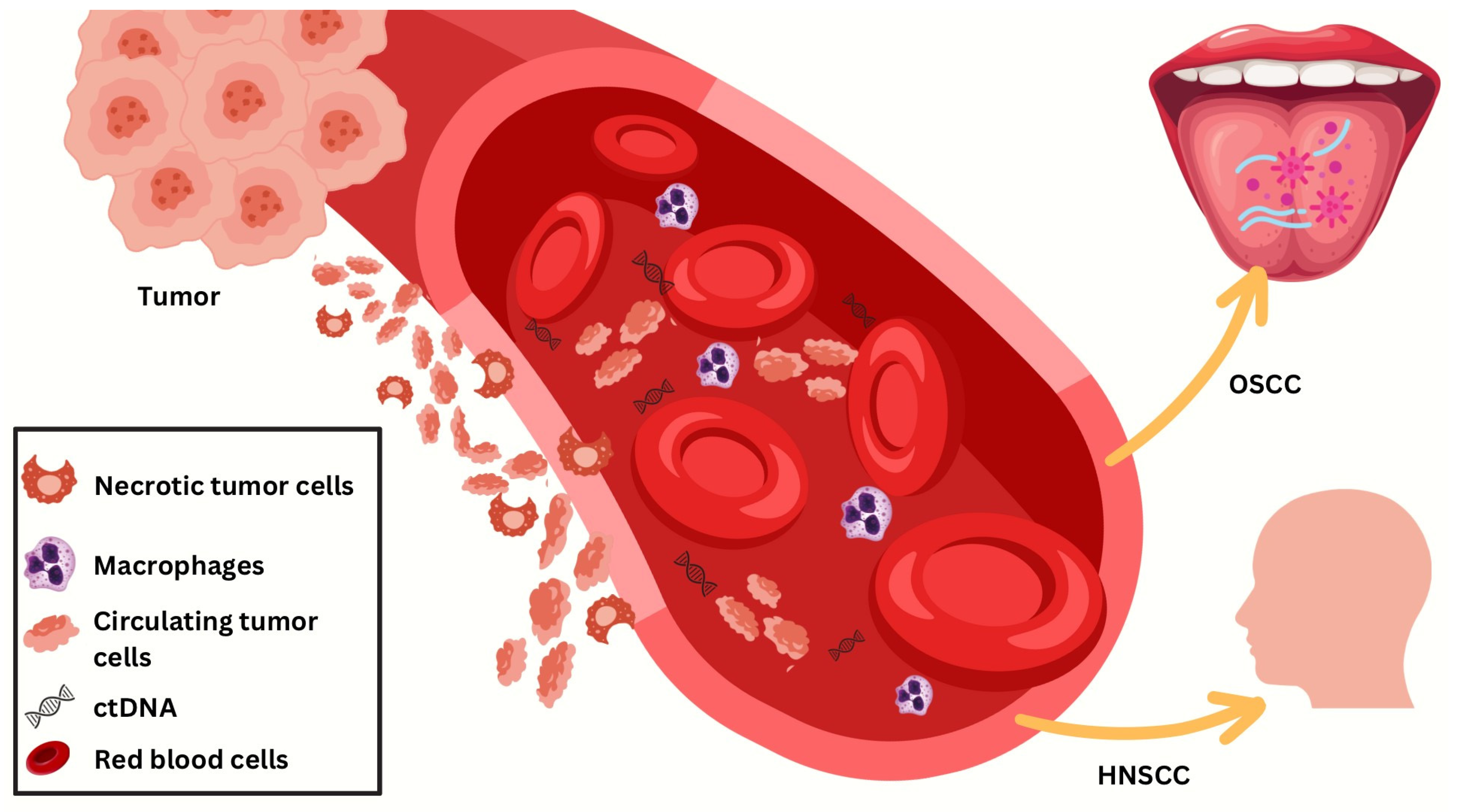
Utilizing Droplet Digital PCR in HPV Diagnostics
3. Tissue-Based Biomarkers
Optimizing HPV Biomarker Algorithms for Accurate OPSCC Prognosis
4. Emerging Technologies Revolutionizing HPV-Positive HNSCC Diagnosis
4.1. CRISPR-Based Diagnostic Tools
4.2. Next-Generation Sequencing in HPV-Related Diagnostics
4.3. Nanodiagnostics and Biosensor Technologies
5. Biomarker-Targeted Treatment Strategies in HPV-Positive and HPV-Negative HNSCC
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bhat, G.R.; Hyole, R.G.; Li, J. Head and neck cancer: Current challenges and future perspectives. Adv. Cancer Res. 2021, 152, 67–102. [Google Scholar] [CrossRef]
- Chow, L.Q.M. Head and neck cancer. N. Engl. J. Med. 2020, 382, 60–72. [Google Scholar] [CrossRef]
- Gormley, M.; Creaney, G.; Schache, A.; Ingarfield, K.; Conway, D.I. Reviewing the Epidemiology of Head and Neck Cancer: Definitions, Trends and Risk Factors. Br. Dent. J. 2022, 233, 780–786. [Google Scholar] [CrossRef]
- Rettig, E.M.; Sethi, R.K.V. Cancer of the Oropharynx and the Association with Human Papillomavirus. Hematol./Oncol. Clin. N. Am. 2021, 35, 913–931. [Google Scholar] [CrossRef]
- Zhang, M.; Luo, Z.; Liu, S.; Zhang, X.; Wang, L.; Wang, C. Identification of an 8 HPV-related RNA signature as a novel prognostic biomarker for squamous cell carcinoma of the head and neck. Medicine 2024, 103, e36448. [Google Scholar] [CrossRef]
- Leemans, C.R.; Snijders, P.J.F.; Brakenhoff, R.H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer 2018, 18, 269–282. [Google Scholar] [CrossRef]
- Oton-Gonzalez, L.; Rotondo, J.C.; Lanzillotti, C.; Mazzoni, E.; Bononi, I.; Iaquinta, M.R.; Cerritelli, L.; Malagutti, N.; Ciorba, A.; Bianchini, C.; et al. Serum HPV16 E7 oncoprotein is a recurrence marker of oropharyngeal squamous cell carcinomas. Cancers 2021, 13, 3370. [Google Scholar] [CrossRef]
- Qian, X.; Nguyen, D.T.; Dong, Y.; Sinikovic, B.; Kaufmann, A.M.; Myers, J.N.; Albers, A.E.; Graviss, E.A. Prognostic score predicts survival in HPV-negative head and neck squamous cell cancer patients. Int. J. Biol. Sci. 2019, 15, 1336–1344. [Google Scholar] [CrossRef]
- Eberly, H.W.; Sciscent, B.Y.; Lorenz, F.J.; Rettig, E.M.; Goyal, N. Current and Emerging Diagnostic, Prognostic, and Predictive Biomarkers in Head and Neck Cancer. Biomedicines 2024, 12, 415. [Google Scholar] [CrossRef]
- Ludwig, S.; Sharma, P.; Wise, P.; Sposto, R.; Hollingshead, D.; Lamb, J.; Lang, S.; Fabbri, M.; Whiteside, T.L. mRNA and miRNA profiles of exosomes from cultured tumor cells reveal biomarkers specific for HPV16-positive and HPV16-negative head and neck cancer. Int. J. Mol. Sci. 2020, 21, 8570. [Google Scholar] [CrossRef]
- Ferris, R.L.; Spanos, W.C.; Leidner, R.; Gonçalves, A.; Martens, U.M.; Kyi, C.; Sharfman, W.; Chung, C.H.; Devriese, L.A.; Gauthier, H.; et al. Neoadjuvant nivolumab for patients with resectable HPV-positive and HPV-negative squamous cell carcinomas of the head and neck in the checkmate 358 trial. J. Immunother. Cancer 2021, 9, e002568. [Google Scholar] [CrossRef]
- Welters, M.J.P.; Ma, W.; Santegoets, S.J.A.M.; Goedemans, R.; Ehsan, I.; Jordanova, E.S.; van Ham, V.J.; van Unen, V.; Koning, F.; van Egmond, S.I.; et al. Intratumoral HPV16-specific T cells constitute a type I-oriented tumor microenvironment to improve survival in HPV16-driven oropharyngeal cancer. Clin. Cancer Res. 2018, 24, 634–647. [Google Scholar] [CrossRef]
- Bosetti, C.; Carioli, G.; Santucci, C.; Bertuccio, P.; Gallus, S.; Garavello, W.; Negri, E.; La Vecchia, C. Global Trends in Oral and Pharyngeal Cancer Incidence and Mortality. Int. J. Cancer 2020, 147, 1040–1049. [Google Scholar] [CrossRef]
- Conway, D.I.; Purkayastha, M.; Chestnutt, I.G. The Changing Epidemiology of Oral Cancer: Definitions, Trends, and Risk Factors. Br. Dent. J. 2018, 225, 867–873. [Google Scholar] [CrossRef]
- Fakhry, C.; Krapcho, M.; Eisele, D.W.; D’Souza, G. Head and Neck Squamous Cell Cancers in the United States Are Rare and the Risk Now Is Higher among White Individuals Compared with Black Individuals. Cancer 2018, 124, 2125–2133. [Google Scholar] [CrossRef]
- Arantes, L.M.R.B.; De Carvalho, A.C.; Melendez, M.E.; Lopes Carvalho, A. Serum, plasma and saliva biomarkers for head and neck cancer. Expert Rev. Mol. Diagn. 2018, 18, 85–112. [Google Scholar] [CrossRef]
- Kaczor-Urbanowicz, K.E.; Wei, F.; Rao, S.L.; Kim, J.; Shin, H.; Cheng, J.; Tu, M.; Wong, D.T.W.; Kim, Y. Clinical validity of saliva and novel technology for cancer detection. Biochim. Biophys. Acta, Rev. Cancer 2019, 1872, 49–59. [Google Scholar] [CrossRef]
- Chera, B.S.; Kumar, S.; Beaty, B.T.; Marron, D.; Jefferys, S.; Green, R.; Goldman, E.C.; Amdur, R.; Sheets, N.; Dagan, R.; et al. Rapid clearance profile of plasma circulating tumor HPV type 16 DNA during chemoradiotherapy correlates with disease control in HPV-associated oropharyngeal cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 4682–4690. [Google Scholar] [CrossRef] [PubMed]
- Damerla, R.R.; Lee, N.Y.; You, D.; Soni, R.; Shah, R.; Reyngold, M.; Katabi, N.; Wu, V.; McBride, S.M.; Tsai, C.J.; et al. Detection of early human papillomavirus-associated cancers by liquid biopsy. JCO Precis. Oncol. 2019, 3, 1–17. [Google Scholar] [CrossRef]
- Chen, X.; Gole, J.; Gore, A.; He, Q.; Lu, M.; Min, J.; Yuan, Z.; Yang, X.; Jiang, Y.; Zhang, T.; et al. Non-invasive early detection of cancer four years before conventional diagnosis using a blood test. Nat. Commun. 2020, 11, 3475. (In English) [Google Scholar] [CrossRef] [PubMed]
- Lennon, A.M.; Buchanan, A.H.; Kinde, I.; Warren, A.; Honushefsky, A.; Cohain, A.T.; Ledbetter, D.H.; Sanfilippo, F.; Sheridan, K.; Rosica, D.; et al. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science 2020, 369, eabb9601. (In English) [Google Scholar] [CrossRef]
- Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Swanton, C.; Seiden, M.V. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2020, 31, 745–759. (In English) [Google Scholar] [CrossRef]
- Gydush, G.; Nguyen, E.; Bae, J.H.; Blewett, T.; Rhoades, J.; Reed, S.C.; Shea, D.; Xiong, K.; Liu, R.; Yu, F.; et al. Massively parallel enrichment of low-frequency alleles enables duplex sequencing at low depth. Nat. Biomed. Eng. 2022, 6, 257–266. (In English) [Google Scholar] [CrossRef] [PubMed]
- U.S. Preventive Services Task Force, Grade A and B, Category Cancer. Available online: https://uspreventiveservicestaskforce.org/uspstf/topic_search_results?topic_status=All&grades%5B%5D=A&grades%5B%5D=B&category%5B%5D=15&searchterm (accessed on 28 April 2024).
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. (In English) [Google Scholar] [CrossRef]
- Hubbell, E.; Clarke, C.A.; Aravanis, A.M.; Berg, C.D. Modeled Reductions in Late-stage Cancer with a Multi-Cancer Early Detection Test. Cancer Epidemiol. Biomark. Prev. 2021, 30, 460–468. (In English) [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. (In English) [Google Scholar] [CrossRef]
- Clarke, C.A.; Hubbell, E.; Kurian, A.W.; Colditz, G.A.; Hartman, A.R.; Gomez, S.L. Projected Reductions in Absolute Cancer-Related Deaths from Diagnosing Cancers Before Metastasis, 2006–2015. Cancer Epidemiol. Biomark. Prev. 2020, 29, 895–902. (In English) [Google Scholar] [CrossRef] [PubMed]
- Veyer, D.; Wack, M.; Mandavit, M.; Garrigou, S.; Hans, S.; Bonfils, P.; Tartour, E.; Bélec, L.; Wang-Renault, S.F.; Laurent-Puig, P.; et al. HPV circulating tumoral DNA quantification by droplet-based digital PCR: A promising predictive and prognostic biomarker for HPV-associated oropharyngeal cancers. Int. J. Cancer 2020, 147, 1222–1227. [Google Scholar] [CrossRef] [PubMed]
- Berger, B.M.; Hanna, G.J.; Posner, M.R.; Genden, E.M.; Lautersztain, J.; Naber, S.P.; Del Vecchio Fitz, C.; Kuperwasser, C. Detection of occult recurrence using circulating tumor tissue modified viral HPV DNA among patients treated for HPV-driven oropharyngeal carcinoma. Clin. Cancer Res. 2022, 28, 4292–4301. [Google Scholar] [CrossRef]
- Chera, B.S.; Kumar, S.; Shen, C.; Amdur, R.; Dagan, R.; Green, R.; Goldman, E.; Weiss, J.; Grilley-Olson, J.; Patel, S.; et al. Plasma circulating tumor HPV DNA for the surveillance of cancer recurrence in HPV-associated oropharyngeal cancer. J. Clin. Oncol. 2020, 38, 1050–1058. [Google Scholar] [CrossRef]
- Yom, S.S.; Torres-Saavedra, P.; Caudell, J.J.; Waldron, J.N.; Gillison, M.L.; Xia, P.; Truong, M.T.; Kong, C.; Jordan, R.; Subramaniam, R.M.; et al. Reduced-dose radiation therapy for HPV-associated oropharyngeal carcinoma (NRG Oncology HN002). J. Clin. Oncol. 2021, 39, 956–965. [Google Scholar] [CrossRef]
- Yom, S.S.; Torres-Saavedra, P.A.; Kuperwasser, C.; Kumar, S.; Gupta, P.B.; Ha, P.; Cao, H.; Lee, N.Y.; Jordan, R.; Wong, S.J.; et al. Association of plasma tumor tissue modified viral HPV DNA (TTMV) with tumor burden, treatment type, and outcome: A translational analysis from NRG-HN002. J. Clin. Oncol. 2022, 40, 6006. [Google Scholar]
- Pfister, D.G.; Spencer, S.; Adelstein, D.; Adkins, D.; Anzai, Y.; Brizel, D.M.; Bruce, J.Y.; Busse, P.M.; Caudell, J.J.; Cmelak, A.J.; et al. Head and neck cancers, version 2.2020, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2020, 18, 873–898. [Google Scholar] [CrossRef]
- Cheung, P.K.F.; Chin, R.Y.; Eslick, G.D. Detecting residual/recurrent head neck squamous cell carcinomas using PET or PET/CT: Systematic review and meta-analysis. Otolaryngol.–Head. Neck Surg. 2016, 154, 421–432. [Google Scholar] [CrossRef]
- de Ridder, M.; Gouw, Z.A.R.; Navran, A.; Hamming-Vrieze, O.; Jasperse, B.; van den Brekel, M.W.M.; Zuur, C.L.; Hoekstra, O.S.; Comans, E.F.I.; van Velden, F.H.P.; et al. FDG-PET/CT improves detection of residual disease and reduces the need for examination under anaesthesia in oropharyngeal cancer patients treated with (chemo-)radiation. Eur. Arch. Oto-Rhino-Laryngol. 2019, 276, 1447–1455. [Google Scholar] [CrossRef]
- Ho, A.S.; Tsao, G.J.; Chen, F.W.; Shen, T.; Kaplan, M.J.; Colevas, A.D.; Fischbein, N.J.; Iagaru, A.; Laramore, G.E.; Quon, A.; et al. Impact of positron emission tomography/computed tomography surveillance at 12 and 24 months for detecting head and neck cancer recurrence. Cancer 2013, 119, 1349–1356. [Google Scholar] [CrossRef]
- Li, W.; Chen, J.; Liang, B.; Li, Z.; Li, J.; Yuan, X.; Zhang, M.; Liu, Y.; Peng, Y.; Zhang, F.; et al. Long-term monitoring of dynamic changes in plasma EBV DNA for improved prognosis prediction of nasopharyngeal carcinoma. Cancer Med. 2021, 10, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.W.M.; Lee, V.H.F.; Ng, W.T.; Strojan, P.; Saba, N.F.; Rinaldo, A.; Genden, E.M.; Ferlito, A.; Kowalski, L.P.; Mendenhall, W.M.; et al. A systematic review and recommendations on the use of plasma EBV DNA for nasopharyngeal carcinoma. Eur. J. Cancer 2021, 153, 109–122. [Google Scholar] [CrossRef]
- Colevas, A.D. HPV DNA as a Biomarker in Oropharyngeal Cancer: A Step in the Right Direction. Clin. Cancer Res. 2022, 28, 4171–4172. [Google Scholar] [CrossRef] [PubMed]
- Ekanayake Weeramange, C.; Liu, Z.; Hartel, G.; Li, Y.; Vasani, S.; Langton-Lockton, J.; Kenny, L.; Morris, L.; Frazer, I.; Tang, K.D.; et al. Salivary high-risk human papillomavirus (HPV) DNA as a biomarker for HPV-driven head and neck cancers. J. Mol. Diagn. J. Mod. Dynam. 2021, 23, 1334–1342. [Google Scholar] [CrossRef] [PubMed]
- Bettampadi, D.; Villa, L.L.; Ponce, E.L.; Salmeron, J.; Sirak, B.A.; Abrahamsen, M.; Rathwell, J.A.; Reich, R.R.; Giuliano, A.R. Oral human papillomavirus prevalence and type distribution by country (Brazil, Mexico and the United States) and age among HPV infection in men study participants. Int. J. Cancer 2020, 146, 3026–3033. [Google Scholar] [CrossRef] [PubMed]
- Martin-Gomez, L.; Fulp, W.J.; Schell, M.J.; Sirak, B.; Abrahamsen, M.; Isaacs-Soriano, K.A.; Lorincz, A.; Wenig, B.; Chung, C.H.; Caudell, J.J.; et al. Oral gargle-tumor biopsy human papillomavirus (HPV) agreement and associated factors among oropharyngeal squamous cell carcinoma (OPSCC) cases. Oral Oncol. 2019, 92, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Martin-Gomez, L.; Fulp, W.J.; Schell, M.J.; Sirak, B.; Abrahamsen, M.; Isaacs-Soriano, K.A.; Lorincz, A.; Wenig, B.; Chung, C.H.; Caudell, J.J.; et al. Performance of oral HPV DNA, oral HPV mRNA and circulating tumor HPV DNA in the detection of HPV-related oropharyngeal cancer and cancer of unknown primary. Int. J. Cancer 2022, 150, 174–186. [Google Scholar] [CrossRef]
- Fakhry, C.; Blackford, A.L.; Neuner, G.; Xiao, W.; Jiang, B.; Agrawal, A.; Gillison, M.L. Association of oral human papillomavirus DNA persistence with cancer progression after primary treatment for oral cavity and oropharyngeal squamous cell carcinoma. JAMA Oncol. 2019, 5, 985–992. [Google Scholar] [CrossRef] [PubMed]
- Brakenhoff, R.H.; Wagner, S.; Klussmann, J.P. Molecular patterns and biology of HPV-associated HNSCC. Recent Results Cancer Res. 2017, 206, 37–56. [Google Scholar] [CrossRef]
- D’Souza, G.; Clemens, G.; Troy, T.; Castillo, R.G.; Struijk, L.; Waterboer, T.; Bender, N.; Pierorazio, P.M.; Haddad, R.I.; Zevallos, J.P.; et al. Evaluating the Utility and Prevalence of HPV Biomarkers in Oral Rinses and Serology for HPV-Related Oropharyngeal Cancer. Cancer Prev. Res. 2019, 12, 689–700. [Google Scholar] [CrossRef]
- Sano, D.; Oridate, N. The molecular mechanism of human papillomavirus-induced carcinogenesis in head and neck squamous cell carcinoma. Int. J. Clin. Oncol. 2016, 21, 819–826. [Google Scholar] [CrossRef]
- Solomon, B.; Young, R.J.; Rischin, D. Head and neck squamous cell carcinoma: Genomics and emerging biomarkers for immunomodulatory cancer treatments. Semin. Cancer Biol. 2018, 52, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Lechner, M.; Fenton, T.R. The genomics, epigenomics, and transcriptomics of HPV-associated oropharyngeal cancer–understanding the basis of a rapidly evolving disease. Adv. Genet. 2016, 93, 1–56. [Google Scholar] [CrossRef]
- Gaździcka, J.; Gołąbek, K.; Strzelczyk, J.K.; Ostrowska, Z. Epigenetic modifications in head and neck cancer. Biochem. Genet. 2020, 58, 213–244. [Google Scholar] [CrossRef]
- Hill, M.; Tran, N. miRNA interplay: Mechanisms and consequences in cancer. Dis. Model. Mech. 2021, 14, dmm.047662. [Google Scholar] [CrossRef] [PubMed]
- Emmett, S.; Whiteman, D.C.; Panizza, B.J.; Antonsson, A. An update on cellular MicroRNA expression in human papillomavirus-associated head and neck squamous cell carcinoma. Oncology 2018, 95, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Kalfert, D.; Pesta, M.; Kulda, V.; Topolcan, O.; Ryska, A.; Celakovsky, P.; Laco, J.; Ludvikova, M. MicroRNA profile in site-specific head and neck squamous cell cancer. Anticancer Res. 2015, 35, 2455–2463. [Google Scholar] [PubMed]
- Emmett, S.E.; Stark, M.S.; Pandeya, N.; Panizza, B.; Whiteman, D.C.; Antonsson, A. MicroRNA expression is associated with human papillomavirus status and prognosis in mucosal head and neck squamous cell carcinomas. Oral Oncol. 2021, 113, 105136. [Google Scholar] [CrossRef] [PubMed]
- Nunvar, J.; Pagacova, L.; Vojtechova, Z.; Azevedo, N.T.D.; Smahelova, J.; Salakova, M.; Tachezy, R.; Betka, J.; Klozar, J.; Grega, M.; et al. Lack of conserved miRNA deregulation in HPV-induced squamous cell carcinomas. Biomolecules 2021, 11, 764. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Yang, M.; Li, S.; Zhu, W.; Chen, M.; Pan, J.; Long, D.; Liu, Z.; Zhang, C. Expression and molecular regulation of non-coding RNAs in HPV-positive head and neck squamous cell carcinoma. Front. Oncol. 2023, 13, 1122982. [Google Scholar] [CrossRef] [PubMed]
- Gougousis, S.; Mouchtaropoulou, E.; Besli, I.; Vrochidis, P.; Skoumpas, I.; Constantinidis, I. HPV-related oropharyngeal cancer and biomarkers based on epigenetics and microbiome profile. Front. Cell Dev. Biol. 2020, 8, 625330. [Google Scholar] [CrossRef]
- Lajer, C.B.; Garnæs, E.; Friis-Hansen, L.; Norrild, B.; Therkildsen, M.H.; Glud, M.; Rossing, M.; Lajer, H.; Svane, D.; Skotte, L.; et al. The role of miRNAs in human papilloma virus (HPV)-associated cancers: Bridging between HPV-related head and neck cancer and cervical cancer. Br. J. Cancer 2012, 106, 1526–1534. [Google Scholar] [CrossRef]
- Lajer, C.B.; Nielsen, F.C.; Friis-Hansen, L.; Norrild, B.; Borup, R.; Garnæs, E.; Rossing, M.; Specht, L.; Therkildsen, M.H.; Nauntofte, B.; et al. Different miRNA signatures of oral and pharyngeal squamous cell carcinomas: A prospective translational study. Br. J. Cancer 2011, 104, 830–840. [Google Scholar] [CrossRef]
- Vojtechova, Z.; Sabol, I.; Salakova, M.; Smahelova, J.; Zavadil, J.; Turek, L.; Grega, M.; Klozar, J.; Prochazka, B.; Tachezy, R.; et al. Comparison of the miRNA profiles in HPV-positive and HPV-negative tonsillar tumors and a model system of human keratinocyte clones. BMC Cancer 2016, 16, 382. [Google Scholar] [CrossRef]
- McCombie, W.R.; McPherson, J.D.; Mardis, E.R. Next-generation sequencing technologies. Cold Spring Harb. Perspect. Med. 2019, 9, a036798. [Google Scholar] [CrossRef]
- Liu, Z.H.; Chen, L.D.; He, Y.B.; Xu, B.; Wang, K.B.; Sun, G.X.; Zhang, Z.H. Levelsof expression levels and clinical significance of miR-503 and miR-375 in patients with esophageal squamous cell carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3799–3805. [Google Scholar] [CrossRef]
- House, R.; Majumder, M.; Janakiraman, H.; Ogretmen, B.; Kato, M.; Erkul, E.; Longnecker, R.; Kellermayer, R.; Lee, H.; Sampson, L.; et al. Smoking-induced control of miR-133a-3p alters the expression of EGFR and HuR in HPV-infected oropharyngeal cancer. PLoS ONE 2018, 13, e0205077. [Google Scholar] [CrossRef] [PubMed]
- Weiss, B.G.; Anczykowski, M.Z.; Ihler, F.; Bertlich, M.; Spiegel, J.L.; Haubner, F.; Przypadlo, C.M.; Heindl, L.M.; Hoffmann, T.K.; Sauter, A.; et al. MicroRNA-182-5p and microRNA-205-5p as potential biomarkers for prognostic stratification of p16-positive oropharyngeal squamous cell carcinoma. Cancer Biomark. 2022, 33, 331–347. [Google Scholar] [CrossRef]
- Bersani, C.; Mints, M.; Tertipis, N.; Haeggblom, L.; Näsman, A.; Romanitan, M.; Marklund, L.; Dalianis, T.; Ramqvist, T.; Munck-Wikland, E.; et al. MicroRNA-155, -185 and -193b as biomarkers in human papillomavirus positive and negative tonsillar and base of tongue squamous cell carcinoma. Oral Oncol. 2018, 82, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Chen, H.; Deng, Z.; Long, D.; Xu, L.; Liu, Z. DGCR8/miR-106 axis enhances radiosensitivity of head and neck squamous cell carcinomas by downregulating RUNX3. Front. Med. 2020, 7, 582097. [Google Scholar] [CrossRef]
- Casarotto, M.; Fanetti, G.; Guerrieri, R.; Palazzari, E.; Lupato, V.; Steffan, A.; Baggio, V.; Del Vecchio, C.; Giorgi, C.; Barzan, L.; et al. Beyond MicroRNAs: Emerging role of other non-coding RNAs in HPV-driven cancers. Cancers 2020, 12, 1246. [Google Scholar] [CrossRef]
- Castro-Oropeza, R.; Piña-Sánchez, P. Epigenetic and transcriptomic regulation landscape in HPV+ cancers: Biological and clinical implications. Front. Genet. 2022, 13, 886613. [Google Scholar] [CrossRef]
- Luo, X.J.; Zheng, M.; Cao, M.X.; Zhang, W.L.; Huang, M.C.; Dai, L.; Wu, S.Y.; Liu, M.Z.; Liao, X.B.; Wang, H.Y.; et al. Distinguishable prognostic miRNA signatures of head and neck squamous cell cancer with or without HPV infection. Front. Oncol. 2020, 10, 614487. [Google Scholar] [CrossRef]
- Salazar-Ruales, C.; Arguello, J.V.; López-Cortés, A.; Cabrera-Andrade, A.; García-Cárdenas, J.M.; Guevara-Ramírez, P.; Leone, P.E.; Paz-Y-Miño, C.; Ortiz, M.; Cabrera, A.; et al. Salivary MicroRNAs for early detection of head and neck squamous cell carcinoma: A case-control study in the high altitude mestizo Ecuadorian population. BioMed Res. Int. 2018, 2018, 9792730. [Google Scholar] [CrossRef] [PubMed]
- Sannigrahi, M.K.; Sharma, R.; Singh, V.; Panda, N.K.; Rattan, V.; Khullar, M. Role of host miRNA hsa-miR-139-3p in HPV-16-Induced carcinomas. Clin. Cancer Res. 2017, 23, 3884–3895. [Google Scholar] [CrossRef] [PubMed]
- Orosz, E.; Gombos, K.; Petrevszky, N.; Csonka, D.; Haber, I.; Kaszas, B.; Gergely, L.; Csereklyei, M.; Bajzik, G.; Polgar, C.; et al. Visualization of mucosal field in HPV positive and negative oropharyngeal squamous cell carcinomas: Combined genomic and radiology based 3D model. Sci. Rep. 2020, 10, 40. [Google Scholar] [CrossRef]
- He, H.; Liu, X.; Liu, Y.; Zhang, M.; Lai, Y.; Hao, Y.; Wang, Q.; Shi, D.; Wang, N.; Luo, X.G.; et al. Human Papillomavirus E6/E7 and Long Noncoding RNA TMPOP2 Mutually Upregulated Gene Expression in Cervical Cancer Cells. J. Virol. 2019, 93, e01808-18. [Google Scholar] [CrossRef]
- Shen, L.; Li, N.; Zhou, Q.; Li, Z.; Shen, L. Development and validation of an autophagy-related LncRNA prognostic signature in head and neck squamous cell carcinoma. Front. Oncol. 2021, 11, 743611. [Google Scholar] [CrossRef]
- Chen, L.; Zhou, Y.; Sun, Q.; Zhou, J.; Pan, H.; Sui, X. Regulation of autophagy by MiRNAs and their emerging roles in tumorigenesis and cancer treatment. Int. Rev. Cell Mol. Biol. 2017, 334, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Aranda-Rivera, A.K.; Cruz-Gregorio, A.; Briones-Herrera, A.; Pedraza-Chaverri, J. Regulation of autophagy by high- and low-risk human papillomaviruses. Rev. Med. Virol. 2021, 31, e2169. [Google Scholar] [CrossRef] [PubMed]
- Vahabi, M.; Pulito, C.; Sacconi, A.; Donzelli, S.; D’Andrea, M.; Manciocco, V.; Pellini, R.; Pacini, L.; Sanguineti, G.; Strigari, L.; et al. miR-96-5p targets PTEN expression affecting radio-chemosensitivity of HNSCC cells. J. Exp. Clin. Cancer Res. 2019, 38, 141. [Google Scholar] [CrossRef]
- Long, D.; Xu, L.; Deng, Z.; Guo, D.; Zhang, Y.; Liu, Z.; Wang, Y.; Shi, H.; Zhang, L.; Liu, J.; et al. HPV16 E6 enhances the radiosensitivity in HPV-positive human head and neck squamous cell carcinoma by regulating the miR-27a-3p/SMG1 axis. Infect. Agent. Cancer 2021, 16, 56. [Google Scholar] [CrossRef]
- Inoue, H.; Hirasaki, M.; Kogashiwa, Y.; Kuba, K.; Ebihara, Y.; Nakahira, M.; Saito, K.; Onitsuka, T.; Nakashima, T. Predicting the radiosensitivity of HPV-negative oropharyngeal squamous cell carcinoma using miR-130b. Acta Otolaryngol. 2021, 141, 640–645. [Google Scholar] [CrossRef]
- Fu, E.; Liu, T.; Yu, S.; Chen, X.; Song, L.; Lou, H.; Sun, Y.; Shi, Q.; Zhao, Y.; Li, J.; et al. M2 macrophages reduce the radiosensitivity of head and neck cancer by releasing HB−EGF. Oncol. Rep. 2020, 44, 698–710. [Google Scholar] [CrossRef]
- Feng, Q.; Zhang, H.; Yao, D.; Chen, W.D.; Wang, Y.D. Emerging Role of Non-Coding RNAs in Esophageal Squamous Cell Carcinoma. Int. J. Mol. Sci. 2019, 21, 258. [Google Scholar] [CrossRef]
- Dai, D.; Feng, X.D.; Zhu, W.Q.; Bao, Y.N. LncRNA BLACAT1 regulates the viability, migration and invasion of oral squamous cell carcinoma cells by targeting miR-142-5p. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10313–10323. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Du, J.; Tang, J.; Tan, B. Integrated analysis of ceRNA regulatory network associated with tumor stage in cervical cancer. Front. Genet. 2021, 12, 618753. [Google Scholar] [CrossRef]
- Kopczyńska, M.; Kolenda, T.; Guglas, K.; Sobocińska, J.; Teresiak, A.; Bliźniak, R.; Mackiewicz, A.; Lamperska, K. PRINS lncRNA is a new biomarker candidate for HPV infection and prognosis of head and neck squamous cell carcinomas. Diagnostics 2020, 10, 762. [Google Scholar] [CrossRef]
- Chen, X.; Liu, Y.; Liu, H.; Wang, Z.W.; Zhu, X. Unraveling diverse roles of noncoding RNAs in various human papillomavirus negative cancers. Pharmacol. Ther. 2022, 238, 108188. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, T.; Li, G.; Cao, Z. The exploration of new therapeutic targets for HPV-negative head and neck squamous cell cancer through the construction of a ceRNA network and immune microenvironment analysis. J. Cell Biochem. 2020, 121, 3426–3437. [Google Scholar] [CrossRef]
- Haque, S.U.; Niu, L.; Kuhnell, D.; Hendershot, J.; Biesiada, J.; Niu, W.; Xu, L.; Sun, W.; Feng, C.; Peng, S.; et al. Differential expression and prognostic value of long non-coding RNA in HPV-negative head and neck squamous cell carcinoma. Head. Neck 2018, 40, 1555–1564. [Google Scholar] [CrossRef]
- Yang, Y.; Feng, L.; Wang, R.; Ma, H.; He, S.; Fang, J. Integrated analysis of lncRNA-associated ceRNA network in p16-positive and p16-negative head and neck squamous cell carcinoma. Medicine 2022, 101, e26120. [Google Scholar] [CrossRef]
- Mainguené, J.; Vacher, S.; Kamal, M.; Hamza, A.; Masliah-Planchon, J.; Baulande, S.; Nicolas, A.; Couturier, J.; Pierron, G.; Le Tourneau, C.; et al. Human papilloma virus integration sites and genomic signatures in head and neck squamous cell carcinoma. Mol. Oncol. 2022, 16, 3001–3016. [Google Scholar] [CrossRef]
- Zhong, Z.; Hong, M.; Chen, X.; Xi, Y.; Xu, Y.; Kong, D.; Deng, J.; Li, Y.; Hu, R.; Sun, C.; et al. Transcriptome analysis reveals the link between lncRNA-mRNA co-expression network and tumor immune microenvironment and overall survival in head and neck squamous cell carcinoma. BMC Med. Genom. 2020, 13, 57. [Google Scholar] [CrossRef]
- Guo, Y.; Pan, W.K.; Wang, Z.W.; Su, W.H.; Xu, K.; Jia, H.; Li, M.M.; Liu, H.; Zhang, B.; Li, Q.; et al. Identification of novel biomarkers for predicting prognosis and immunotherapy response in head and neck squamous cell carcinoma based on ceRNA network and immune infiltration analysis. BioMed Res. Int. 2021, 2021, 4532438. [Google Scholar] [CrossRef]
- Song, L.; Xie, H.; Tong, F.; Yan, B.; Zhang, S.; Fu, E.; Liu, H.; Lou, H.; Li, X.; Shi, Q.; et al. Association of lnc-IL17RA-11 with increased radiation sensitivity and improved prognosis of HPV-positive HNSCC. J. Cell Biochem. 2019, 120, 17438–17448. [Google Scholar] [CrossRef]
- Meng, X.; Lou, Q.Y.; Yang, W.Y.; Wang, Y.R.; Chen, R.; Wang, L.; Xu, T.; Zhang, L. The role of non-coding RNAs in drug resistance of oral squamous cell carcinoma and therapeutic potential. Cancer Commun. 2021, 41, 981–1006. [Google Scholar] [CrossRef]
- Zhang, H.; Si, J.; Yue, J.; Ma, S. The mechanisms and reversal strategies of tumor radioresistance in esophageal squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2021, 147, 1275–1286. [Google Scholar] [CrossRef]
- Ma, X.; Sheng, S.; Wu, J.; Jiang, Y.; Gao, X.; Cen, X.; Zhang, Y.; Wang, Z.; Chen, G.; Hu, Y.; et al. LncRNAs as an intermediate in HPV16 promoting myeloid-derived suppressor cell recruitment of head and neck squamous cell carcinoma. Oncotarget 2017, 8, 42061–42075. [Google Scholar] [CrossRef]
- Dias, T.R.; Santos, J.M.O.; Estêvão, D.; Costa, N.R.; Mestre, V.F.; Medeiros-Fonseca, B.; Marques, R.; Gil da Costa, R.M.; Lopes, C.; Oliveira, P.A.; et al. Expression of LncRNAs in HPV-induced carcinogenesis and cancer cachexia: A study in K14-HPV16 mice. Anticancer Res. 2022, 42, 2443–2460. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Hussen, B.M.; Shaterabadi, D.; Abak, A.; Shoorei, H.; Taheri, M.; Mokhtari, M.J.; Mohammadi-Yeganeh, S.; Savadi-Shiraz, E.; Hashemzadeh-Chaleshtori, M.; et al. The interaction between human papilloma viruses related cancers and non-coding RNAs. Pathol. Res. Pract. 2022, 234, 153939. [Google Scholar] [CrossRef]
- Salinas-Montalvo, A.M.; Supramaniam, A.; McMillan, N.A.; Idris, A. RNA-Based gene targeting therapies for human papillomavirus driven cancers. Cancer Lett. 2021, 523, 111–120. [Google Scholar] [CrossRef]
- Huang, Z.L.; Chen, R.P.; Zhou, X.T.; Zhan, H.L.; Hu, M.M.; Liu, B.; Zhang, Y.Y.; Lei, Z.; Xie, L.; Chen, M.; et al. Long non-coding RNA MEG3 induces cell apoptosis in esophageal cancer through endoplasmic reticulum stress. Oncol. Rep. 2017, 37, 3093–3099. [Google Scholar] [CrossRef]
- Sannigrahi, M.K.; Sharma, R.; Panda, N.K.; Khullar, M. Role of non-coding RNAs in head and neck squamous cell carcinoma: A narrative review. Oral. Dis. 2018, 24, 1417–1427. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Luo, T.Q.; Xu, S.S.; Chen, C.Y.; Sun, Y.; Lin, L.; Zhang, X.; Xie, J.; Li, C.; Tang, L.; et al. An immune-related seven-lncRNA signature for head and neck squamous cell carcinoma. Cancer Med. 2021, 10, 2268–2285. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Yang, P.T.; Wang, Z.W.; Xu, K.; Kou, W.H.; Luo, H. Identification of three autophagy-related long non-coding RNAs as a novel head and neck squamous cell carcinoma prognostic signature. Front. Oncol. 2020, 10, 603864. [Google Scholar] [CrossRef]
- Wu, S.; Huang, X.; Tie, X.; Cheng, Y.; Xue, X.; Fan, M. Role and mechanism of action of circular RNA and laryngeal cancer. Pathol. Res. Pract. 2021, 223, 153460. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Zhang, D.H.; Wu, N.; Xiao, J.H.; Wang, X.; Ma, W. ceRNA in cancer: Possible functions and clinical implications. J. Med. Genet. 2015, 52, 710–718. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Jakobsen, T.; Hager, H.; Kjems, J. The emerging roles of circRNAs in cancer and oncology. Nat. Rev. Clin. Oncol. 2022, 19, 188–206. [Google Scholar] [CrossRef]
- Tornesello, M.L.; Faraonio, R.; Buonaguro, L.; Annunziata, C.; Starita, N.; Cerasuolo, A.; Pezzuto, F.; Tornesello, A.L.; Buonaguro, F.M.; Botti, G.; et al. The role of microRNAs, long non-coding RNAs, and circular RNAs in cervical cancer. Front. Oncol. 2020, 10, 150. [Google Scholar] [CrossRef] [PubMed]
- Jun, W.; Shaobo, O.; Zhang, X.; Zhao, S.; Chen, M.; Fan, X.; Cai, Y.; Lan, L. Deregulation of hsa_circ_0001971/miR-186 and hsa_circ_0001874/miR-296 signaling pathways promotes the proliferation of oral squamous carcinoma cells by synergistically activating SHP2/PLK1 signals. Sci. Rep. 2021, 11, 20561. [Google Scholar] [CrossRef]
- Chen, X.; Yu, J.; Tian, H.; Shan, Z.; Liu, W.; Pan, Z.; Ren, X.; Li, A.; Wang, X.; Xie, C.; et al. Circle RNA hsa_circRNA_100290 serves as a ceRNA for miR-378a to regulate oral squamous cell carcinoma cells growth via glucose transporter-1 (GLUT1) and glycolysis. J. Cell Physiol. 2019, 234, 19130–19140. [Google Scholar] [CrossRef]
- Bonelli, P.; Borrelli, A.; Tuccillo, F.M.; Buonaguro, F.M.; Tornesello, M.L. The role of circRNAs in human papillomavirus (HPV)-associated cancers. Cancers 2021, 13, 1173. [Google Scholar] [CrossRef]
- Zhao, S.Y.; Wang, J.; Ouyang, S.B.; Huang, Z.K.; Liao, L. Salivary circular RNAs Hsa_Circ_0001874 and Hsa_Circ_0001971 as novel biomarkers for the diagnosis of oral squamous cell carcinoma. Cell Physiol. Biochem. 2018, 47, 2511–2521. [Google Scholar] [CrossRef]
- Zhao, W.; Cui, Y.; Liu, L.; Qi, X.; Liu, J.; Ma, S.; Zhou, Q.; Cao, Q.; Nie, J.; Li, Y.; et al. Splicing factor derived circular RNA circUHRF1 accelerates oral squamous cell carcinoma tumorigenesis via feedback loop. Cell Death Differ. 2020, 27, 919–933. [Google Scholar] [CrossRef] [PubMed]
- Cristóbal, I.; Caramés, C.; Rubio, J.; Sanz-Alvarez, M.; Luque, M.; Madoz-Gúrpide, J.; García-Foncillas, J. Functional and clinical impact of CircRNAs in oral cancer. Cancers 2020, 12, 1041. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Dou, M.; Song, X.; Dong, Y.; Liu, S.; Liu, H.; Tao, J.; Li, W.; Li, H.; Xie, R.; et al. The emerging role of the piRNA/piwi complex in cancer. Mol. Cancer 2019, 18, 123. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, A.R.; Qu, Y.; Li, P.X.; Zou, A.E.; Califano, J.A.; Wang-Rodriguez, J.; Gokhale, P.C.; Gachechiladze, M.A.; Ozturk, K.; Rubenstein, M.; et al. Computational methods reveal novel functionalities of PIWI-interacting RNAs in human papillomavirus-induced head and neck squamous cell carcinoma. Oncotarget 2018, 9, 4614–4624. [Google Scholar] [CrossRef] [PubMed]
- Zou, A.E.; Zheng, H.; Saad, M.A.; Rahimy, M.; Ku, J.; Kuo, S.Z.; Honda, T.K.; Wang-Rodriguez, J.; Xuan, Y.; Korrapati, A.; et al. The non-coding landscape of head and neck squamous cell carcinoma. Oncotarget 2016, 7, 51211–51222. [Google Scholar] [CrossRef]
- Firmino, N.; Martinez, V.D.; Rowbotham, D.A.; Enfield, K.S.S.; Bennewith, K.L.; Lam, W.L. HPV status is associated with altered PIWI-interacting RNA expression pattern in head and neck cancer. Oral. Oncol. 2016, 55, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Zhang, X.; Zhang, X.; Tong, D. Expression scoring of a small-nucleolar-RNA signature identified by machine learning serves as a prognostic predictor for head and neck cancer. J. Cell Physiol. 2020, 235, 8071–8084. [Google Scholar] [CrossRef]
- Paver, E.C.; Currie, A.M.; Gupta, R.; Dahlstrom, J.E. Human Papilloma Virus Related Squamous Cell Carcinomas of the Head and Neck: Diagnosis, Clinical Implications and Detection of HPV. Pathology 2020, 52, 179–191. [Google Scholar] [CrossRef]
- Rahimi, S. HPV-Related Squamous Cell Carcinoma of Oropharynx: A Review. J. Clin. Pathol. 2020, 73, 624–629. [Google Scholar] [CrossRef]
- Lewis, J.S. Sinonasal Squamous Cell Carcinoma: A Review with Emphasis on Emerging Histologic Subtypes and the Role of Human Papillomavirus. Head Neck Pathol. 2016, 10, 60–67. [Google Scholar] [CrossRef]
- Švajdler, M.; Němcova, J.; Dubinský, P.; Metelkova, A.; Švajdler, P.; Straka, Ľ.; Saláková, M.; Kment, M.; Michálková, R.; Boudová, L.; et al. Significance of Transcriptionally-Active High-Risk Human Papillomavirus in Sinonasal Squamous Cell Carcinoma: Case Series and a Meta-Analysis. Neoplasma 2021, 67, 1456–1463. [Google Scholar] [CrossRef] [PubMed]
- Prigge, E.; Arbyn, M.; von Knebel Doeberitz, M.; Reuschenbach, M. Diagnostic Accuracy of P16ink4a Immunohistochemistry in Oropharyngeal Squamous Cell Carcinomas: A Systematic Review and Meta-Analysis. Int. J. Cancer 2017, 140, 1186–1198. [Google Scholar] [CrossRef] [PubMed]
- Menegaldo, A.; Schroeder, L.; Holzinger, D.; Tirelli, G.; Cin, E.D.; Tofanelli, M.; Giorgi Rossi, P.; Boscolo-Rizzo, P.; Da Mosto, M.C.; Del Mistro, A.; et al. Detection of HPV16/18 E6 Oncoproteins in Head and Neck Squamous Cell Carcinoma Using a Protein Immunochromatographic Assay. Laryngoscope 2021, 131, 1042–1048. [Google Scholar] [CrossRef]
- Chernesky, M.; Jang, D.; Schweizer, J.; Arias, M.; Doerwald-Munoz, L.; Gupta, M.; Ely, S.; Borkowski, R.; Day, A.; Varley, C.; et al. HPV E6 Oncoproteins and Nucleic Acids in Neck Lymph Node Fine Needle Aspirates and Oral Samples From Patients with Oropharyngeal Squamous Cell Carcinoma. Papillomavirus Res. 2018, 6, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Augustin, J.G.; Lepine, C.; Morini, A.; Brunet, A.; Veyer, D.; Brochard, C.; Lévêque, N.; Mirghani, H.; Pecking, M.; Temam, S.; et al. HPV Detection in Head and Neck Squamous Cell Carcinomas: What Is the Issue? Front. Oncol. 2020, 10, 1751. [Google Scholar] [CrossRef]
- Williams, J.; Kostiuk, M.; Biron, V.L. Molecular Detection Methods in HPV-Related Cancers. Front. Oncol. 2022, 12, 864820. [Google Scholar] [CrossRef] [PubMed]
- Jeannot, E.; Latouche, A.; Bonneau, C.; Calméjane, M.-A.; Beaufort, C.M.; Ruigrok-Ritstier, K.; Bernard-Tessier, A.; Bossard, C.; Malaquin, N.; Rouzier, R.; et al. Circulating HPV DNA as a Marker for Early Detection of Relapse in Patients with Cervical Cancer. Clin. Cancer Res. 2021, 27, 5869–5877. [Google Scholar] [CrossRef]
- Krasniqi, E.; Barba, M.; Venuti, A.; Pizzuti, L.; Cappuzzo, F.; Landi, L.; Carpano, S.; Marchetti, P.; Villa, A.; Vizza, E.; et al. Circulating HPV DNA in the Management of Oropharyngeal and Cervical Cancers: Current Knowledge and Future Perspectives. J. Clin. Med. 2021, 10, 1525. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Taylor, S.C.; Laperriere, G.; Germain, H. Droplet Digital PCR Versus qPCR for Gene Expression Analysis with Low Abundant Targets: From Variable Nonsense to Publication Quality Data. Sci. Rep. 2017, 7, 2409. [Google Scholar] [CrossRef]
- Li, H.; Bai, R.; Zhao, Z.; Tao, L.; Ma, M.; Ji, Z.; Li, X.; Zhang, H.; Sun, J.; Wang, J.; et al. Application of Droplet Digital PCR to Detect the Pathogens of Infectious Diseases. Biosci. Rep. 2018, 38, BSR20181170. [Google Scholar] [CrossRef]
- Malin, K.; Louise, B.M.; Gisela, H.; Mats, K.G.; Gabriella, L.-L. Optimization of Droplet Digital PCR Assays for the Type-Specific Detection and Quantification of Five HPV Genotypes, Including Additional Data on Viral Loads of Nine Different HPV Genotypes in Cervical Carcinomas. J. Virol. Methods 2021, 294, 114193. [Google Scholar] [CrossRef] [PubMed]
- Hayden, J.P.; Wiggins, A.; Sullivan, T.; Kalantzakos, T.; Hooper, K.; Moinzadeh, A.; Rieger-Christ, K. Use of Droplet Digital Polymerase Chain Reaction to Identify Biomarkers for Differentiation of Benign and Malignant Renal Masses. Cancers 2024, 16, 787. [Google Scholar] [CrossRef] [PubMed]
- Larsson, G.L.; Helenius, G. Digital Droplet PCR (ddPCR) for the Detection and Quantification of HPV 16, 18, 33 and 45—A Short Report. Cell. Oncol. 2017, 40, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Rotondo, J.C.; Oton-Gonzalez, L.; Mazziotta, C.; Lanzillotti, C.; Iaquinta, M.R.; Tognon, M.; Martini, F.; Contini, C. Simultaneous Detection and Viral DNA Load Quantification of Different Human Papillomavirus Types in Clinical Specimens by the High Analytical Droplet Digital PCR Method. Front. Microbiol. 2020, 11, 591452. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.C.H.; Wang, H.M.; Wu, M.H.; Chang, K.P.; Chang, P.H.; Liao, C.T.; Chen, I.H.; Yen, T.C.; Lin, C.Y.; Lin, J.T.; et al. Review of emerging biomarkers in head and neck squamous cell carcinoma in the era of immunotherapy and targeted therapy. Head Neck 2019, 41, 19–45. [Google Scholar] [CrossRef] [PubMed]
- Barber, B.R.; Biron, V.L.; Klimowicz, A.C.; Puttagunta, L.; Côté, D.W.J.; Seikaly, H. Molecular predictors of locoregional and distant metastases in oropharyngeal squamous cell carcinoma. J. Otolaryngol. Head Neck Surg. 2013, 42, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Mehrad, M.; Zhao, H.; Gao, G.; Wang, X.; Lewis, J.S. Transcriptionally-active human papillomavirus is consistently retained in the distant metastases of primary oropharyngeal carcinomas. Head Neck Pathol. 2014, 8, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.S., Jr. Human papillomavirus testing in head and neck squamous cell carcinoma in 2020: Where are we now and where are we going? Head Neck Pathol. 2020, 14, 321–329. [Google Scholar] [CrossRef]
- Galati, L.; Chiocca, S.; Duca, D.; Tagliabue, M.; Simoens, C.; Gheit, T.; Arbyn, M.; Tommasino, M. HPV and head and neck cancers: Towards early diagnosis and prevention. Tumor Virus Res. 2022, 14, 200245. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mes, S.W.; Heideman, D.A.M.; Bloemena, E.; Brink, A.; Bogaarts, M.; Leemans, C.R.; Brakenhoff, R.H. Development and validation of a novel and rapid molecular detection method for high-risk human papillomavirus in formalin-fixed, paraffin-embedded tumor tissue. J. Mol. Diagn. J. Mod. Dynam. 2020, 22, 262–271. [Google Scholar] [CrossRef] [PubMed]
- von Knebel Doeberitz, M. The causal role of human papillomavirus infections in non-anogenital cancers. It’s time to ask for the functional evidence. Int. J. Cancer 2016, 139, 9–11. [Google Scholar] [CrossRef] [PubMed]
- Chera, B.S.; Amdur, R.J.; Green, R.; Shen, C.; Gupta, G.; Tan, X.; Knowles, M.; Fried, D.; Hayes, N.; Weiss, J.; et al. Phase II trial of de-intensified chemoradiotherapy for human papillomavirus-associated oropharyngeal squamous cell carcinoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2019, 37, 2661–2669. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Hasegawa, M.; Aoki, K.; Matayoshi, S.; Kiyuna, A.; Yamashita, Y.; Uehara, T.; Agena, S.; Maeda, H.; Suzuki, M.; et al. A comprehensive evaluation of human papillomavirus positive status and p16INK4a overexpression as a prognostic biomarker in head and neck squamous cell carcinoma. Int. J. Oncol. 2014, 45, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Sabatini, M.E.; Chiocca, S. Human papillomavirus as a driver of head and neck cancers. Br. J. Cancer 2020, 122, 306–314. [Google Scholar] [CrossRef]
- Simoens, C.; Gorbaslieva, I.; Gheit, T.; Tommasino, M.; Vorsters, A.; Lefevre, K.; Dorny, P.; Vanden Broeck, D.; Weyn, C.; Praet, M.; et al. HPV DNA genotyping, HPV E6*I mRNA detection, and p16(INK4a)/Ki-67 staining in Belgian head and neck cancer patient specimens, collected within the HPV-AHEAD study. Cancer Epidemiol. 2021, 72, 101925. [Google Scholar] [CrossRef]
- Shinn, J.R.; Davis, S.J.; Lang-Kuhs, K.A.; Harris, J.P.; Chen, M.M.; Walter, L.C.; Habboushe, J.; Thakar, S.D.; Hara, W.; El-Sayed, I.H.; et al. Oropharyngeal squamous cell carcinoma with discordant p16 and HPV mRNA results: Incidence and characterization in a large, contemporary United States cohort. Am. J. Surg. Pathol. 2021, 45, 951–961. [Google Scholar] [CrossRef]
- Gheit, T.; Anantharaman, D.; Holzinger, D.; Alemany, L.; Tous, S.; Lucas, E.; Prabhu, P.R.; Pawlita, M.; Ridder, R.; Rehm, S.; et al. Role of mucosal high-risk human papillomavirus types in head and neck cancers in central India. Int. J. Cancer 2017, 141, 143–151. [Google Scholar] [CrossRef]
- Ghouneimy, A.; Ali, Z.; Aman, R.; Jiang, W.; Aouida, M.; Mahfouz, M. CRISPR-Based Multiplex Detection of Human Papillomaviruses for One-Pot Point-of-Care Diagnostics. ACS Synth. Biol. 2024, 13, 837–850. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar]
- Choudhary, M.L.; Anand, S.P.; Tikhe, S.A.; Walimbe, A.M.; Potdar, V.A.; Chadha, M.S.; Mishra, A.C. Comparison of the conventional multiplex RT-PCR, real time RT-PCR and Luminex xTAG® RVP fast assay for the detection of respiratory viruses. J. Med. Virol. 2016, 88, 51–57. [Google Scholar] [CrossRef]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded Dnase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef]
- Li, L.; Li, S.; Wu, N.; Wu, J.; Wang, G.; Zhao, G.; Wang, J. HOLMESv2: A CRISPR-Cas12b- Assisted Platform for Nucleic Acid Detection and DNA Methylation Quantitation. ACS Synth. Biol. 2019, 8, 2228–2237. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef]
- Tian, T.; Qiu, Z.; Jiang, Y.; Zhu, D.; Zhou, X. Exploiting the orthogonal CRISPR-Cas12a/Cas13a trans-cleavage for dual-gene virus detection using a handheld device. Biosens. Bioelectron. 2022, 196, 113701. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef] [PubMed]
- Dmytrenko, O.; Neumann, G.C.; Hallmark, T.; Keiser, D.J.; Crowley, V.M.; Vialetto, E.; Mougiakos, I.; Wandera, K.G.; Domgaard, H.; Weber, J.; et al. Cas12a2 elicits abortive infection through RNA-triggered destruction of dsDNA. Nature 2023, 613, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, M.I.; Makhawi, A.M. SHERLOCK and DETECTR: CRISPR-Cas Systems as Potential Rapid Diagnostic Tools for Emerging Infectious Diseases. J. Clin. Microbiol. 2021, 59, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Newbigging, A.M.; Tao, J.; Cao, Y.; Peng, H.; Le, C.; Wu, J.; Pang, B.; Li, J.; Tyrrell, D.L.; et al. CRISPR technology incorporating amplification strategies: Molecular assays for nucleic acids, proteins, and small molecules. Chem. Sci. 2021, 12, 4683–4698. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mabey, D.; Peeling, R.W.; Ustianowski, A.; Perkins, M.D. Diagnostics for the developing world. Nat. Rev. Microbiol. 2004, 2, 231–240. [Google Scholar] [CrossRef]
- Gong, J.; Zhang, G.; Wang, W.; Liang, L.; Li, Q.; Liu, M.; Xue, L.; Tang, G. A simple and rapid diagnostic method for 13 types of high-risk human papillomavirus (HR-HPV) detection using CRISPR-Cas12a technology. Sci. Rep. 2021, 11, 12800. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Subica, A.M. CRISPR in Public Health: The Health Equity Implications and Role of Community in Gene-Editing Research and Applications. Am. J. Public. Health 2023, 113, 874–882. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kim, N.; Kim, H.K.; Lee, S.; Seo, J.H.; Choi, J.W.; Park, J.; Min, S.; Yoon, S.; Cho, S.R.; Kim, H.H. Prediction of the Sequence-specific Cleavage Activity of Cas9 Variants. Nat. Biotechnol. 2020, 38, 1328–1336. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Dai, Z.; Dai, X. C-RNNCrispr: Prediction of CRISPR/Cas9 sgRNA activity using convolutional and recurrent neural networks. Comput. Struct. Biotechnol. J. 2020, 18, 344–354. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef] [PubMed]
- Satam, H.; Joshi, K.; Mangrolia, U.; Waghoo, S.; Zaidi, G.; Rawool, S.; Thakare, R.P.; Banday, S.; Mishra, A.K.; Das, G.; et al. Next-Generation Sequencing Technology: Current Trends and Advancements. Biology 2023, 12, 997. [Google Scholar] [CrossRef] [PubMed]
- Costain, G.; Cohn, R.D.; Scherer, S.W.; Marshall, C.R. Genome sequencing as a diagnostic test. Can. Med. Assoc. J. 2021, 193, E1626–E1629. [Google Scholar] [CrossRef] [PubMed]
- Andersen, K.; Holm, K.; Tranberg, M.; Pedersen, C.L.; Bønløkke, S.; Steiniche, T.; Andersen, B.; Stougaard, M. Targeted Next Generation Sequencing for Human Papillomavirus Genotyping in Cervical Liquid-Based Cytology Samples. Cancers 2022, 14, 652. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, J.-W.; Shrestha, L.; Green, G.; Leier, A.; Marquez-Lago, T.T. The hitchhikers’ guide to RNA sequencing and functional analysis. Brief. Bioinform. 2023, 24, bbac529. [Google Scholar] [CrossRef]
- Stark, R.; Grzelak, M.; Hadfield, J. RNA sequencing: The teenage years. Nat. Rev. Genet. 2019, 20, 631–656. [Google Scholar] [CrossRef]
- Van den Bossche, V.; Zaryouh, H.; Vara-Messler, M.; Vignau, J.; Machiels, J.P.; Wouters, A.; Schmitz, S.; Corbet, C. Microenvironment-driven intratumoral heterogeneity in head and neck cancers: Clinical challenges and opportunities for precision medicine. Drug Resist. Updat. 2022, 60, 100806. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.U. The role of next-generation sequencing in hematologic malignancies. Blood Res. 2024, 59, 11. [Google Scholar] [CrossRef] [PubMed]
- Singhal, J.; Verma, S.; Kumar, S.; Mehrotra, D. Recent Advances in Nano-Bio-Sensing Fabrication Technology for the Detection of Oral Cancer. Mol. Biotechnol. 2021, 63, 339–362. [Google Scholar] [CrossRef] [PubMed]
- Bartosik, M.; Moranova, L.; Izadi, N.; Strmiskova, J.; Sebuyoya, R.; Holcakova, J.; Hrstka, R. Advanced technologies towards improved HPV diagnostics. J. Med. Virol. 2024, 96, e29409. [Google Scholar] [CrossRef] [PubMed]
- Safari, M.; Moghaddam, A.; Salehi Moghaddam, A.; Absalan, M.; Kruppke, B.; Ruckdäschel, H.; Khonakdar, H.A. Carbon-based biosensors from graphene family to carbon dots: A viewpoint in cancer detection. Talanta 2023, 258, 124399. [Google Scholar] [CrossRef] [PubMed]
- Laraib, U.; Sargazi, S.; Rahdar, A.; Khatami, M.; Pandey, S. Nanotechnology-based approaches for effective detection of tumor markers: A comprehensive state-of-the-art review. Int. J. Biol. Macromol. 2022, 195, 356–383. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.S.; Ou, B.R.; Lung, F.D. Developing a Biosensor-Based Immunoassay to Detect HPV E6 Oncoprotein in the Saliva Rinse Fluid of Oral Cancer Patients. J. Pers. Med. 2022, 12, 594. [Google Scholar] [CrossRef] [PubMed]
- Shigeishi, H. Association between human papillomavirus and oral cancer: A literature review. Int. J. Clin. Oncol. 2023, 28, 982–989. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Barajas, M.G.; Jave-Suárez, L.F.; Ramírez-López, I.G.; García-Chagollán, M.; Zepeda-Nuño, J.S.; Ramírez-de-Arellano, A.; Ortiz-Lazareno, P.C.; Villegas-Pineda, J.C.; Pereira-Suárez, A.L. HPV-Negative and HPV-Positive Oral Cancer Cells Stimulate the Polarization of Neutrophils towards Different Functional Phenotypes In Vitro. Cancers 2023, 15, 5814. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lyu, X.; Zhang, M.; Li, G.; Jiang, Y.; Qiao, Q. PD-1 and PD-L1 Expression Predicts Radiosensitivity and Clinical Outcomes in Head and Neck Cancer and is Associated with HPV Infection. J. Cancer 2019, 10, 937–948. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, H.; Zheng, X.; Li, L.; Huang, L.; Huang, W.; Ma, Y. Peptide-Based Therapeutic HPV Cancer Vaccine Synthesized via Bacterial Outer Membrane Vesicles. Int. J. Nanomed. 2023, 18, 4541–4554. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mock, A.; Plath, M.; Moratin, J.; Tapken, M.J.; Jäger, D.; Krauss, J.; Fröhling, S.; Hess, J.; Zaoui, K. EGFR and PI3K Pathway Activities Might Guide Drug Repurposing in HPV-Negative Head and Neck Cancers. Front. Oncol. 2021, 11, 678966. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Marquard, F.E.; Jücker, M. PI3K/AKT/mTOR signaling as a molecular target in head and neck cancer. Biochem. Pharmacol. 2020, 172, 113729. [Google Scholar] [CrossRef] [PubMed]
- Ruffin, A.T.; Li, H.; Vujanovic, L.; Zandberg, D.P.; Ferris, R.L.; Bruno, T.C. Improving head and neck cancer therapies by immunomodulation of the tumor microenvironment. Nat. Rev. Cancer. 2023, 23, 173–188. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krsek, A.; Baticic, L.; Sotosek, V.; Braut, T. The Role of Biomarkers in HPV-Positive Head and Neck Squamous Cell Carcinoma: Towards Precision Medicine. Diagnostics 2024, 14, 1448. https://doi.org/10.3390/diagnostics14131448
Krsek A, Baticic L, Sotosek V, Braut T. The Role of Biomarkers in HPV-Positive Head and Neck Squamous Cell Carcinoma: Towards Precision Medicine. Diagnostics. 2024; 14(13):1448. https://doi.org/10.3390/diagnostics14131448
Chicago/Turabian StyleKrsek, Antea, Lara Baticic, Vlatka Sotosek, and Tamara Braut. 2024. "The Role of Biomarkers in HPV-Positive Head and Neck Squamous Cell Carcinoma: Towards Precision Medicine" Diagnostics 14, no. 13: 1448. https://doi.org/10.3390/diagnostics14131448
APA StyleKrsek, A., Baticic, L., Sotosek, V., & Braut, T. (2024). The Role of Biomarkers in HPV-Positive Head and Neck Squamous Cell Carcinoma: Towards Precision Medicine. Diagnostics, 14(13), 1448. https://doi.org/10.3390/diagnostics14131448





