The Protection Level of S-RBD SARS-CoV-2 Immunoglobulin G Antibodies Using the Chemiluminescent Immunoassay Compared to the Surrogate Virus Neutralization Test Method
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Ethical Clearance
2.3. Chemiluminescent Immunoassay Anti-SARS-CoV-2 IgG Antibody Test
2.4. SARS-CoV-2 Neutralization Antibody Detection Kit
2.5. Statistical Analysis
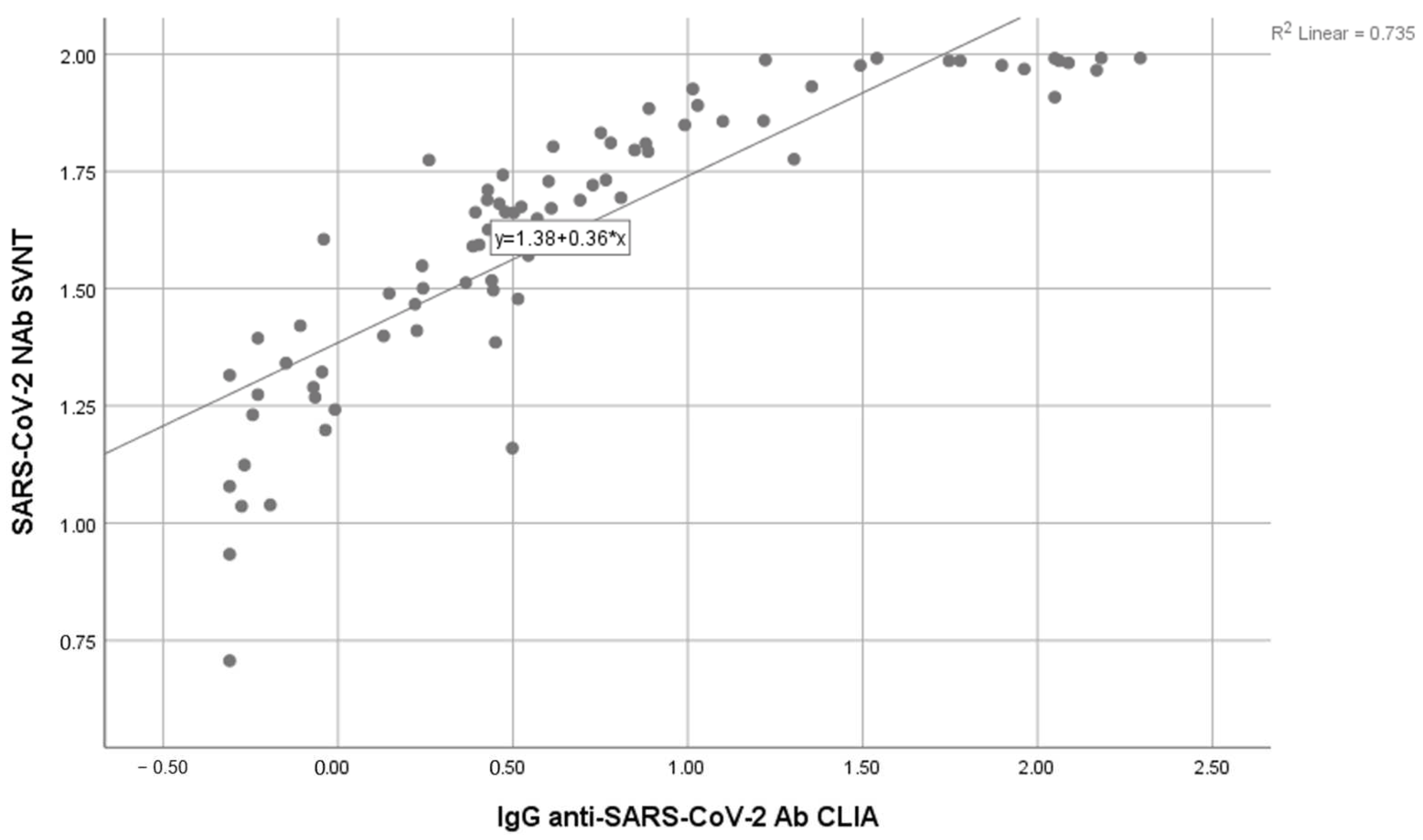
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P.; et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef] [PubMed]
- Azkur, A.K.; Akdis, M.; Azkur, D.; Sokolowska, M.; Van De Veen, W.; Brüggen, M.; O’Mahony, L.; Gao, Y.; Nadeau, K.; Akdis, C.A.; et al. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy 2020, 75, 1564–1581. [Google Scholar] [CrossRef] [PubMed]
- Kenny, G.; O’Reilly, S.; Wrigley Kelly, N.; Negi, R.; Gaillard, C.; Alalwan, D.; Saini, G.; Alrawahneh, T.; Francois, N.; Angeliadis, M.; et al. Distinct receptor binding domain IgG thresholds predict protective host immunity across SARS-CoV-2 variants and time. Nat. Commun. 2023, 14, 7015. [Google Scholar] [CrossRef] [PubMed]
- Sil, B.K.; Jahan, N.; Haq, M.d.A.; Oishee, M.J.; Ali, T.; Khandker, S.S.; Kobatake, E.; Mie, M.; Khondoker, M.U.; Jamiruddin, M.R.; et al. Development and performance evaluation of a rapid in-house ELISA for retrospective serosurveillance of SARS-CoV-2. PLoS ONE 2021, 16, e0246346. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Su, B.; Guo, X.; Sun, W.; Deng, Y.; Bao, L.; Zhu, Q.; Zhang, X.; Zheng, Y.; Geng, C.; et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell 2020, 182, 73–84.e16. [Google Scholar] [CrossRef]
- Embregts, C.W.E.; Verstrepen, B.; Langermans, J.A.M.; Böszörményi, K.P.; Sikkema, R.S.; de Vries, R.D.; Hoffmann, D.; Wernike, K.; Smit, L.A.M.; Zhao, S.; et al. Evaluation of a multi-species SARS-CoV-2 surrogate virus neutralization test. One Health 2021, 13, 100313. [Google Scholar] [CrossRef]
- Tan, C.W.; Chia, W.N.; Qin, X.; Liu, P.; Chen, M.I.C.; Tiu, C.; Hu, Z.; Chen, V.C.-W.; Young, B.E.; Sia, W.R.; et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2–spike protein–protein interaction. Nat. Biotechnol. 2020, 38, 1073–1078. [Google Scholar] [CrossRef]
- Lynch, K.L.; Zhou, S.; Kaul, R.; Walker, R.; Wu, A.H. Evaluation of Neutralizing Antibodies against SARS-CoV-2 Variants after Infection and Vaccination Using a Multiplexed Surrogate Virus Neutralization Test. Clin. Chem. 2022, 68, 702–712. [Google Scholar] [CrossRef] [PubMed]
- Kolesov, D.E.; Sinegubova, M.V.; Dayanova, L.K.; Dolzhikova, I.V.; Vorobiev, I.I.; Orlova, N.A. Fast and Accurate Surrogate Virus Neutralization Test Based on Antibody-Mediated Blocking of the Interaction of ACE2 and SARS-CoV-2 Spike Protein RBD. Diagnostics 2022, 12, 393. [Google Scholar] [CrossRef]
- Valcourt, E.J.; Manguiat, K.; Robinson, A.; Chen, J.C.Y.; Dimitrova, K.; Philipson, C.; Lamoureux, L.; McLachlan, E.; Schiffman, Z.; Drebot, M.A.; et al. Evaluation of a commercially-available surrogate virus neutralization test for severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). Diagn. Microbiol. Infect. Dis. 2021, 99, 115294. [Google Scholar] [CrossRef]
- Siemens Healthcare Diagnostics Inc. SARS-CoV-2 IgG (sCOVG) Assay for the Detection of IgG Antibodies to SARS-CoV-2; Siemens Healthcare Diagnostics Inc.: Munich, Germany, 2021. [Google Scholar]
- GenScript. cPass. SARS-CoV-2 Neutralization Antibody Detection Kit Instruction for Use; GenScript: Piscataway, NJ, USA, 2022. [Google Scholar]
- Qi, S.; Ngwa, C.; Morales Scheihing, D.A.; Al Mamun, A.; Ahnstedt, H.W.; Finger, C.E.; Colpo, G.D.; Sharmeen, R.; Kim, Y.; Choi, H.A.; et al. Sex differences in the immune response to acute COVID-19 respiratory tract infection. Biol. Sex Differ. 2021, 12, 66. [Google Scholar] [CrossRef]
- Zeng, F.; Dai, C.; Cai, P.; Wang, J.; Xu, L.; Li, J.; Hu, G.; Wang, Z.; Zheng, F.; Wang, L. A comparison study of SARS-CoV-2 IgG antibody between male and female COVID-19 patients: A possible reason underlying different outcome between gender. Infectious Diseases (except HIV/AIDS). J. Med. Virol. 2020, 92, 2050–2054. [Google Scholar] [CrossRef]
- Bayram, A.; Demirbakan, H.; Günel Karadeniz, P.; Erdoğan, M.; Koçer, I. Quantitation of antibodies against SARS-CoV-2 spike protein after two doses of CoronaVac in healthcare workers. J. Med. Virol. 2021, 93, 5560–5567. [Google Scholar] [CrossRef]
- Li, Z.; Xiang, T.; Liang, B.; Deng, H.; Wang, H.; Feng, X.; Quan, X.; Wang, X.; Li, S.; Yang, X.; et al. Characterization of SARS-CoV-2-Specific Humoral and Cellular Immune Responses Induced by Inactivated COVID-19 Vaccines in a Real-World Setting. Front. Immunol. 2021, 12, 802858. [Google Scholar] [CrossRef]
- Mukherjee, S.; Pahan, K. Is COVID-19 Gender-sensitive? J. Neuroimmune Pharmacol. 2021, 16, 38–47. [Google Scholar] [CrossRef]
- Hou, Y.; Chen, M.; Bian, Y.; Hu, Y.; Chuan, J.; Zhong, L.; Zhu, Y.; Tong, R. Insights into vaccines for elderly individuals: From the impacts of immunosenescence to delivery strategies. Npj Vaccines 2024, 9, 77. [Google Scholar] [CrossRef]
- Ferrara, P.; Ponticelli, D.; Agüero, F.; Caci, G.; Vitale, A.; Borrelli, M.; Schiavone, B.; Antonazzo, I.; Mantovani, L.; Tomaselli, V.; et al. Does smoking have an impact on the immunological response to COVID-19 vaccines? Evidence from the VASCO study and need for further studies. Public Health 2022, 203, 97–99. [Google Scholar] [CrossRef] [PubMed]
- Soegiarto, G.; Wulandari, L.; Purnomosari, D.; Dhia Fahmita, K.; Ikhwan Gautama, H.; Tri Hadmoko, S.; Prasetyo, M.E.; Mahdi, B.A.; Arafah, N.; Prasetyaningtyas, D.; et al. Hypertension is associated with antibody response and breakthrough infection in health care workers following vaccination with inactivated SARS-CoV-2. Vaccine 2022, 40, 4046–4056. [Google Scholar] [CrossRef]
- Soetedjo, N.N.M.; Iryaningrum, M.R.; Lawrensia, S.; Permana, H. Antibody response following SARS-CoV-2 vaccination among patients with type 2 diabetes mellitus: A systematic review. Diabetes Metab. Syndr. Clin. Res. Rev. 2022, 16, 102406. [Google Scholar] [CrossRef]
- He, Y.F.; Ouyang, J.; Hu, X.D.; Wu, N.; Jiang, Z.G.; Bian, N.; Wang, J. Correlation between COVID-19 vaccination and diabetes mellitus: A systematic review. World J. Diabetes 2023, 14, 892–918. [Google Scholar] [CrossRef]
- Chauvin, C.; Retnakumar, S.V.; Bayry, J. Obesity negatively impacts maintenance of antibody response to COVID-19 vaccines. Cell Rep. Med. 2023, 4, 101117. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.; Alahmad, B.; Al-Shammari, A.A.; Alterki, A.; Hammad, M.; Cherian, P.; Alkhairi, I.; Sindhu, S.; Thanaraj, T.A.; Mohammad, A.; et al. Previous COVID-19 Infection and Antibody Levels After Vaccination. Front. Public Health 2021, 9, 778243. [Google Scholar] [CrossRef]
- Zhang, R.; Khong, K.W.; Leung, K.Y.; Liu, D.; Fan, Y.; Lu, L.; Chan, P.-C.; Chen, L.; To, K.K.-W.; Chen, H.; et al. Antibody Response of BNT162b2 and CoronaVac Platforms in Recovered Individuals Previously Infected by COVID-19 against SARS-CoV-2 Wild Type and Delta Variant. Vaccines 2021, 9, 1442. [Google Scholar] [CrossRef]
- Lau, C.S.; Oh, M.L.H.; Phua, S.K.; Liang, Y.L.; Li, Y.; Huo, J.; Huang, Y.; Zhang, B.; Xu, S.; Aw, T.C. Kinetics of the Neutralizing and Spike SARS-CoV-2 Antibodies following the Sinovac Inactivated Virus Vaccine Compared to the Pfizer mRNA Vaccine in Singapore. Antibodies 2022, 11, 38. [Google Scholar] [CrossRef]
- Liu, K.T.; Han, Y.J.; Wu, G.H.; Huang, K.Y.A.; Huang, P.N. Overview of Neutralization Assays and International Standard for Detecting SARS-CoV-2 Neutralizing Antibody. Viruses 2022, 14, 1560. [Google Scholar] [CrossRef]
- Tiwari, A.K.; Negi, G.; Jaiswal, R.M.; Aggarwal, G.; Yadav, N.; Kumar, V.; Kulathu, K. Correlation of sample-to-cut-off ratio of anti-SARS-CoV-2 IgG antibody chemiluminescent assay with neutralization activity: A prospective multi-centric study in India. ISBT Sci. Ser. 2021, 16, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Saito, K.; Ai, T.; Nojiri, S.; Khasawneh, A.; Paran, F.J.; Horiuchi, Y.; Takei, S.; Yamamoto, T.; Wakita, M.; et al. Performance evaluation of the Ortho VITROS SARS-CoV-2 Spike-Specific Quantitative IgG test by comparison with the surrogate virus neutralizing antibody test and clinical assessment. PLoS ONE 2023, 18, e0279779. [Google Scholar] [CrossRef]
- Kitagawa, Y.; Imai, K.; Matsuoka, M.; Fukada, A.; Kubota, K.; Sato, M.; Takada, T.; Noguchi, S.; Tarumoto, N.; Maesaki, S.; et al. Evaluation of the correlation between the access SARS-CoV-2 IgM and IgG II antibody tests with the SARS-CoV-2 surrogate virus neutralization test. J. Med. Virol. 2022, 94, 335–341. [Google Scholar] [CrossRef]
- Ye, Q.; Zhang, T.; Lu, D. Potential false-positive reasons for SARS-CoV-2 antibody testing and its solution. J. Med. Virol. 2021, 93, 4242–4246. [Google Scholar] [CrossRef] [PubMed]
- Hicks, J.; Klumpp-Thomas, C.; Kalish, H.; Shunmugavel, A.; Mehalko, J.; Denson, J.P.; Snead, K.R.; Drew, M.; Corbett, K.S.; Graham, B.S.; et al. Serologic Cross-Reactivity of SARS-CoV-2 with Endemic and Seasonal Betacoronaviruses. J. Clin. Immunol. 2021, 41, 906–913. [Google Scholar] [CrossRef]
- Lokida, D.; Karyana, M.; Kosasih, H.; Mardian, Y.; Sugiyono, R.I.; Arlinda, D.; Lukman, N.; Salim, G.; Butar Butar, D.P.; Naysilla, A.M.; et al. Performance and correlation of ten commercial immunoassays for the detection of SARS-CoV-2 antibodies. Heliyon 2022, 8, e12614. [Google Scholar] [CrossRef] [PubMed]
- Vilibic-Cavlek, T.; Bogdanic, M.; Borko, E.; Hruskar, Z.; Zilic, D.; Ferenc, T.; Tabain, I.; Barbic, L.; Vujica Ferenc, M.; Ferencak, I.; et al. Detection of SARS-CoV-2 Antibodies: Comparison of Enzyme Immunoassay, Surrogate Neutralization and Virus Neutralization Test. Antibodies 2023, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Pieri, M.; Infantino, M.; Manfredi, M.; Nuccetelli, M.; Grossi, V.; Lari, B.; Tomassetti, F.; Sarubbi, S.; Russo, E.; Amedei, A.; et al. Performance evaluation of four surrogate Virus Neutralization Tests (sVNTs) in comparison to the in vivo gold standard test. Front. Biosci-Landmark 2022, 27, 074. [Google Scholar] [CrossRef] [PubMed]
- Shibata, S.; Ishiguro, T.; Kobayashi, Y.; Koike, M.; Numano, T.; Shimizu, Y.; Takayanagi, N. High incidence of false-positive results of IgG antibody against SARS-CoV-2 with rapid immunochromatographic antibody test due to human common cold coronavirus infection. Respir. Med. Case Rep. 2020, 31, 101180. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Rusling, J.F. COVID-19 Antibody Tests and Their Limitations. ACS Sens. 2021, 6, 593–612. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jia, Y.; Ji, Y.; Cong, X.; Liu, Y.; Yang, R.; Kong, X.; Shi, Y.; Zhu, L.; Wang, Z.; et al. Inactivated Vaccines Against SARS-CoV-2: Neutralizing Antibody Titers in Vaccine Recipients. Front. Microbiol. 2022, 13, 816778. [Google Scholar] [CrossRef]
- Gillot, C.; Favresse, J.; David, C.; Maloteau, V.; Dogne, J.M.; Douxfils, J. An Evaluation of a SARS-CoV-2 Pseudovirus Neutralization Test and A Comparison to a SARS-CoV-2 Surrogate Virus Neutralization Test in a COVID-19 Long-Term Follow-Up Cohort. Microbiol. Res. 2024, 15, 422–430. [Google Scholar] [CrossRef]

| Characteristics | Frequency (n = 79) | Percentage (%) | |
|---|---|---|---|
| Age, (years) | |||
| Median | 48 | ||
| Min–Max | 21–76 | ||
| Sex | |||
| Male | 34 | 43 | |
| Female | 45 | 57 | |
| History of Smoking | |||
| No | 74 | 93.7 | |
| Yes | 5 | 6.3 | |
| Comorbidity | |||
| History of Chronic Illness | 19 | 24.1 | |
| Hypertension | 12 | 15.2 | |
| Type 2 Diabetes Mellitus | 5 | 6.3 | |
| Asthma Bronchiale | 3 | 3.8 | |
| Chronic Obstructive Pulmonary Disease | 1 | 1.3 | |
| Body Mass Index (kg/m2) | |||
| Underweight (<18.5) | 1 | 1.3 | |
| Normal weight (18.5–24.9) | 33 | 41.8 | |
| Overweight (25–29.9) | 34 | 43 | |
| Obese (≥30) | 11 | 10.1 | |
| Complete COVID-19 Vaccination * | 58 | 73.4 | |
| Previous COVID-19 Infection | 2 | 2.5 | |
| IgG anti-SARS-CoV-2 Ab CLIA (U/L) | |||
| Median | 3.18 | ||
| Min–Max | 0.49–196.66 | ||
| SARS-CoV-2 SVNT (%) | |||
| Median | 46.91 | ||
| Min–Max | 5.09–98.19 |
| SVNT | ||||||
|---|---|---|---|---|---|---|
| S-RBD SARS-CoV-2 IgG CLIA (BAU/mL) | Protective (≥30%) | Non-Protective (<30%) | Sensitivity (%) | Specificity (%) | Positive Predictive Value (%) | Negative Predictive Value (%) |
| ≥37.29 | 55 | 2 | 90.5 | |||
| <37.29 | 2 | 20 | 96.5 | 96.5 | 90.9 | |
| Total | 57 | 22 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Indrati, A.R.; Horian, E.; Dewi, N.S.; Suraya, N.; Tiara, M.R.; Djauhari, H.; Alisjahbana, B. The Protection Level of S-RBD SARS-CoV-2 Immunoglobulin G Antibodies Using the Chemiluminescent Immunoassay Compared to the Surrogate Virus Neutralization Test Method. Diagnostics 2024, 14, 1776. https://doi.org/10.3390/diagnostics14161776
Indrati AR, Horian E, Dewi NS, Suraya N, Tiara MR, Djauhari H, Alisjahbana B. The Protection Level of S-RBD SARS-CoV-2 Immunoglobulin G Antibodies Using the Chemiluminescent Immunoassay Compared to the Surrogate Virus Neutralization Test Method. Diagnostics. 2024; 14(16):1776. https://doi.org/10.3390/diagnostics14161776
Chicago/Turabian StyleIndrati, Agnes Rengga, Erinca Horian, Nina Susana Dewi, Nida Suraya, Marita Restie Tiara, Hofiya Djauhari, and Bachti Alisjahbana. 2024. "The Protection Level of S-RBD SARS-CoV-2 Immunoglobulin G Antibodies Using the Chemiluminescent Immunoassay Compared to the Surrogate Virus Neutralization Test Method" Diagnostics 14, no. 16: 1776. https://doi.org/10.3390/diagnostics14161776





