A Novel Ensemble Framework for Multi-Classification of Brain Tumors Using Magnetic Resonance Imaging
Abstract
1. Introduction
1.1. Motivation
- Existing studies have generally applied the ensemble technique by majority voting on a few predetermined CNN models. To the best of our knowledge, there are no studies in the literature on determining the base models and the weights to which they will contribute.
- Even if the CNN models proposed in existing studies are optimized, they perform limited feature extraction from the dataset. For example, features extracted from a scratch CNN model or a few predetermined CNN models fall into this group. Feature extraction should be diversified with CNN models with different architectures.
1.2. Contributions
- We introduce a new ensemble strategy for gathering the best performance. The most appropriate CNN models were iteratively identified and combined with ensemble learning at optimum weights to classify three brain tumor types accurately.
- Utilized a PSO-based algorithm to find the optimum weights that enhance the performance of ensemble CNN models.
- The proposed PSO-Ensemble framework utilizes three different datasets and demonstrates outstanding performance, as supported by extensive experimental results.
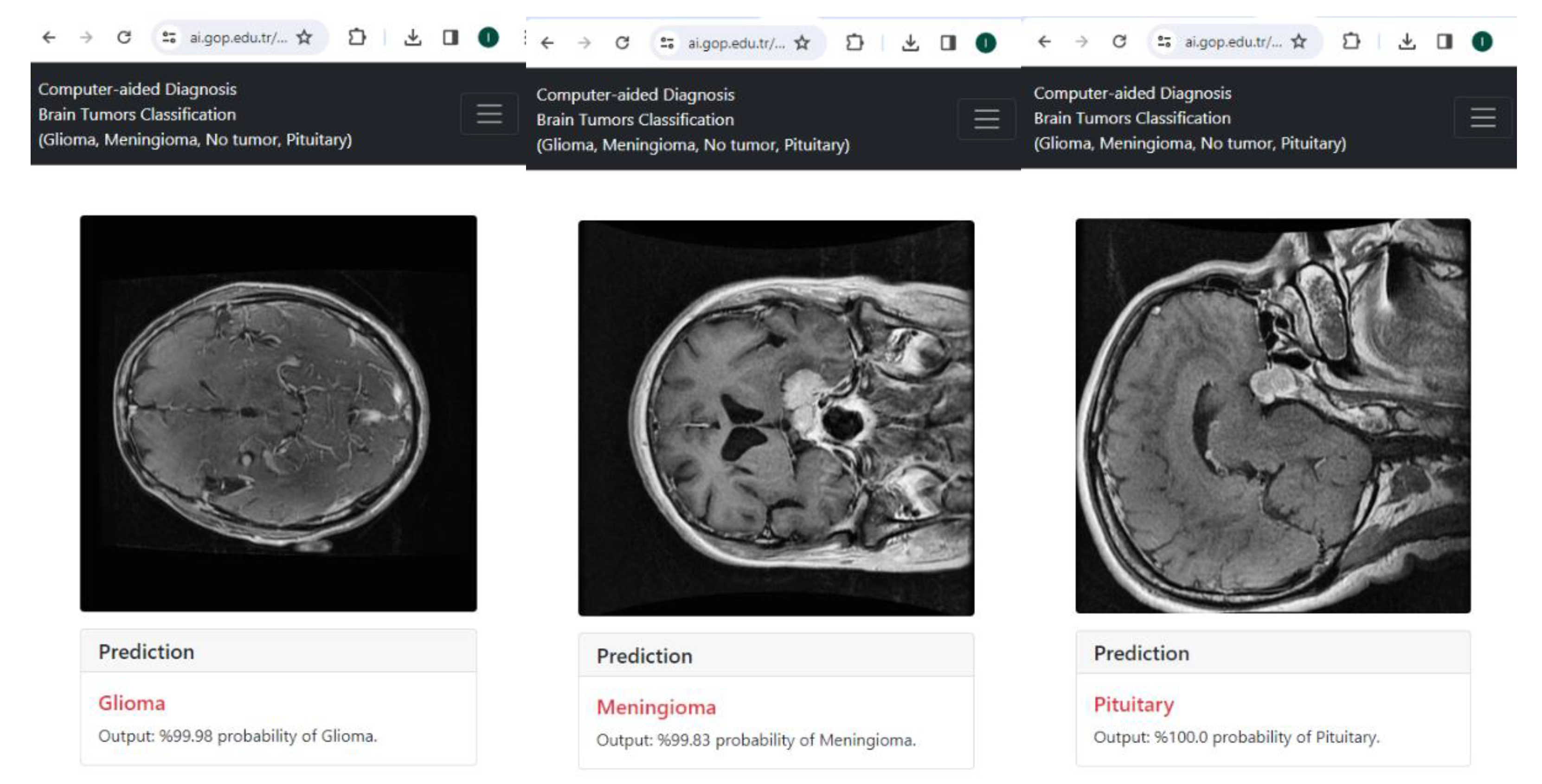
- Existing studies have generally not presented the use of their models. The framework proposed in this study is integrated into the online system and available for use (https://ai.gop.edu.tr/bt, accessed on 8 February 2024).
2. Related Works
3. Materials and Methods
3.1. Dataset
3.2. Transfer Learning
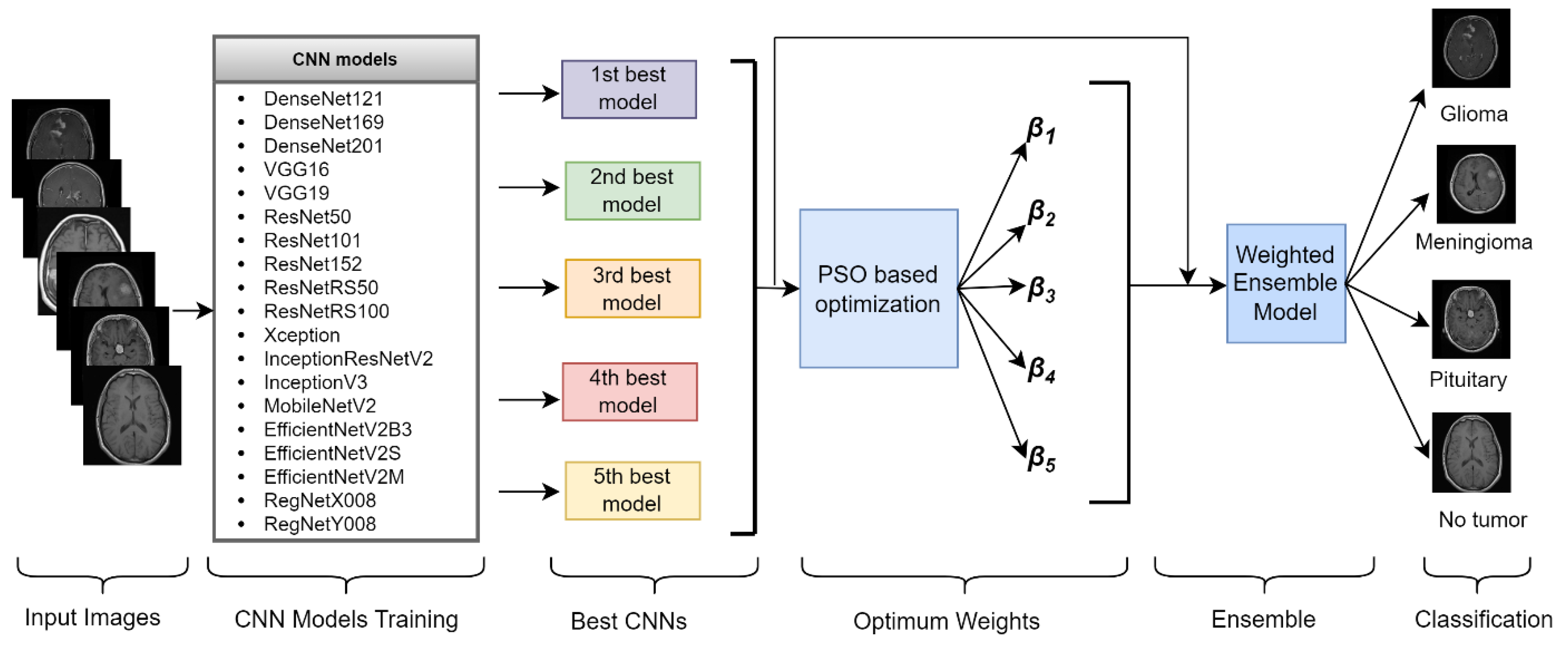
3.3. Proposed Framework
| Algorithm 1 PSO-based weighted ensemble learning algorithm |
| Obtain prediction probabilities (Pi) for each model; initial values of βi are determined randomly for each particle, number of particles:= 100, maxIteration: = 1000 while i < maxIteration for particle in swarm do: for m in models do: #Calculate final probabilities via Equation (3) newPredictions += particle[m] × modelPredictions[m] #Calculate objective value (loss) via Equation (4) loss_score = log_loss(y, newPredictions) results.append(loss_score) end for for j in swarm do if results[j] < individualBestResult[j] then individualBestResult[j]: = results[j] end if #Find minimum objective value and βi in particles if min(results) < bestGlobalObjectiveValue then bestGlobalObjectiveValue: = min(results) bestβi: = βi end if Update βi in each particle according to Equations (1) and (2) Adjust βi in each particle to satisfy Equation (5) i: = i + 1 end while |
3.4. Performance Metrics
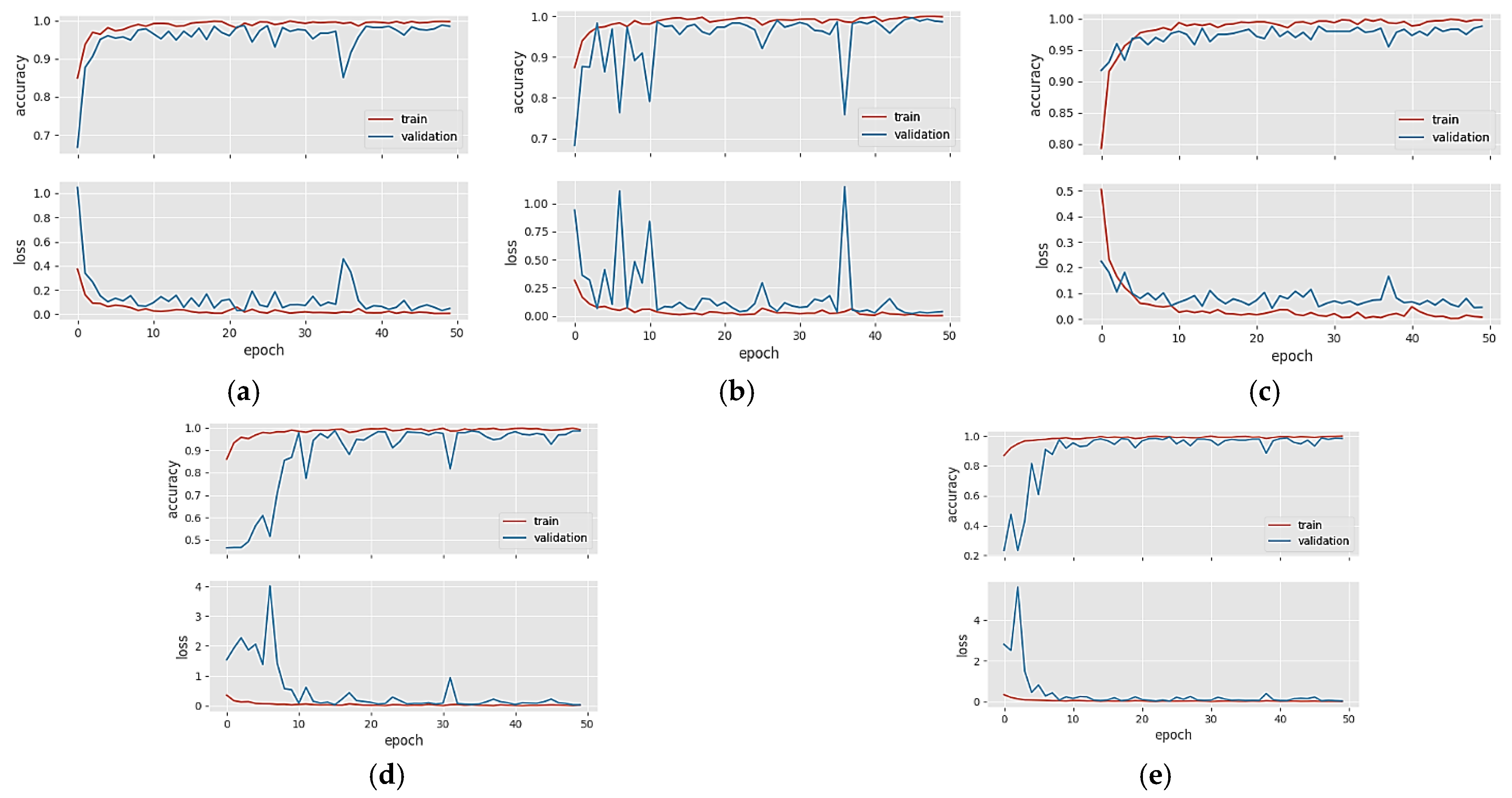
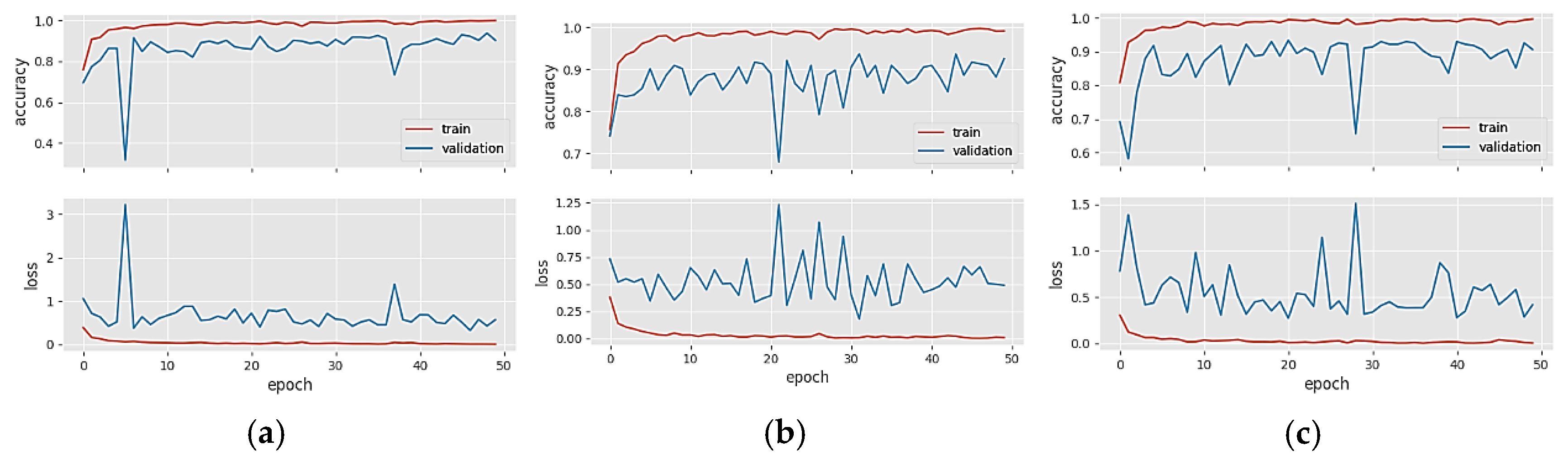
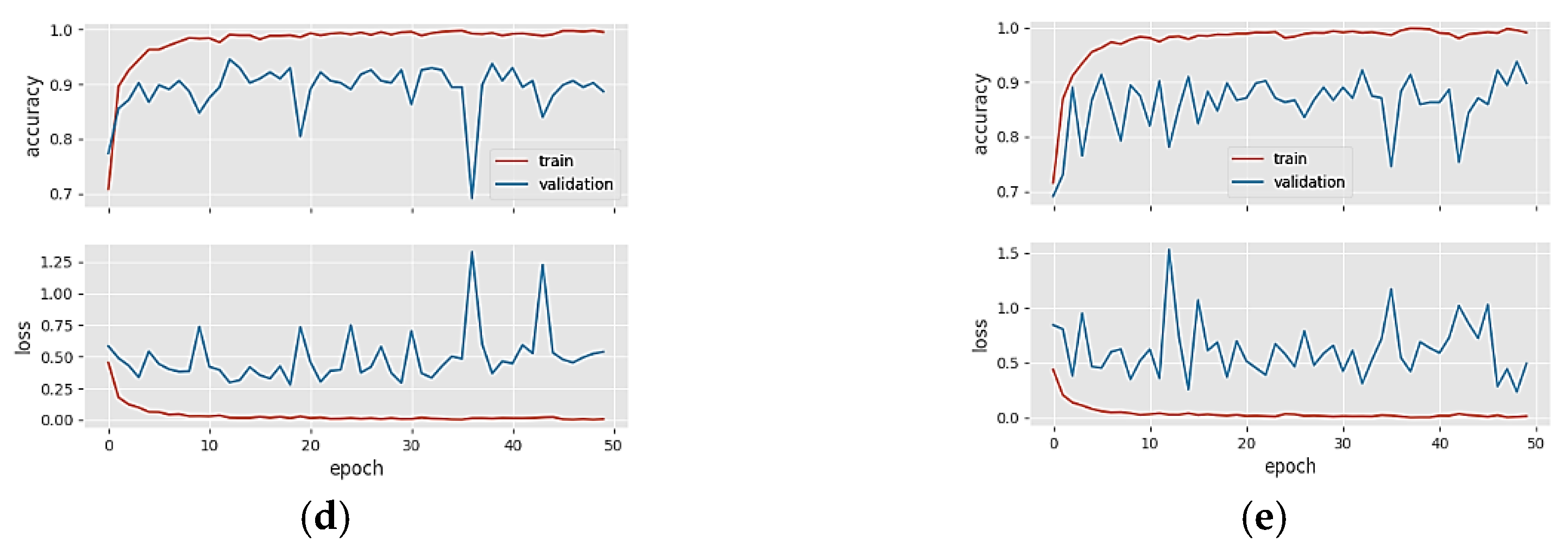
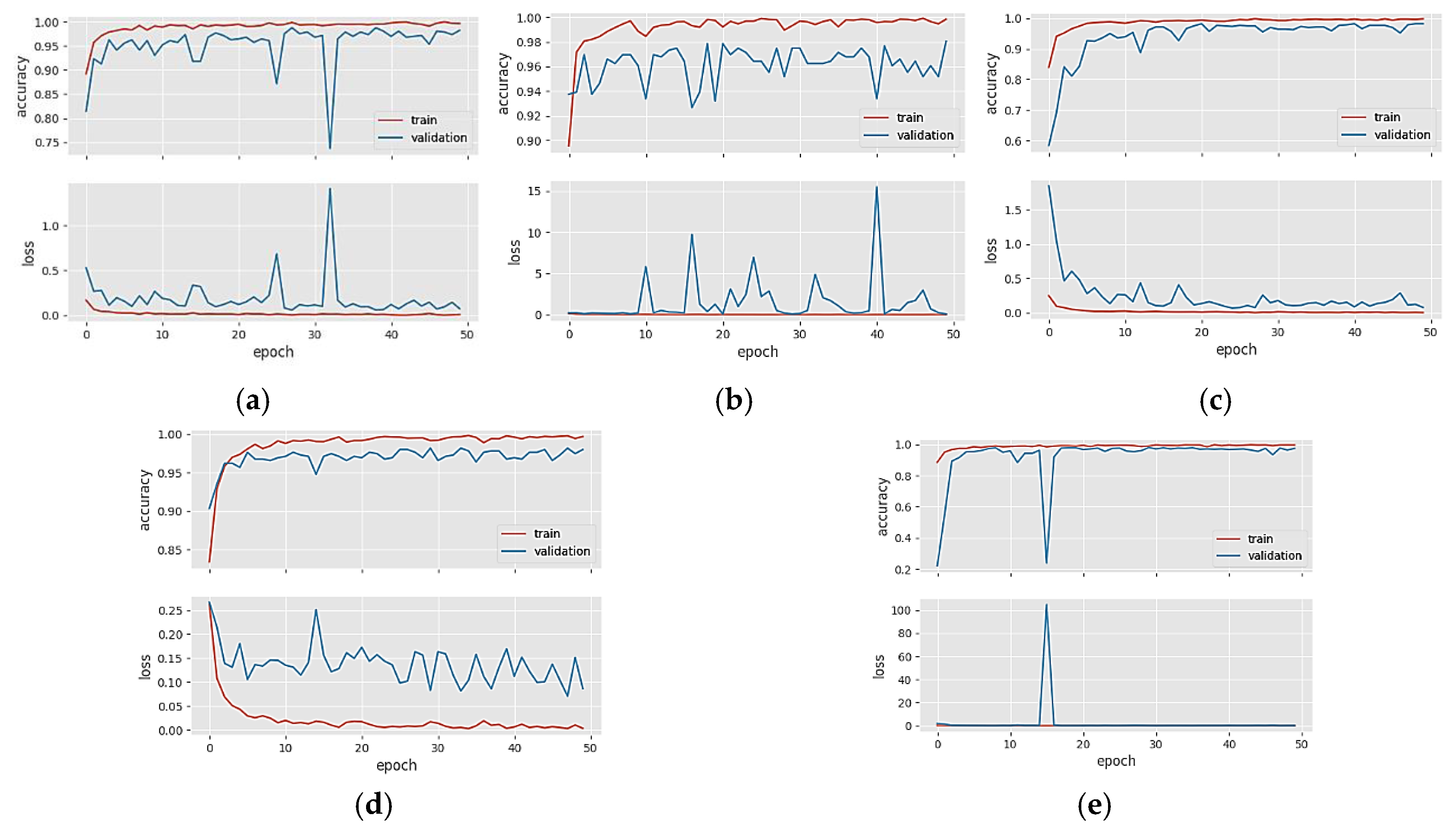
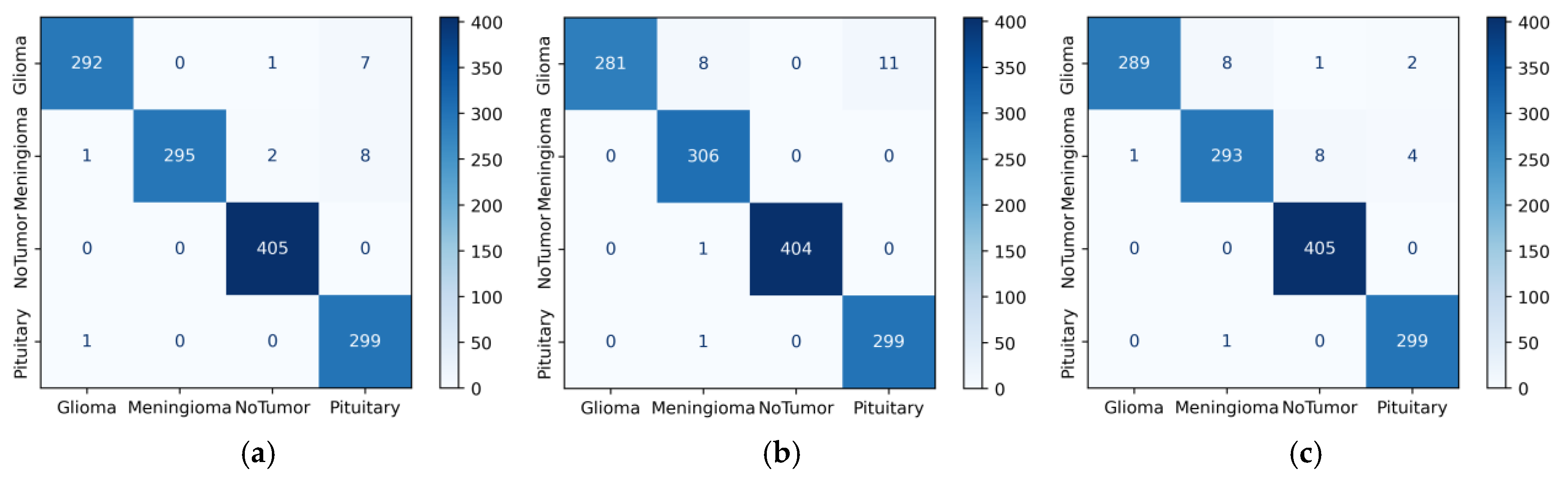
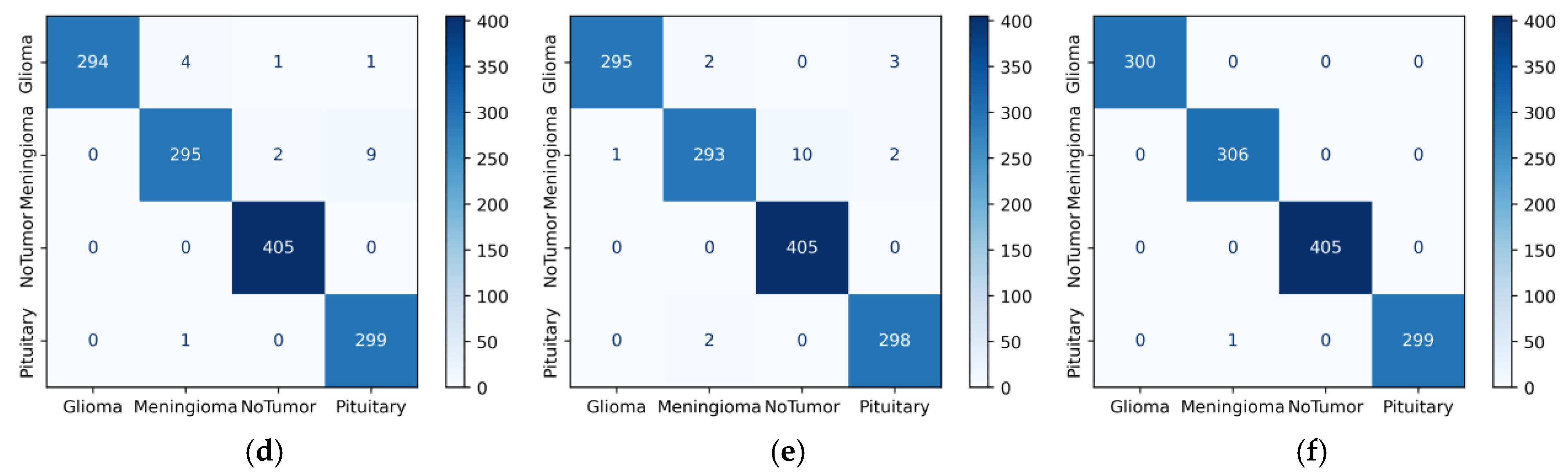
4. Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aurna, N.F.; Yousuf, M.A.; Taher, K.A.; Azad, A.K.M.; Moni, M.A. A classification of MRI brain tumor based on two stage feature level ensemble of deep CNN models. Comput. Biol. Med. 2022, 146, 105539. [Google Scholar] [CrossRef] [PubMed]
- Mehnatkesh, H.; Jalali, S.M.J.; Khosravi, A.; Nahavandi, S. An intelligent driven deep residual learning framework for brain tumor classification using MRI images. Expert Syst. Appl. 2023, 213, 119087. [Google Scholar] [CrossRef]
- Gupta, R.K.; Bharti, S.; Kunhare, N.; Sahu, Y.; Pathik, N. Brain tumor detection and classification using cycle generative adversarial networks. Interdiscip. Sci. Comput. Life Sci. 2022, 14, 485–502. [Google Scholar] [CrossRef] [PubMed]
- Nayak, D.R.; Padhy, N.; Mallick, P.K.; Bagal, D.K.; Kumar, S. Brain tumor classification using noble deep learning approach with parametric optimization through metaheuristics approaches. Computers 2022, 11, 10. [Google Scholar] [CrossRef]
- Kaya, M.; Çetin-Kaya, Y. Seamless computation offloading for mobile applications using an online learning algorithm. Computing 2021, 103, 771–799. [Google Scholar] [CrossRef]
- Nguyen, T.D.; Le, D.-T.; Bum, J.; Kim, S.; Song, S.J.; Choo, H. Retinal disease diagnosis using deep learning on ultra-wide-field fundus images. Diagnostics 2024, 14, 105. [Google Scholar] [CrossRef] [PubMed]
- Kaya, M. Feature fusion-based ensemble CNN learning optimization for automated detection of pediatric pneumonia. Biomed. Signal Process. Control 2024, 87, 105472. [Google Scholar] [CrossRef]
- Noor, M.B.T.; Zenia, N.Z.; Kaiser, M.S.; Mamun, S.A.; Mahmud, M. Application of deep learning in detecting neurological disorders from magnetic resonance images: A survey on the detection of Alzheimer’s disease, Parkinson’s disease and schizophrenia. Brain Inform. 2020, 7, 11. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Zhang, C.; Bengio, S.; Hardt, M.; Recht, B.; Vinyals, O. Understanding deep learning (still) requires rethinking generalization. Commun. ACM 2021, 64, 107–115. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the International Conference on Machine Learning, Lille, France, 6–11 July 2015; pp. 448–456. [Google Scholar]
- Kumar, A.; Kim, J.; Lyndon, D.; Fulham, M.; Feng, D. An ensemble of fine-tuned convolutional neural networks for medical image classification. IEEE J. Biomed. Health Inform. 2016, 21, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, J.; Eberhart, R. Particle swarm optimization. In Proceedings of the ICNN’95-International Conference on Neural Networks, Perth, Australia, 27 November–1 December 1995; pp. 1942–1948. [Google Scholar]
- Mahmud, M.; Kaiser, M.S.; McGinnity, T.M.; Hussain, A. Deep learning in mining biological data. Cogn. Comput. 2021, 13, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, M.; Kaiser, M.S.; Hussain, A.; Vassanelli, S. Applications of deep learning and reinforcement learning to biological data. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 2063–2079. [Google Scholar] [CrossRef] [PubMed]
- Ayadi, W.; Elhamzi, W.; Charfi, I.; Atri, M. Deep CNN for brain tumor classification. Neural Process. Lett. 2021, 53, 671–700. [Google Scholar] [CrossRef]
- Raza, A.; Ayub, H.; Khan, J.A.; Ahmad, I.; Salama, A.S.; Daradkeh, Y.I.; Javeed, D.; Ur Rehman, A.; Hamam, H. A hybrid deep learning-based approach for brain tumor classification. Electronics 2022, 11, 1146. [Google Scholar] [CrossRef]
- Khan, M.S.I.; Rahman, A.; Debnath, T.; Karim, M.R.; Nasir, M.K.; Band, S.S.; Mosavi, A.; Dehzangi, I. Accurate brain tumor detection using deep convolutional neural network. Comput. Struct. Biotechnol. J. 2022, 20, 4733–4745. [Google Scholar] [CrossRef]
- Rahman, T.; Islam, M.S. MRI brain tumor detection and classification using parallel deep convolutional neural networks. Meas. Sens. 2023, 26, 100694. [Google Scholar] [CrossRef]
- Asif, S.; Zhao, M.; Tang, F.; Zhu, Y. An enhanced deep learning method for multi-class brain tumor classification using deep transfer learning. Multimed. Tools Appl. 2023, 82, 31709–31736. [Google Scholar] [CrossRef]
- Saurav, S.; Sharma, A.; Saini, R.; Singh, S. An attention-guided convolutional neural network for automated classification of brain tumor from MRI. Neural Comput. Appl. 2023, 35, 2541–2560. [Google Scholar] [CrossRef]
- Akter, A.; Nosheen, N.; Ahmed, S.; Hossain, M.; Yousuf, M.A.; Almoyad, M.A.A.; Hasan, K.F.; Moni, M. A Robust clinical applicable CNN and U-Net based algorithm for MRI classification and segmentation for brain tumor. Expert Syst. Appl. 2024, 238, 122347. [Google Scholar] [CrossRef]
- Deepak, S.; Ameer, P.M. Brain tumor classification using deep CNN features via transfer learning. Comput. Biol. Med. 2019, 111, 103345. [Google Scholar] [CrossRef] [PubMed]
- Swati, Z.N.K.; Zhao, Q.; Kabir, M.; Ali, F.; Ali, Z.; Ahmed, S.; Lu, J. Brain tumor classification for MR images using transfer learning and fine-tuning. Comput. Med. Imaging Graph. 2019, 75, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Ismael, S.A.A.; Mohammed, A.; Hefny, H. An enhanced deep learning approach for brain cancer MRI images classification using residual networks. Artif. Intell. Med. 2020, 102, 101779. [Google Scholar] [CrossRef] [PubMed]
- Mehrotra, R.; Ansari, M.A.; Agrawal, R.; Anand, R.S. A transfer learning approach for AI-based classification of brain tumors. Mach. Learn. Appl. 2020, 2, 100003. [Google Scholar] [CrossRef]
- Rasool, M.; Ismail, N.A.; Boulila, W.; Ammar, A.; Samma, H.; Yafooz, W.M.; Emara, A.H.M. A hybrid deep learning model for brain tumour classification. Entropy 2022, 24, 799. [Google Scholar] [CrossRef]
- Badjie, B.; Ülker, E.D. A deep transfer learning based architecture for brain tumor classification using MR Images. Inf. Technol. Control 2022, 51, 332–344. [Google Scholar] [CrossRef]
- Alnowami, M.; Taha, E.; Alsebaeai, S.; Muhammad Anwar, S.; Alhawsawi, A. MR image normalization dilemma and the accuracy of brain tumor classification model. J. Radiat. Res. Appl. Sci. 2022, 15, 33–39. [Google Scholar] [CrossRef]
- Talukder, M.A.; Islam, M.M.; Uddin, M.A.; Akhter, A.; Pramanik, M.A.J.; Aryal, S.; Almoyad, M.A.A.; Hasan, K.F.; Moni, M.A. An efficient deep learning model to categorize brain tumor using reconstruction and fine-tuning. Expert Syst. Appl. 2023, 230, 120534. [Google Scholar] [CrossRef]
- Zulfiqar, F.; Ijaz Bajwa, U.; Mehmood, Y. Multi-class classification of brain tumor types from MR images using EfficientNets. Biomed. Signal Process. Control 2023, 84, 104777. [Google Scholar] [CrossRef]
- Alanazi, M.F.; Ali, M.U.; Hussain, S.J.; Zafar, A.; Mohatram, M.; Irfan, M.; Albarrak, A.M. Brain tumor/mass classification framework using magnetic-resonance-imaging-based isolated and developed transfer deep-learning model. Sensors 2022, 22, 372. [Google Scholar] [CrossRef]
- Gómez-Guzmán, M.A.; Jiménez-Beristain, L.; García-Guerrero, E.E.; López-Bonilla, O.R.; Tamayo-Pérez, U.J.; Esqueda-Elizondo, J.J.; Palomino-Vizcaino, K.; Inzunza-González, E. Classifying brain tumors on magnetic resonance imaging by using convolutional neural networks. Electronics 2023, 12, 955. [Google Scholar] [CrossRef]
- Rezaei, K.; Agahi, H.; Mahmoodzadeh, A. A weighted voting classifiers ensemble for the brain tumors classification in mr images. IETE J. Res. 2020, 5, 3829–3842. [Google Scholar] [CrossRef]
- Noreen, N.; Palaniappan, S.; Qayyum, A.; Ahmad, I.; Alassafi, M.O. Brain tumor classification based on fine-tuned models and the ensemble method. Comput. Mater. Contin. 2021, 67, 3967–3982. [Google Scholar] [CrossRef]
- Patil, S.; Kirange, D. Ensemble of deep learning models for brain tumor detection. Procedia Comput. Sci. 2023, 218, 2468–2479. [Google Scholar] [CrossRef]
- Khan, F.; Ayoub, S.; Gulzar, Y.; Majid, M.; Reegu, F.A.; Mir, M.S.; Soomro, A.B.; Elwasila, O. MRI-based effective ensemble frameworks for predicting human brain tumor. J. Imaging 2023, 9, 163. [Google Scholar] [CrossRef] [PubMed]
- Tandel, G.S.; Tiwari, A.; Kakde, O.G.; Gupta, N.; Saba, L.; Suri, J.S. Role of ensemble deep learning for brain tumor classification in multiple magnetic resonance imaging sequence data. Diagnostics 2023, 13, 481. [Google Scholar] [CrossRef]
- Kang, J.; Ullah, Z.; Gwak, J. Mri-based brain tumor classification using ensemble of deep features and machine learning classifiers. Sensors 2021, 21, 2222. [Google Scholar] [CrossRef]
- Ait Amou, M.; Xia, K.; Kamhi, S.; Mouhafid, M. A Novel MRI diagnosis method for brain tumor classification based on cnn and Bayesian optimization. Healthcare 2022, 10, 494. [Google Scholar] [CrossRef]
- Devi, R.L. Detection and automated classification of brain tumor types in MRI images using a convolutional neural network with grid search optimization. In Proceedings of the Fifth International Conference on I-SMAC (IoT in Social, Mobile, Analytics and Cloud) (I-SMAC), Palladam, India, 11–13 November 2021; pp. 1280–1284. [Google Scholar]
- Dehkordi, A.A.; Hashemi, M.; Neshat, M.; Mirjalili, S.; Sadiq, A.S. Brain tumor detection and classification using a new evolutionary convolutional neural network. arXiv 2022, arXiv:2204.12297. [Google Scholar] [CrossRef]
- Bashkandi, A.H.; Sadoughi, K.; Aflaki, F.; Alkhazaleh, H.A.; Mohammadi, H.; Jimenez, G. Combination of political optimizer, particle swarm optimizer, and convolutional neural network for brain tumor detection. Biomed. Signal Process. Control 2023, 81, 104434. [Google Scholar] [CrossRef]
- Wu, P.; Shen, J. Brain tumor diagnosis based on convolutional neural network improved by a new version of political optimizer. Biomed. Signal Process. Control 2023, 85, 104853. [Google Scholar] [CrossRef]
- Anaraki, A.K.; Ayati, M.; Kazemi, F. Magnetic resonance imaging-based brain tumor grades classification and grading via convolutional neural networks and genetic algorithms. Biocybern. Biomed. Eng. 2019, 39, 63–74. [Google Scholar] [CrossRef]
- Bacanin, N.; Bezdan, T.; Venkatachalam, K.; Al-Turjman, F. Optimized convolutional neural network by firefly algorithm for magnetic resonance image classification of glioma brain tumor grade. J. Real-Time Image Process. 2021, 18, 1085–1098. [Google Scholar] [CrossRef]
- Bezdan, T.; Milosevic, S.; Venkatachalam, K.; Zivkovic, M.; Bacanin, N.; Strumberger, I. Optimizing convolutional neural network by hybridized elephant herding optimization algorithm for magnetic resonance image classification of glioma brain tumor grade. In Proceedings of the Zooming Innovation in Consumer Technologies Conference (ZINC), Novi Sad, Serbia, 26–27 May 2021; pp. 171–176. [Google Scholar]
- Kothandaraman, V. Binary swallow swarm optimization with convolutional neural network brain tumor classifier for magnetic resonance imaging images. Concurr. Comput. Pract. Exp. 2023, 35, e7661. [Google Scholar] [CrossRef]
- Rammurthy, D.; Mahesh, P.K. Whale Harris hawks optimization based deep learning classifier for brain tumor detection using MRI images. J. King Saud. Univ.-Comput. Inf. Sci. 2022, 34, 3259–3272. [Google Scholar] [CrossRef]
- Chawla, R.; Beram, S.M.; Murthy, C.R.; Thiruvenkadam, T.; Bhavani, N.P.G.; Saravanakumar, R.; Sathishkumar, P.J. Brain tumor recognition using an integrated bat algorithm with a convolutional neural network approach. Meas. Sens. 2022, 24, 100426. [Google Scholar] [CrossRef]
- Sharif, M.I.; Li, J.P.; Khan, M.A.; Kadry, S.; Tariq, U. M3BTCNet: Multi model brain tumor classification using metaheuristic deep neural network features optimization. Neural Comput. Appl. 2022, 36, 95–110. [Google Scholar] [CrossRef]
- Xu, L.; Mohammadi, M. Brain tumor diagnosis from MRI based on Mobilenetv2 optimized by contracted fox optimization algorithm. Heliyon 2024, 10, e23866. [Google Scholar] [CrossRef]
- Jun, C. Brain Tumor Dataset. 2017. Available online: https://figshare.com/articles/dataset/brain_tumor_dataset/1512427 (accessed on 5 May 2023).
- Sartaj, B. Brain Tumor Classification (MRI). 2020. Available online: https://www.kaggle.com/datasets/sartajbhuvaji/brain-tumor-classification-mri (accessed on 5 May 2023).
- Nickparvar, M. Brain Tumor MRI Dataset. Available online: https://www.kaggle.com/datasets/masoudnickparvar/brain-tumor-mri-dataset?select=Training (accessed on 5 May 2023).
- Hamada, A. Br35H Brain Tumor Detection 2020 Dataset. 2020. Available online: https://www.kaggle.com/ahmedhamada0/brain-tumor-detection/metadata (accessed on 5 May 2023).
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Tajbakhsh, N.; Shin, J.Y.; Gurudu, S.R.; Hurst, R.T.; Kendall, C.B.; Gotway, M.B.; Liang, J. Convolutional neural networks for medical image analysis: Full training or fine tuning? IEEE Trans. Med. Imaging 2016, 35, 1299–1312. [Google Scholar] [CrossRef] [PubMed]
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How transferable are features in deep neural networks? In Proceedings of the Advances in Neural Information Processing Systems 27: Annual Conference on Neural Information Processing Systems 2014, Montreal, QC, Canada, 8–13 December 2014. [Google Scholar]
- Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Powers, D. Precision, Recall and F-Measure to ROC, Informedness, Markedness & Correlation. J. Mach. Learn. Technol. 2011, 2.1, 37–63. [Google Scholar]
- Hassija, V.; Chamola, V.; Mahapatra, A.; Singal, A.; Goel, D.; Huang, K.; Scardapane, S.; Spinelli, I.; Mahmud, M.; Hussain, A. Interpreting black-box models: A review on explainable artificial intelligence. Cogn. Comput. 2023, 16, 45–74. [Google Scholar] [CrossRef]
- Jahan, S.; Taher, K.A.; Kaiser, M.S.; Mahmud, M.; Rahman, M.S.; Hosen, A.S.; Ra, I.H. Explainable AI-based Alzheimer’s prediction and management using multimodal data. PLoS ONE 2022, 18, e0294253. [Google Scholar] [CrossRef] [PubMed]
- Viswan, V.; Shaffi, N.; Mahmud, M.; Subramanian, K.; Hajamohideen, F. Explainable artificial intelligence in Alzheimer’s disease classification: A systematic review. Cogn. Comput. 2024, 16, 1–44. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Alyasseri, Z.A.A.; Al-Betar, M.A.; Doush, I.A.; Awadallah, M.A.; Abasi, A.K.; Makhadmeh, S.N.; Alomari, O.A.; Abdulkareem, K.H.; Adam, A.; Damasevicius, R.; et al. Review on COVID-19 diagnosis models based on machine learning and deep learning approaches. Expert Syst. 2022, 39, e12759. [Google Scholar] [CrossRef] [PubMed]

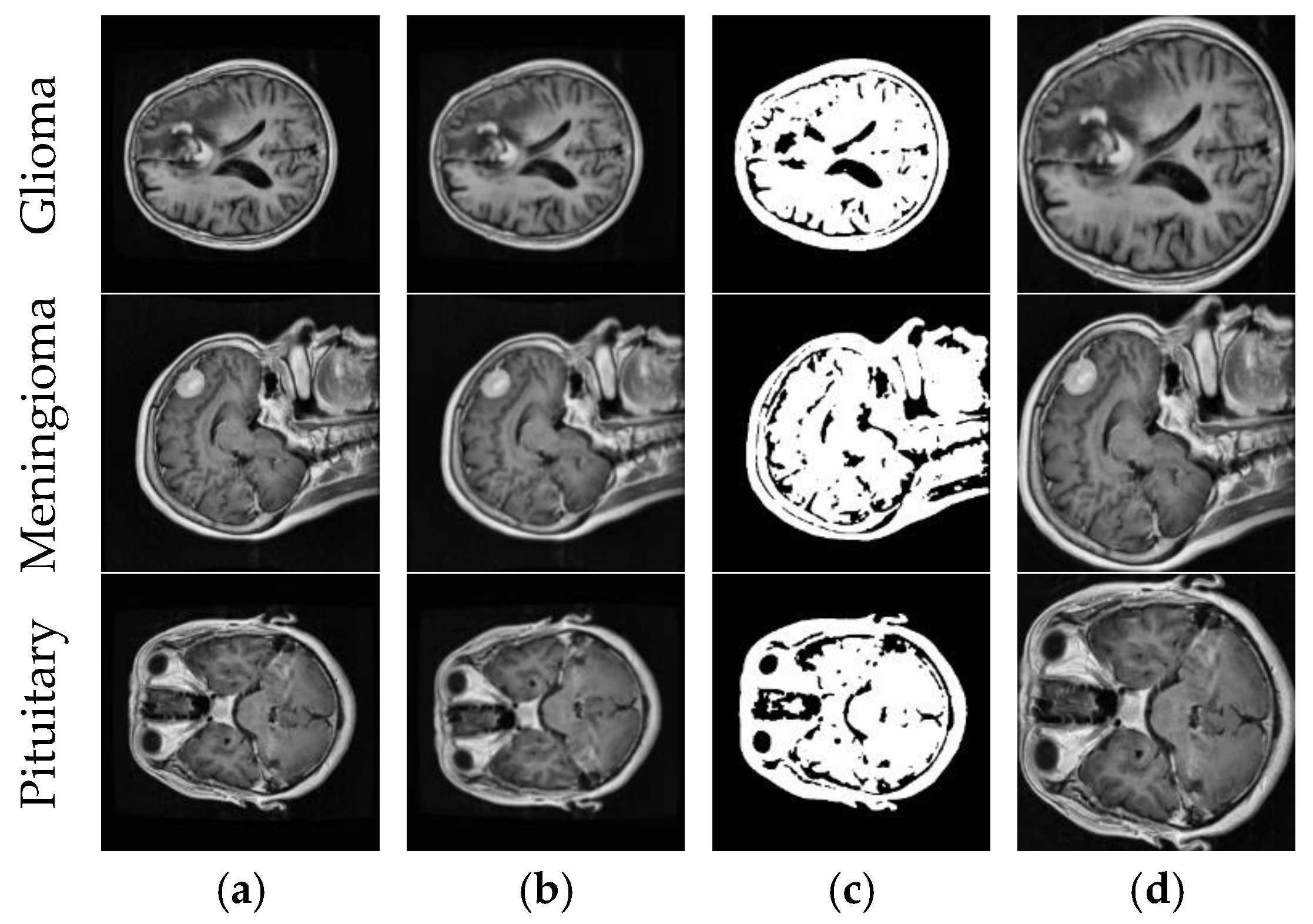


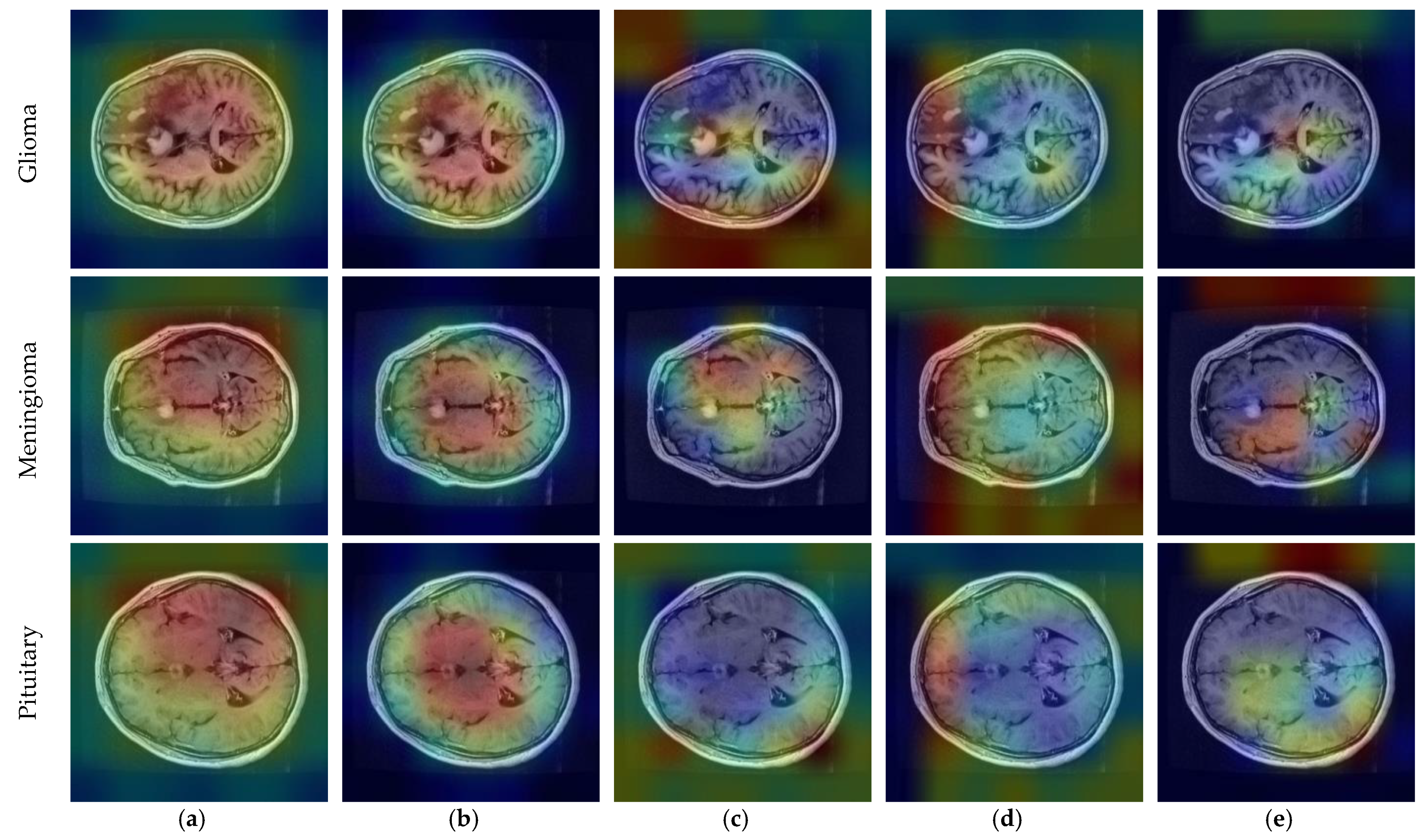
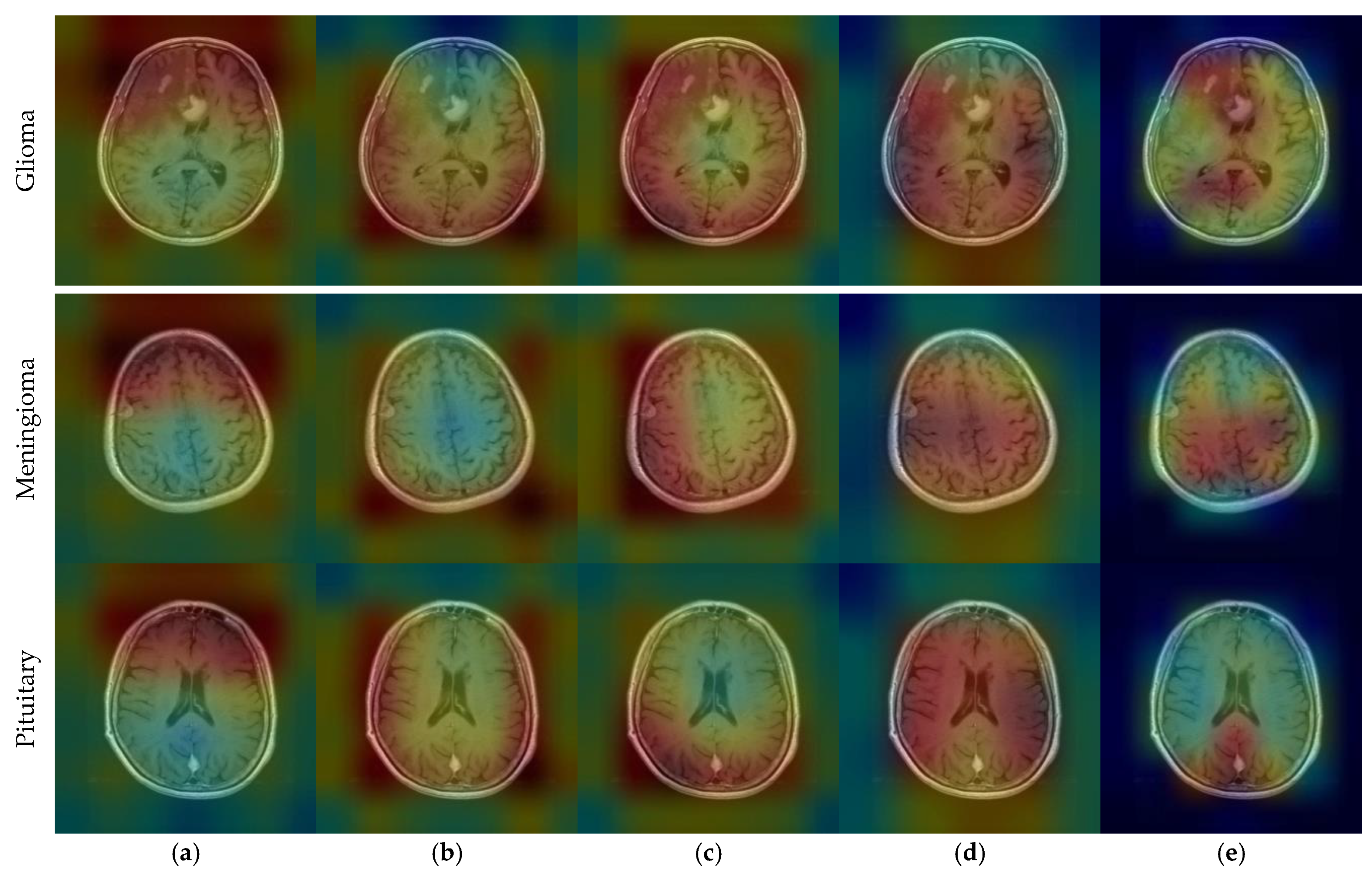

| Method | Reference | Year | Dataset | Classification Type | Accuracy (%) |
|---|---|---|---|---|---|
| Scratch Model | Ayadi et al. [19] | 2021 | Figshare MRI | Multi | 94.74 |
| Radiopaedia | 93.71 | ||||
| Rembrandt | 95 | ||||
| Raza et al. [20] | 2022 | CE-MRI | Multi | 99.67 | |
| Khan et al. [21] | 2022 | Figshare MRI | Multi | 97.8 | |
| Harvard Medical | Binary | 100 | |||
| Rahman and Islam [22] | 2023 | Figshare MRI | Multi | 97.60 | |
| Kaggle-Nickparvar | 98.12 | ||||
| Asif et al. [23] | 2023 | Figshare MRI | Multi | 99.67 | |
| Kaggle -Sartaj | 95.87 | ||||
| Saurav et al. [24] | 2023 | BT-Small-2C | Binary | 96.08 | |
| BT-Large-2C | 99.83 | ||||
| BT-Large-3C | Multi | 97.23 | |||
| BT-Large-4C | 95.71 | ||||
| Akter et al. [25] | 2024 | Dataset-a | Binary | 96.7 | |
| Dataset-b | 89.4 | ||||
| Dataset-c | 97.7 | ||||
| Dataset-d | 95.2 | ||||
| Merged Dataset-1 | 98.7 | ||||
| Merged Dataset-2 | 97.6 | ||||
| Transfer Learning | Swati et al. [27] | 2019 | CE-MRI | Multi | 94.82 |
| Deepak and Ameer [26] | 2019 | Figshare MRI | Multi | 97.1 | |
| Abdelaziz et al. [28] | 2020 | CE-MRI | Multi | 99 | |
| Mehrotra et al. [29] | 2020 | TCIA | Binary | 99.04 | |
| Rasool et al. [30] | 2022 | Kaggle-Sartaj | Multi | 98.1 | |
| Badjie and Deniz Ülker [31] | 2022 | BraTS2020 | Binary | 99.62 | |
| Alnowami et al. [32] | 2022 | Dataset-1 | Multi | 72.10 | |
| Dataset-2 | 87.02 | ||||
| Dataset-3 | 96.52 | ||||
| Talukder et al. [33] | 2023 | Figshare MRI | Multi | 99.68 | |
| Zulfiqar et al. [34] | 2023 | Figshare MRI | Multi | 98.86 | |
| Alanazi et al. [35] | 2022 | Br35H | Binary | 99.33 | |
| Kaggle-Sartaj | Multi | 96.90 | |||
| Figshare MRI | Multi | 95.75 | |||
| Gomez et al. [36] | 2023 | Kaggle-Nickparvar | Multi | 97.12 | |
| Ensemble Learning | Rezaei et al. [37] | 2020 | MRI Dataset | Multi | 92.46 |
| Noreen et al. [38] | 2021 | MRI dataset | Multi | 94.34 | |
| Patil and Kirange [39] | 2023 | Figshare MRI | Multi | 97.77 | |
| Aurna et al. [1] | 2022 | Figshare MRI | Multi | 99.13 | |
| Kaggle-Sartaj | Multi | 98.96 | |||
| Kang et al. [42] | 2021 | Kaggle-Sartaj | Multi | 93.72 | |
| Khan et al. [40] | 2023 | Figshare MRI | Binary | 95.4 | |
| Tantel et al. [41] | 2023 | T1W | Binary | 94.75 | |
| T2W | 97.98 | ||||
| FLAIR | 98.88 | ||||
| With the help of Optimization Algorithms | Ait-Amou et al. [43] | 2022 | Figshare MRI | Multi | 98.70 |
| Devi [44] | 2021 | Kaggle-Sartaj | Multi | 90.25 | |
| Dehkordi et al. [45] | 2022 | BRATS 2015 | Multi | 97.4 | |
| Bashkandi et al. [46] | 2023 | Br35H | Binary | 97.09 | |
| Wu and Sen [47] | 2023 | Figshare MRI | Multi | 95.98 | |
| Anaraki et al. [48] | 2019 | IXI, REMBRAND, TCGA-LGG | Multi | 90.9 | |
| Figshare MRI | Multi | 94.2 | |||
| Bacanin et al. [49] | 2021 | IXI, REMBRANDT, TCGA-GBM, TCGA-LGG | Multi | 93.3 | |
| Figshare MRI | Multi | 96.5 | |||
| Bezdan et al. [50] | 2021 | IXI, REMBRANDT, TCGA-GBM, TCGA-LGG | Multi | 94.50 | |
| Kothandaraman [51] | 2023 | Figshare MRI | Multi | 96.125 | |
| Rammurthy and Mahesh [52] | 2022 | BRATS | Multi | 81.6 | |
| SimBRATS | 81.6 | ||||
| Chawla et al. [53] | 2022 | Figshare MRI | Multi | 99.5 | |
| Sharif et al. [54] | 2022 | BRATS 2013 | Multi | 99.06 | |
| BRATS 2015 | 98.76 | ||||
| BRATS 2017 | 98.18 | ||||
| BRATS 2018 | 94.6 | ||||
| Xu and Mohammadi [55] | 2024 | Figshare MRI | Multi | 97.32 |
| Hyperparameter | Values |
|---|---|
| Number of fully connected layers | 1, 2, 3 |
| Number of neurons in the fully connected layer | 64, 128, 256, 512, 1024 |
| Dropout rate | 0, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6 |
| Optimizer | Adam, SGD |
| Learning rate | 0.001, 0.0001 |
| CNN Models | Dataset 1 (DS1) | Dataset 2 (DS2) | Dataset 3 (DS3) | |||
|---|---|---|---|---|---|---|
| Accuracy (%) | F1-Score (%) | Accuracy (%) | F1-Score (%) | Accuracy (%) | F1-Score (%) | |
| DenseNet121 | 96.25 | 96.18 | 96.47 | 96.66 | 96.65 | 96.61 |
| DenseNet169 | 94.13 | 93.88 | 96.01 | 96.15 | 97.64 | 97.48 |
| DenseNet201 | 96.08 | 95.96 | 96.93 | 97.01 | 98.48 | 98.38 |
| VGG16 | 91.52 | 90.64 | 82.06 | 81.88 | 97.18 | 96.97 |
| VGG19 | 94.62 | 94.11 | 94.79 | 94.58 | 96.91 | 96.75 |
| ResNet50 | 95.92 | 95.86 | 95.09 | 95.17 | 98.14 | 97.99 |
| ResNet101 | 95.27 | 95.07 | 94.33 | 94.51 | 98.47 | 98.45 |
| ResNet152 | 93.56 | 93.46 | 92.33 | 92.82 | 97.66 | 97.62 |
| ResNetRS50 | 93.15 | 92.79 | 95.55 | 95.67 | 97.64 | 97.44 |
| ResNetRS100 | 95.19 | 95.04 | 96.32 | 96.46 | 97.71 | 97.61 |
| InceptionResNetV2 | 94.37 | 94.19 | 96.17 | 96.12 | 98.44 | 98.28 |
| InceptionV3 | 94.54 | 94.41 | 95.39 | 95.55 | 98.09 | 97.97 |
| Xception | 93.47 | 93.11 | 95.39 | 95.55 | 97.79 | 97.73 |
| MobileNetV2 | 90.22 | 90.59 | 93.87 | 94.05 | 98.09 | 98.02 |
| EfficientNetV2B3 | 88.25 | 87.76 | 93.40 | 93.57 | 97.86 | 97.74 |
| EfficientNetV2S | 95.43 | 95.22 | 93.63 | 93.62 | 97.56 | 97.39 |
| EfficientNetV2M | 88.01 | 87.71 | 95.09 | 95.09 | 95.50 | 95.32 |
| RegNetX008 | 94.69 | 94.43 | 94.94 | 94.91 | 98.63 | 98.54 |
| RegNetY008 | 95.11 | 94.98 | 95.86 | 95.84 | 97.18 | 97.00 |
| CNN Models | Dataset 1 (DS1) | Dataset 2 (DS2) | Dataset 3 (DS3) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Precision (%) | Recall (%) | AUC (%) | Precision (%) | Recall (%) | AUC (%) | Precision (%) | Recall (%) | AUC (%) | |
| DenseNet121 | 96.01 | 96.41 | 97.24 | 96.60 | 96.76 | 97.78 | 97.21 | 96.39 | 97.61 |
| DenseNet169 | 94.05 | 93.92 | 95.48 | 96.21 | 96.14 | 97.39 | 97.66 | 97.42 | 98.32 |
| DenseNet201 | 95.84 | 96.10 | 97.02 | 97.21 | 96.83 | 97.88 | 98.49 | 98.35 | 98.93 |
| VGG16 | 90.39 | 90.97 | 93.35 | 81.82 | 81.99 | 87.96 | 97.12 | 96.93 | 98.0 |
| VGG19 | 95.20 | 93.27 | 95.09 | 94.60 | 94.60 | 96.42 | 97.0 | 96.75 | 97.87 |
| ResNet50 | 95.99 | 95.75 | 96.70 | 94.77 | 95.66 | 97.01 | 98.14 | 97.96 | 98.68 |
| ResNet101 | 95.22 | 94.93 | 96.22 | 94.5 | 94.76 | 96.42 | 98.56 | 98.35 | 98.92 |
| ResNet152 | 92.91 | 94.48 | 95.71 | 93.88 | 92.83 | 95.09 | 97.85 | 97.46 | 98.33 |
| ResNetRS50 | 92.83 | 92.85 | 94.66 | 95.45 | 95.98 | 97.24 | 97.68 | 97.42 | 98.32 |
| ResNetRS100 | 95.13 | 94.97 | 96.23 | 96.63 | 96.30 | 97.12 | 97.81 | 97.57 | 98.40 |
| InceptionResNetV2 | 93.88 | 95.12 | 96.25 | 95.79 | 96.51 | 97.62 | 98.41 | 98.30 | 98.90 |
| InceptionV3 | 94.38 | 94.45 | 95.79 | 95.55 | 95.60 | 97.01 | 98.03 | 97.97 | 98.67 |
| Xception | 93.30 | 93.20 | 94.94 | 95.51 | 95.59 | 97.01 | 97.75 | 97.80 | 98.54 |
| MobileNetV2 | 91.21 | 91.10 | 93.09 | 93.66 | 94.61 | 96.27 | 98.13 | 97.94 | 98.65 |
| EfficientNetV2B3 | 88.50 | 87.24 | 90.41 | 93.90 | 93.41 | 95.57 | 97.79 | 97.71 | 98.50 |
| EfficientNetV2S | 95.42 | 95.16 | 96.37 | 94.68 | 95.43 | 96.84 | 97.47 | 97.44 | 98.32 |
| EfficientNetV2M | 88.58 | 88.14 | 91.11 | 95.07 | 95.22 | 96.78 | 95.61 | 95.13 | 96.80 |
| RegNetX008 | 94.38 | 94.66 | 95.99 | 94.60 | 95.31 | 96.81 | 98.59 | 98.52 | 99.03 |
| RegNetY008 | 94.58 | 95.50 | 96.55 | 95.47 | 96.35 | 97.49 | 97.21 | 96.98 | 98.03 |
| DS1 | Models | DenseNet121 | DenseNet201 | EfficientNetV2S | ResNet50 | ResNet101 |
| Weights | 0.209 | 0.212 | 0.237 | 0.038 | 0.304 | |
| DS2 | Models | DenseNet121 | DenseNet169 | DenseNet201 | InceptionResNetV2 | ResNetRS100 |
| Weights | 0.359 | 0.054 | 0.270 | 0.024 | 0.293 | |
| DS3 | Models | DenseNet201 | InceptionResNetV2 | MobileNetV2 | RegNetX008 | ResNet101 |
| Weights | 0.041 | 0.16 | 0.133 | 0.509 | 0.156 |
| Study | Year | Dataset | Classes | Accuracy (%) | F1-Score (%) |
|---|---|---|---|---|---|
| Ayadi et al. [19] | 2021 | [56] | 3 | 94.74 | 94.19 * |
| Deepak and Ameer [26] | 2019 | [56] | 3 | 97.17 | 97.20 |
| Ait-Amou [43] | 2022 | [56] | 3 | 98.70 | 98.60 |
| Kothandaraman [51] | 2023 | [56] | 3 | 96.125 | 96.097 |
| Wu and Sen [47] | 2023 | [56] | 3 | 95.98 | 89.98 |
| Alanazi et al. [35] | 2022 | [57] | 4 | 95.75 | 95.72 * |
| Saurav et al. [24] | 2022 | [57] | 4 | 95.71 | 95.98 |
| Kang et al. [42] | 2021 | [57] | 4 | 93.72 | - |
| Aurna et al. [1] | 2022 | [58] | 4 | 98.96 | 99.0 |
| Gomez et al. [36] | 2023 | [58] | 4 | 97.12 | 97.28 |
| Proposed Model | 2023 | [56] | 3 | 99.35 | 99.20 |
| [57] | 4 | 98.77 | 98.92 | ||
| [58] | 4 | 99.92 | 99.92 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Çetin-Kaya, Y.; Kaya, M. A Novel Ensemble Framework for Multi-Classification of Brain Tumors Using Magnetic Resonance Imaging. Diagnostics 2024, 14, 383. https://doi.org/10.3390/diagnostics14040383
Çetin-Kaya Y, Kaya M. A Novel Ensemble Framework for Multi-Classification of Brain Tumors Using Magnetic Resonance Imaging. Diagnostics. 2024; 14(4):383. https://doi.org/10.3390/diagnostics14040383
Chicago/Turabian StyleÇetin-Kaya, Yasemin, and Mahir Kaya. 2024. "A Novel Ensemble Framework for Multi-Classification of Brain Tumors Using Magnetic Resonance Imaging" Diagnostics 14, no. 4: 383. https://doi.org/10.3390/diagnostics14040383
APA StyleÇetin-Kaya, Y., & Kaya, M. (2024). A Novel Ensemble Framework for Multi-Classification of Brain Tumors Using Magnetic Resonance Imaging. Diagnostics, 14(4), 383. https://doi.org/10.3390/diagnostics14040383






