An Evolutionary Federated Learning Approach to Diagnose Alzheimer’s Disease Under Uncertainty
Abstract
1. Introduction
1.1. Research Objectives
- Systematically clean and curate datasets, including medical images and demographic data, for accurate Alzheimer’s disease (AD) diagnosis.
- Design a convolutional neural network (CNN) architecture tailored for processing and classifying Alzheimer’s medical imaging data.
- Combine image data with demographic information to improve diagnostic accuracy.
- Develop an Optimized trained belief rule base (BRB) to handle data uncertainties.
- Incorporate the belief rule base (BRB) system in the federated learning framework.
- Compare and evaluate the different (i.e., FedAvg, FedProx, and genetic algorithm) aggregators used in the federated learning model.
1.2. Research Questions
- How to develop a pre-processing pipeline combining medical images and demographic data for enhanced Alzheimer’s disease diagnosis?
- How to design a CNN architecture optimized for Alzheimer’s medical imaging data classification?
- How to integrate image and demographic data to improve diagnostic accuracy?
- How to establish a federated learning framework with a belief rule base to handle data uncertainties?
- How does advanced federated learning for medical applications improve privacy-preserving diagnostic frameworks?
- How can multimodal data handling and uncertainty management in machine learning for medical diagnostics improve accuracy and overcome uncertainty?
2. Related Work
2.1. Machine Learning to Classify AD Using MRI Images
2.2. Federated Learning-Based Solution for Building Alzheimer’s Diagnosis System
2.3. Addressing the Existing Research Gaps
3. Methods
3.1. Description of Dataset Applied in This Research
3.2. Steps of MRI Dataset Preprocessing
3.3. Deep Learning Framework
Modified Convolutional Neural Network
3.4. Federated Learning Architecture
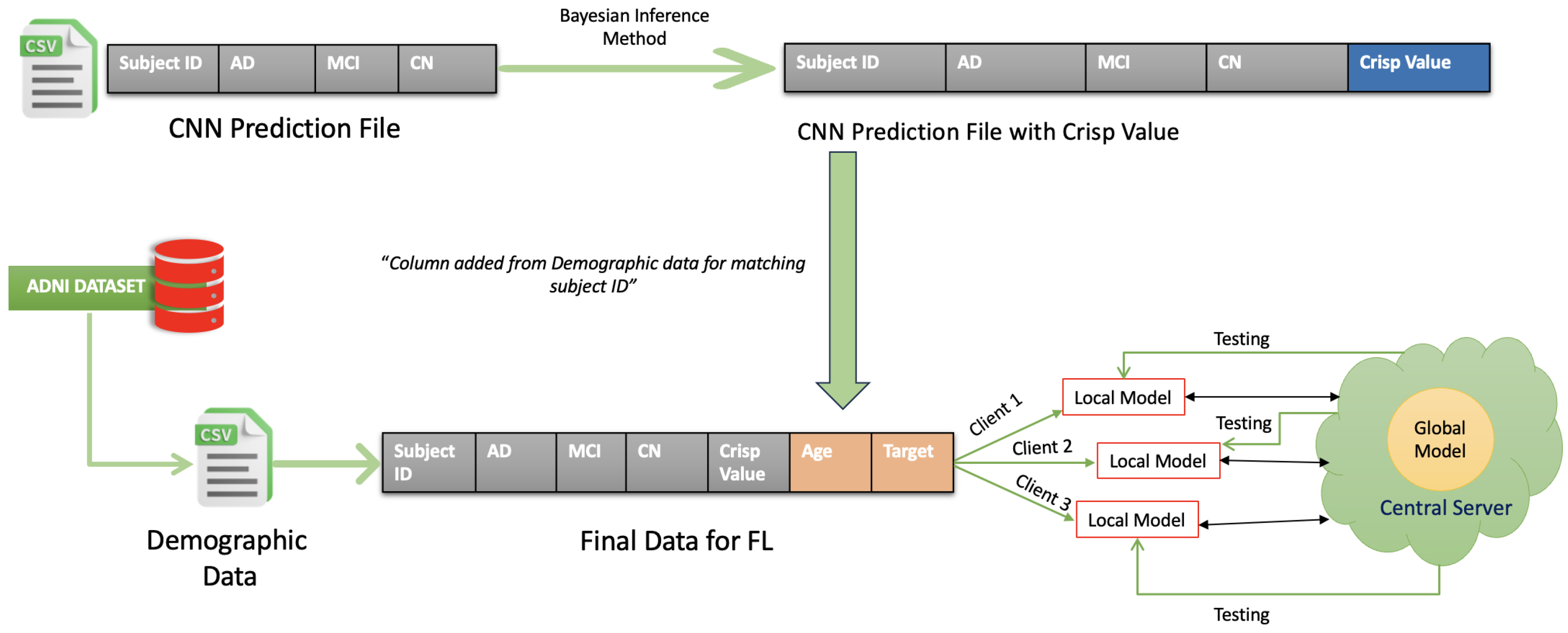
3.4.1. Data Preparation
3.4.2. Local Training with BRB and PSO
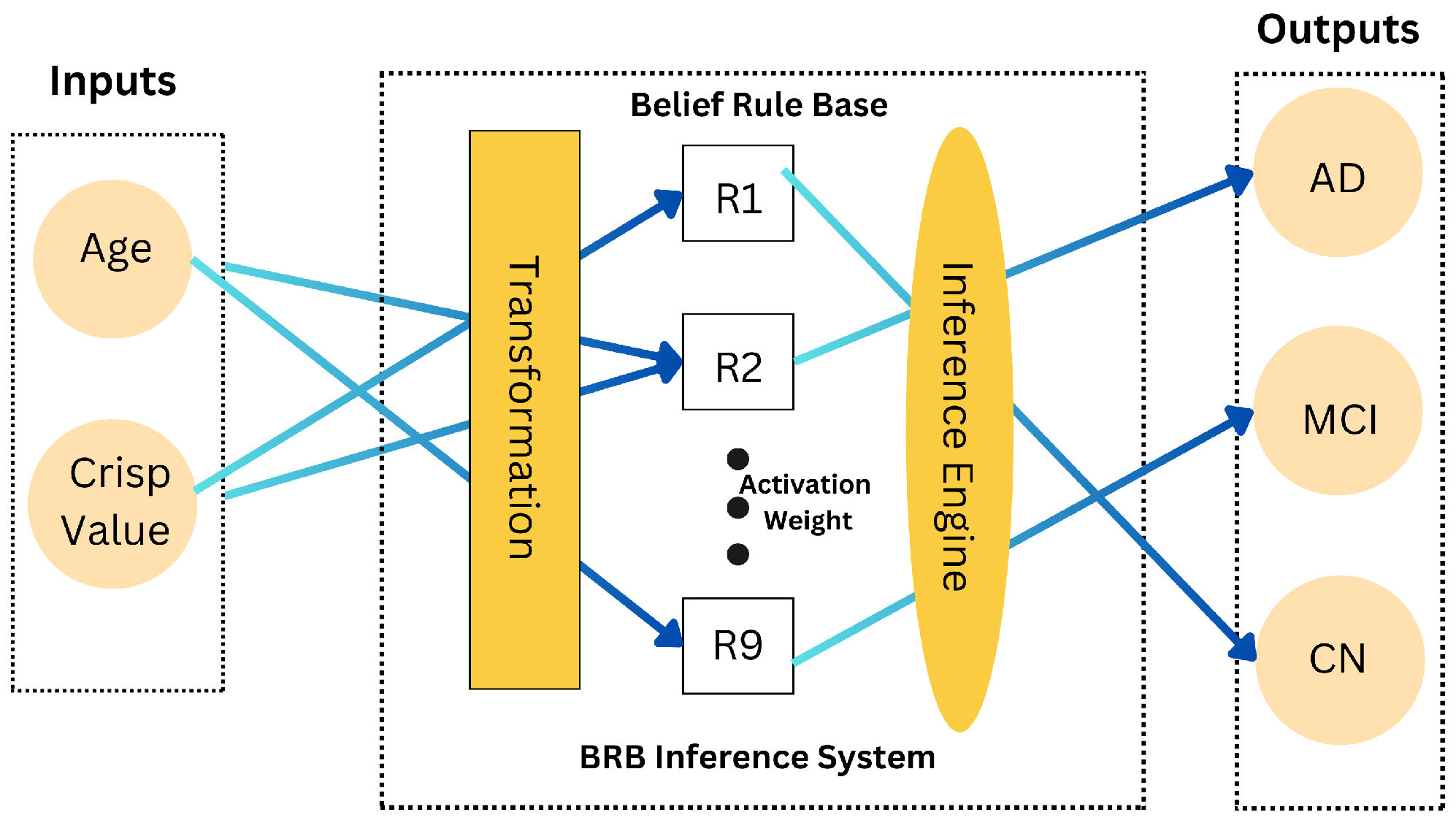
3.4.3. Belief Rule Base (BRB) Mechanism
3.4.4. Local Parameter Optimization
3.4.5. Parameter Sharing and Aggregation
3.4.6. Global Model Update and PSO Optimization
3.4.7. Iterative Optimization with PSO
| Algorithm 1 Federated Learning with Particle Swarm Optimization (PSO) for Alzheimer’s Disease Diagnosis using FedAvg. |
|
4. Results
4.1. Libraries and Packages for System Implementation
4.2. Hyperparameter Setting
4.3. Evaluation Metrics
- n is the number of observations or patients in the dataset.
- represents the actual diagnostic score or label for the i-th patient.
- denotes the predicted diagnostic score or label for the i-th patient made by the model.
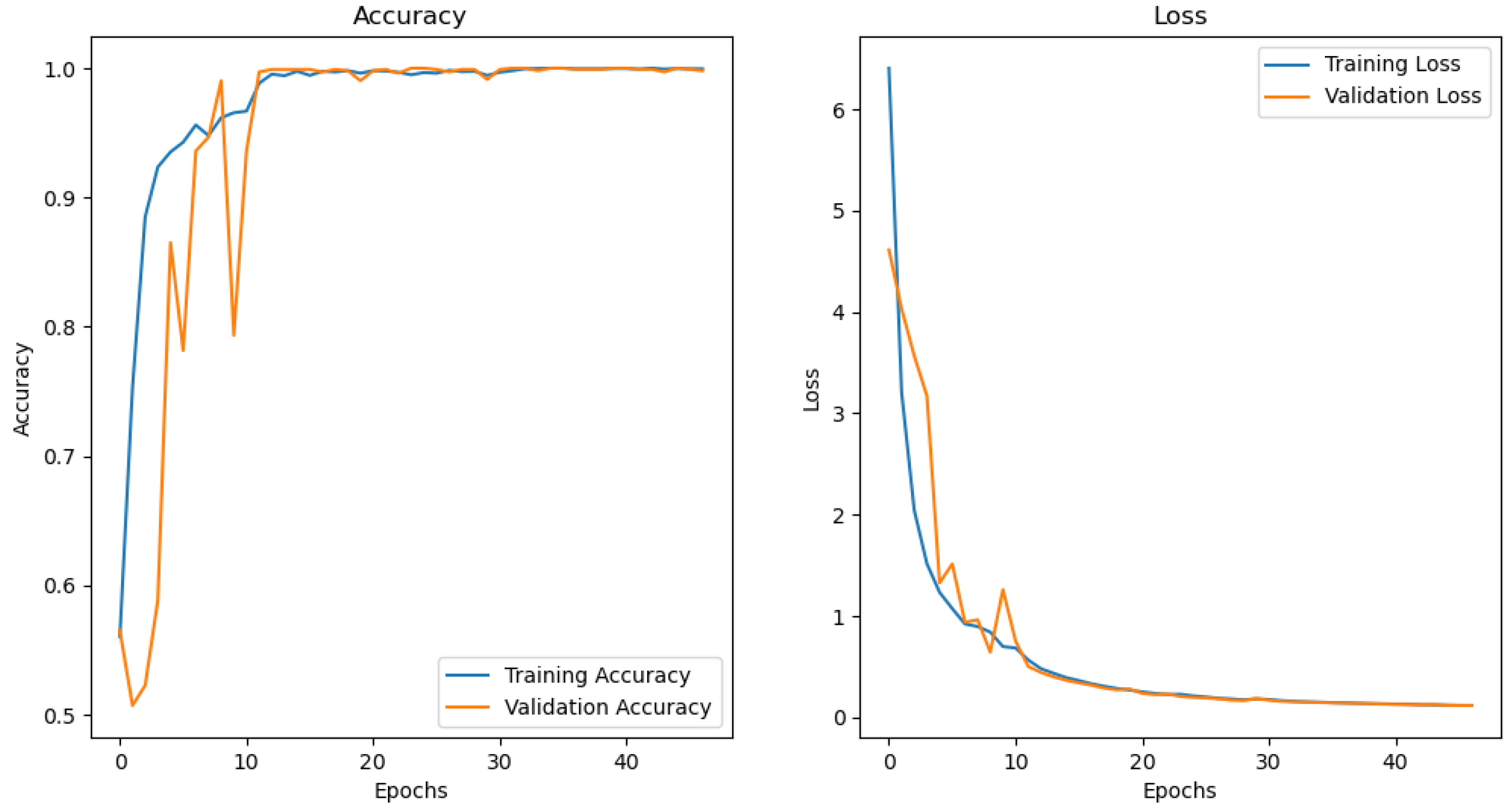
4.4. Performance of CNN for MRI Classification of Alzheimer’s
4.5. Rule Base Overview for Federated Learning Clients and Servers
4.6. Rule Base for Local Training Model on Client Side
4.7. Optimized Rule Base for Global Training Model on Server Side
4.8. Graphical Representation of Accuracy and Loss for the Clients and Servers
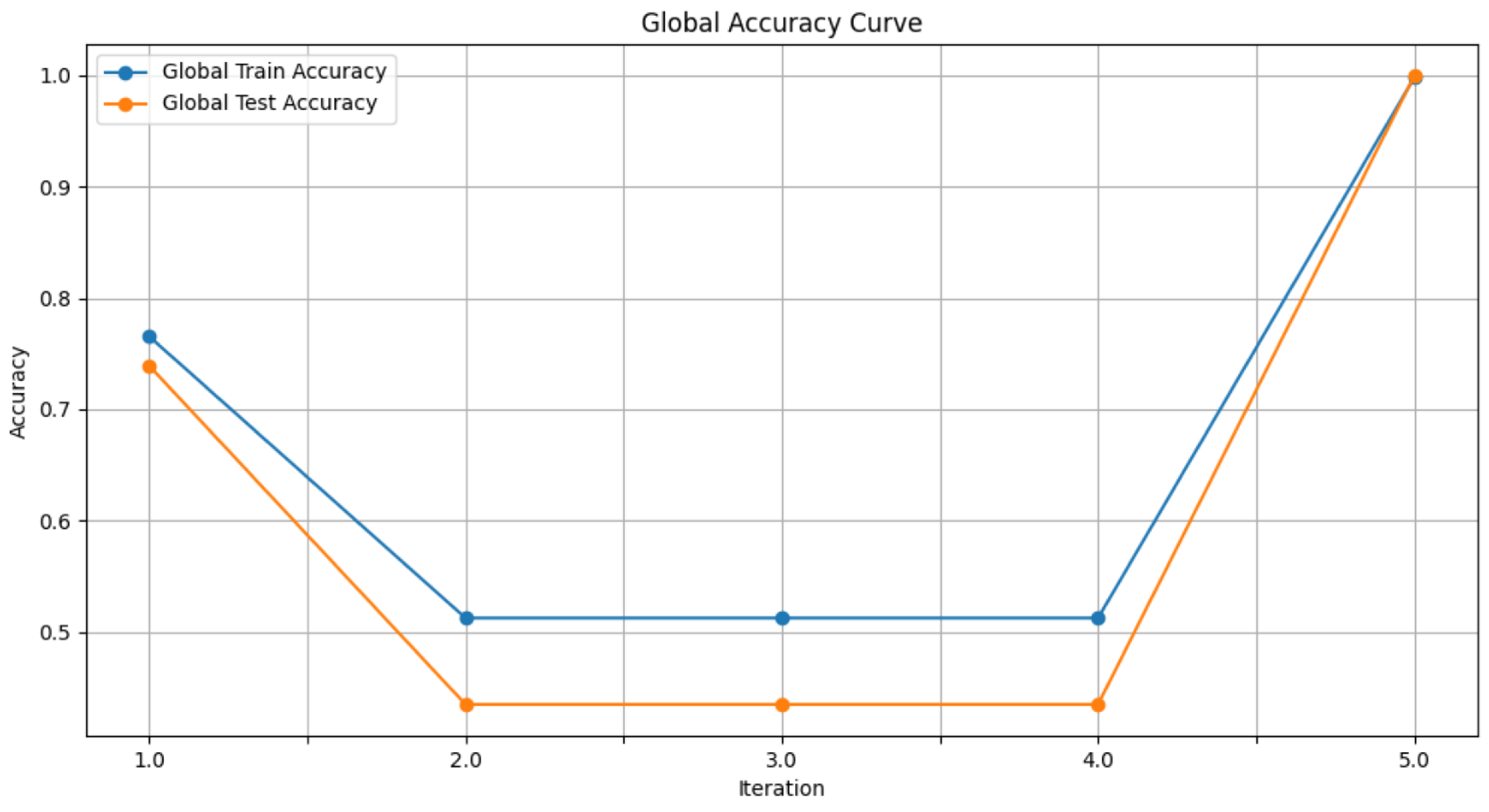
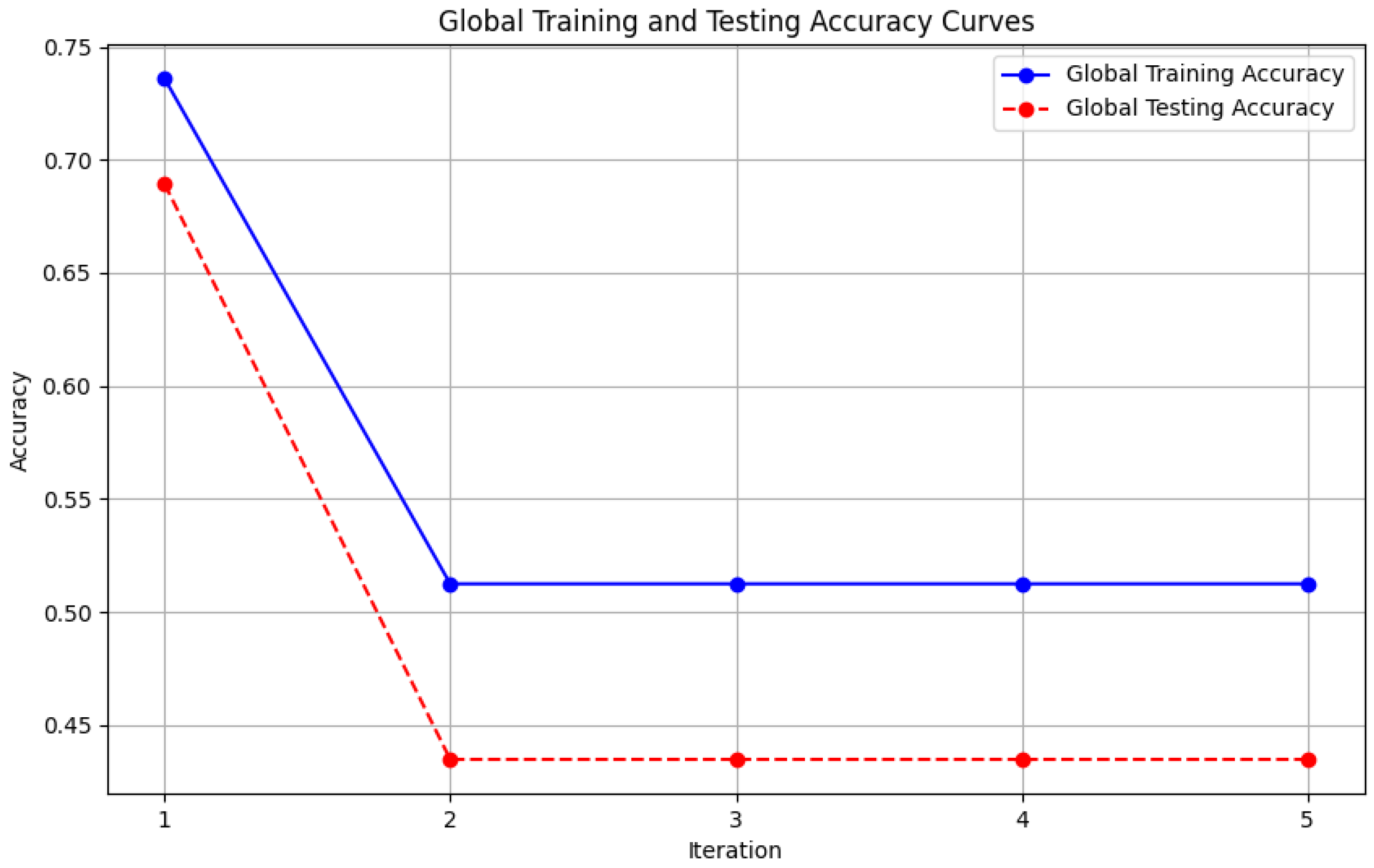
- Figure 6: The graph visualizes the Global Accuracy derived using the FedAvg aggregator in the three iterations. The x-axis represents the number of iterations, and the y-axis shows the accuracy achieved during global training.
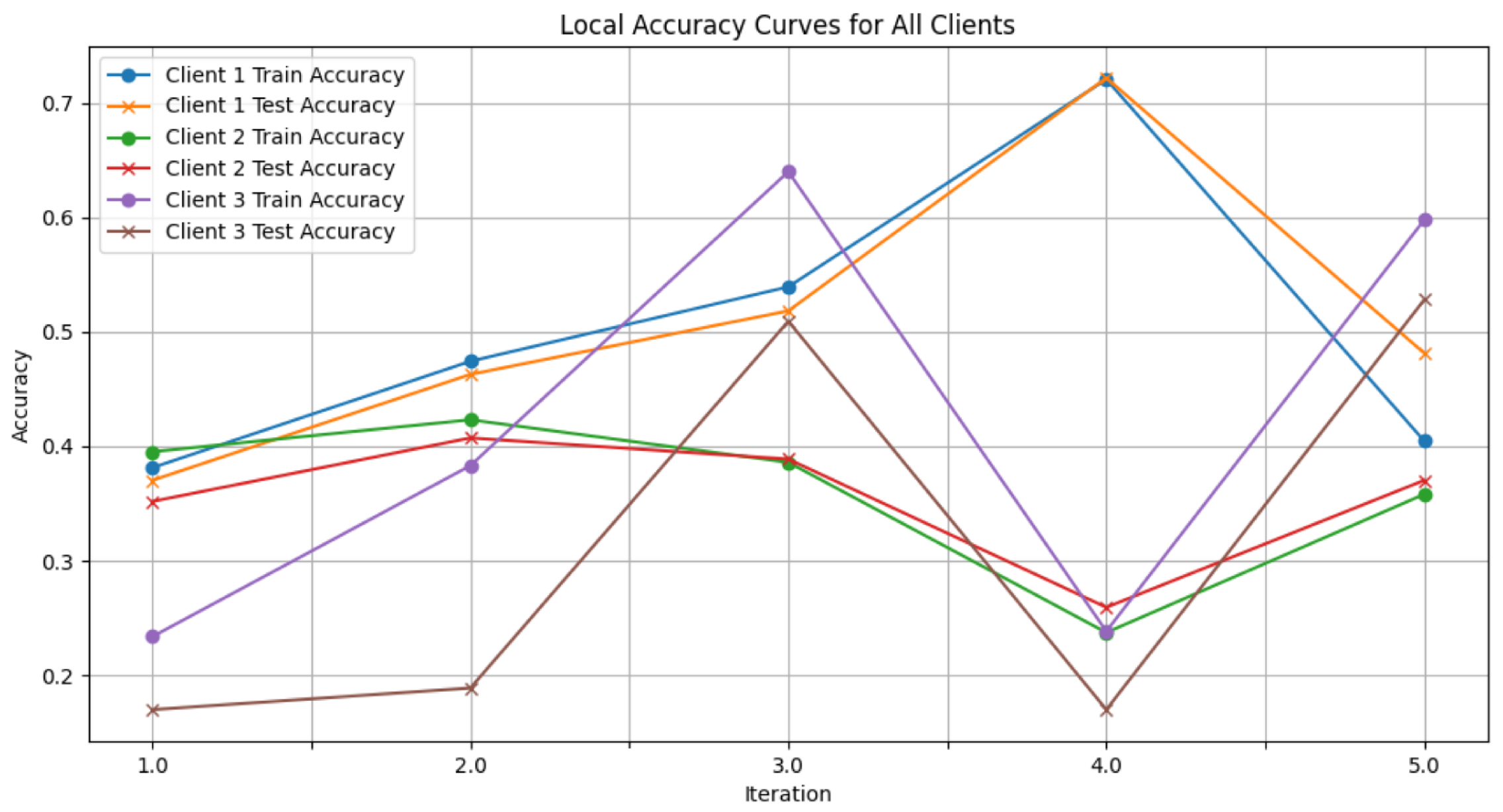
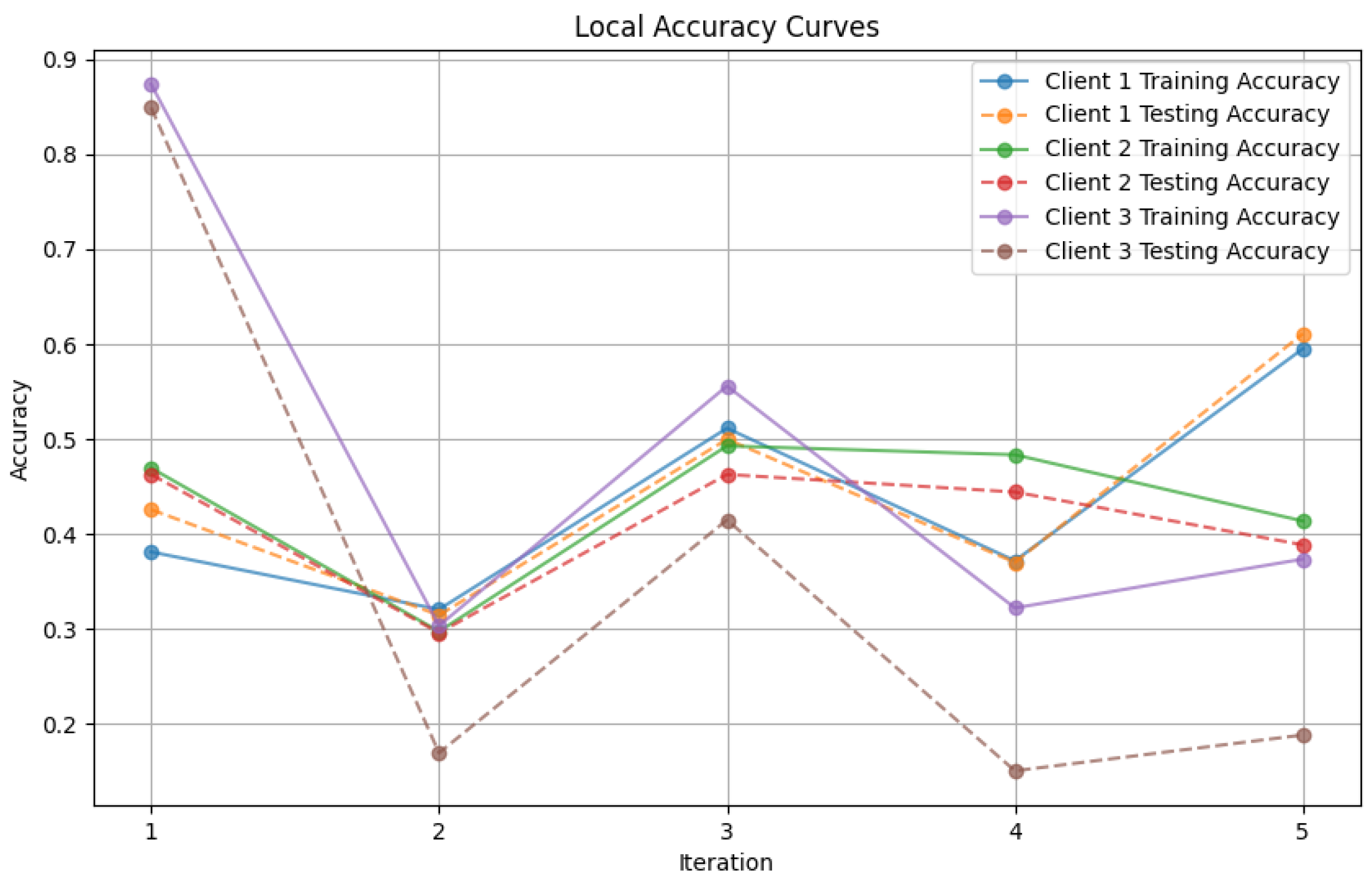
- Figure 7: This plot dictates the Local Client Accuracy from the FedAvg aggregator for each client across the three iterations. The accuracy for each client is plotted independently, demonstrating the performance consistency or variability among the clients.
- Figure 8: This figure presents the Global Accuracy achieved using the Genetic Algorithm aggregator. The plot distinguishes the training versus testing accuracy over the three iterations, illustrating the effectiveness of the Genetic Algorithm in global optimization.
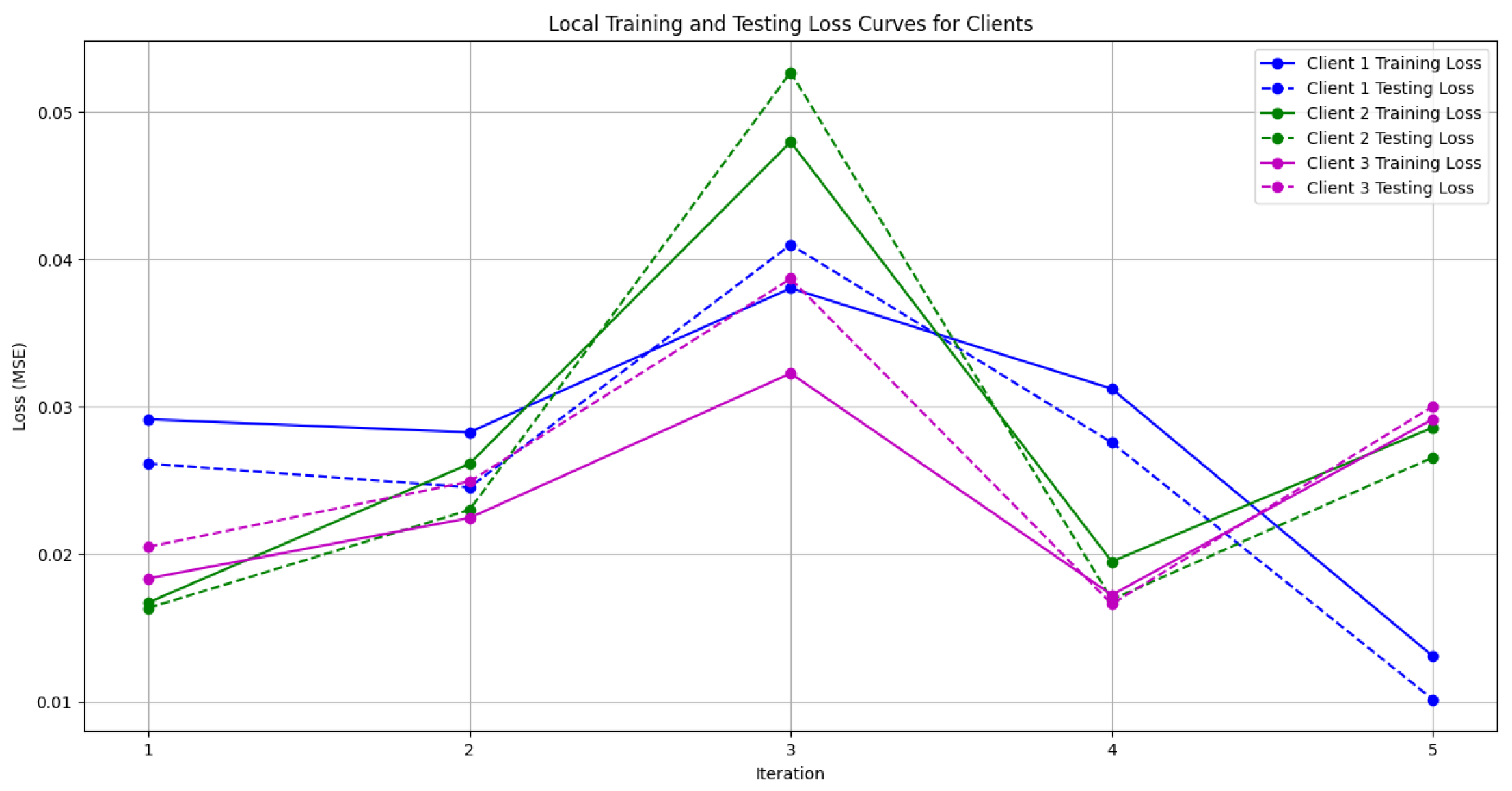
- Figure 10: This plot depicts the Global Loss during training and testing using the Genetic Algorithm aggregator. The y-axis represents the loss, and the x-axis shows the iterations. A reduction in loss over iterations indicates better convergence.
- Figure 11: This figure shows the Local Loss for each client under the Genetic Algorithm aggregator. The figure aids in visualizing how individual clients are optimizing their loss functions.
- Figure 12: This plot shows the Global Accuracy achieved using the FedProx aggregator. It illustrates the effectiveness of FedProx in improving the global model accuracy across iterations.
- Figure 13: Finally, this plot presents the Local Accuracy for each client when using the FedProx aggregator. The comparison among clients is essential to understanding the federated learning model’s performance across distributed data sources.
4.9. Interpretation of Model Performance of Aggregators Used in Federated Learning
4.9.1. FedAvg Aggregator
- Robust Aggregation of Model Updates: The FedAvg method involves averaging the model parameters from clients to reduce the impact of data differences, among them. By blending these updates, the overall model can effectively adapt to datasets resulting in improved performance.
- Effective Handling of Data Heterogeneity: In federated learning setups, there is often a challenge, with IID (Independent and Identically Distributed) data, where various clients possess distinct data distributions. To address this, FedAvg tackles the problem by combining updates from all clients, allowing for a representation of data patterns and minimizing the chances of tailoring to any one client’s specific data.
- Generalization Across Multiple Clients: The variety of data from clients helps the overall model perform effectively in situations. This is shown by the testing accuracy of the model, which is close to 1.0, proving its excellent performance, with different types of data inputs.
4.9.2. FedProx Aggregator
4.9.3. Genetic Algorithm Aggregator
4.10. Comparison with State-of-the-Art Methods
5. Conclusions and Future Research Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Umme Habiba, S.; Debnath, T.; Islam, M.K.; Nahar, L.; Shahadat Hossain, M.; Basnin, N.; Andersson, K. Transfer Learning-Assisted DementiaNet: A Four Layer Deep CNN for Accurate Alzheimer’s Disease Detection from MRI Images. In Proceedings of the International Conference on Brain Informatics, Hoboken, NJ, USA, 1–3 August 2023; Springer: Cham, Switzerland, 2023; pp. 383–394. [Google Scholar]
- Mahmud, T.; Aziz, M.T.; Uddin, M.K.; Barua, K.; Rahman, T.; Sharmen, N.; Shamim Kaiser, M.; Sazzad Hossain, M.; Hossain, M.S.; Andersson, K. Ensemble Learning Approaches for Alzheimer’s Disease Classification in Brain Imaging Data. In Proceedings of the International Conference on Trends in Electronics and Health Informatics, Dhaka, Bangladesh, 20–21 December 2023; Springer: Cham, Switzerland, 2023; pp. 133–147. [Google Scholar]
- Mahmud, T.; Barua, K.; Barua, A.; Das, S.; Basnin, N.; Hossain, M.S.; Andersson, K.; Kaiser, M.S.; Sharmen, N. Exploring Deep Transfer Learning Ensemble for Improved Diagnosis and Classification of Alzheimer’s Disease. In Proceedings of the International Conference on Brain Informatics, Hoboken, NJ, USA, 1–3 August 2023; Springer: Cham, Switzerland, 2023; pp. 109–120. [Google Scholar]
- Hossain, M.S.; Rahaman, S.; Mustafa, R.; Andersson, K. A belief rule-based expert system to assess suspicion of acute coronary syndrome (ACS) under uncertainty. Soft Comput. 2018, 22, 7571–7586. [Google Scholar] [CrossRef]
- Khalil, K.; Khan Mamun, M.M.R.; Sherif, A.; Elsersy, M.S.; Imam, A.A.A.; Mahmoud, M.; Alsabaan, M. A federated learning model based on hardware acceleration for the early detection of alzheimer’s disease. Sensors 2023, 23, 8272. [Google Scholar] [CrossRef] [PubMed]
- Mitrovska, A.; Safari, P.; Ritter, K.; Shariati, B.; Fischer, J.K. Secure federated learning for Alzheimer’s disease detection. Front. Aging Neurosci. 2024, 16, 1324032. [Google Scholar] [CrossRef]
- Mahmud, T.; Barua, K.; Habiba, S.U.; Sharmen, N.; Hossain, M.S.; Andersson, K. An Explainable AI Paradigm for Alzheimer’s Diagnosis Using Deep Transfer Learning. Diagnostics 2024, 14, 345. [Google Scholar] [CrossRef] [PubMed]
- Beheshti, Z.; Shamsuddin, S.M.H.; Beheshti, E.; Yuhaniz, S.S. Enhancement of artificial neural network learning using centripetal accelerated particle swarm optimization for medical diseases diagnosis. Soft Comput. 2014, 18, 2253–2270. [Google Scholar] [CrossRef]
- Qian, C.; Xiong, H.; Li, J. FeDeFo: A Personalized Federated Deep Forest Framework for Alzheimer’s Disease Diagnosis. In Proceedings of the 35th International Conference on Software Engineering and Knowledge Engineering, San Francisco, CA, USA, 1–10 July 2023. [Google Scholar]
- Hossain, M.S.; Ahmed, F.; Andersson, K. A belief rule based expert system to assess tuberculosis under uncertainty. J. Med. Syst. 2017, 41, 43. [Google Scholar] [CrossRef]
- Bordin, V.; Coluzzi, D.; Rivolta, M.W.; Baselli, G. Explainable AI points to white matter hyperintensities for Alzheimer’s disease identification: A preliminary study. In Proceedings of the 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Glasgow, UK, 11–15 July 2022; IEEE: New York, NY, YSA, 2022; pp. 484–487. [Google Scholar]
- De Santi, L.A.; Pasini, E.; Santarelli, M.F.; Genovesi, D.; Positano, V. An Explainable Convolutional Neural Network for the Early Diagnosis of Alzheimer’s Disease from 18F-FDG PET. J. Digit. Imaging 2023, 36, 189–203. [Google Scholar] [CrossRef] [PubMed]
- García-Gutierrez, F.; Díaz-Álvarez, J.; Matias-Guiu, J.A.; Pytel, V.; Matías-Guiu, J.; Cabrera-Martín, M.N.; Ayala, J.L. GA-MADRID: Design and validation of a machine learning tool for the diagnosis of Alzheimer’s disease and frontotemporal dementia using genetic algorithms. Med. Biol. Eng. Comput. 2022, 60, 2737–2756. [Google Scholar] [CrossRef]
- Yang, C.; Rangarajan, A.; Ranka, S. Visual explanations from deep 3D convolutional neural networks for Alzheimer’s disease classification. In Proceedings of the AMIA Annual Symposium Proceedings, San Francisco, CA, USA, 3–7 November 2018; American Medical Informatics Association: Bethesda, MD, USA, 2018; Volume 2018, p. 1571. [Google Scholar]
- Rieke, J.; Eitel, F.; Weygandt, M.; Haynes, J.D.; Ritter, K. Visualizing convolutional networks for MRI-based diagnosis of Alzheimer’s disease. In Proceedings of the Understanding and Interpreting Machine Learning in Medical Image Computing Applications: First International Workshops, MLCN 2018, DLF 2018, and iMIMIC 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 16–20 September 2018; Proceedings 1. Springer: Cham, Switzerland, 2018; pp. 24–31. [Google Scholar]
- Loveleen, G.; Mohan, B.; Shikhar, B.S.; Nz, J.; Shorfuzzaman, M.; Masud, M. Explanation-driven hci model to examine the mini-mental state for alzheimer’s disease. Acm Trans. Multimed. Comput. Commun. Appl. 2023, 20, 1–16. [Google Scholar] [CrossRef]
- Yu, L.; Xiang, W.; Fang, J.; Chen, Y.P.P.; Zhu, R. A novel explainable neural network for Alzheimer’s disease diagnosis. Pattern Recognit. 2022, 131, 108876. [Google Scholar] [CrossRef]
- Kim, M.; Kim, J.; Qu, J.; Huang, H.; Long, Q.; Sohn, K.A.; Kim, D.; Shen, L. Interpretable temporal graph neural network for prognostic prediction of Alzheimer’s disease using longitudinal neuroimaging data. In Proceedings of the 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Houston, TX, USA, 9–12 December 2021; IEEE: New York, NY, YSA, 2021; pp. 1381–1384. [Google Scholar]
- Marwa, E.G.; Moustafa, H.E.D.; Khalifa, F.; Khater, H.; AbdElhalim, E. An MRI-based deep learning approach for accurate detection of Alzheimer’s disease. Alex. Eng. J. 2023, 63, 211–221. [Google Scholar]
- Ghazal, T.M.; Abbas, S.; Munir, S.; Khan, M.; Ahmad, M.; Issa, G.F.; Zahra, S.B.; Khan, M.A.; Hasan, M.K. Alzheimer Disease Detection Empowered with Transfer Learning. Comput. Mater. Contin. 2022, 70, 5005–5019. [Google Scholar] [CrossRef]
- AlSaeed, D.; Omar, S.F. Brain MRI analysis for Alzheimer’s disease diagnosis using CNN-based feature extraction and machine learning. Sensors 2022, 22, 2911. [Google Scholar] [CrossRef] [PubMed]
- Hamdi, M.; Bourouis, S.; Rastislav, K.; Mohmed, F. Evaluation of neuro images for the diagnosis of Alzheimer’s disease using deep learning neural network. Front. Public Health 2022, 10, 834032. [Google Scholar]
- Helaly, H.A.; Badawy, M.; Haikal, A.Y. Deep learning approach for early detection of Alzheimer’s disease. Cogn. Comput. 2021, 14, 1711–1727. [Google Scholar] [CrossRef]
- Mohammed, B.A.; Senan, E.M.; Rassem, T.H.; Makbol, N.M.; Alanazi, A.A.; Al-Mekhlafi, Z.G.; Almurayziq, T.S.; Ghaleb, F.A. Multi-method analysis of medical records and MRI images for early diagnosis of dementia and Alzheimer’s disease based on deep learning and hybrid methods. Electronics 2021, 10, 2860. [Google Scholar] [CrossRef]
- Pradhan, A.; Gige, J.; Eliazer, M. Detection of Alzheimer’s disease (AD) in MRI images using deep learning. Int. J. Eng. Res. Technol. 2021, 10. [Google Scholar]
- Salehi, A.W.; Baglat, P.; Sharma, B.B.; Gupta, G.; Upadhya, A. A CNN model: Earlier diagnosis and classification of Alzheimer disease using MRI. In Proceedings of the 2020 International Conference on Smart Electronics and Communication (ICOSEC), Trichy, India, 10–12 September 2020; IEEE: New York, NY, YSA, 2020; pp. 156–161. [Google Scholar]
- Suganthe, R.C.; Latha, R.S.; Geetha, M.; Sreekanth, G.R. Diagnosis of Alzheimer’s disease from brain magnetic resonance imaging images using deep learning algorithms. Adv. Electr. Comput. Eng. 2020, 20, 57–64. [Google Scholar] [CrossRef]
- Hussain, E.; Hasan, M.; Hassan, S.Z.; Azmi, T.H.; Rahman, M.A.; Parvez, M.Z. Deep learning based binary classification for alzheimer’s disease detection using brain mri images. In Proceedings of the 2020 15th IEEE Conference on Industrial Electronics and Applications (ICIEA), Kristiansand, Norway, 9–13 November 2020; IEEE: New York, NY, YSA, 2020; pp. 1115–1120. [Google Scholar]
- Basaia, S.; Agosta, F.; Wagner, L.; Canu, E.; Magnani, G.; Santangelo, R.; Filippi, M.; The Alzheimer’s Disease Neuroimaging Initiative. Automated classification of Alzheimer’s disease and mild cognitive impairment using a single MRI and deep neural networks. Neuroimage Clin. 2019, 21, 101645. [Google Scholar] [CrossRef]
- Ji, H.; Liu, Z.; Yan, W.Q.; Klette, R. Early diagnosis of Alzheimer’s disease using deep learning. In Proceedings of the 2nd International Conference on Control and Computer Vision, Jeju, Republic of Korea, 15–18 June 2019; pp. 87–91. [Google Scholar]
- Islam, J.; Zhang, Y. A novel deep learning based multi-class classification method for Alzheimer’s disease detection using brain MRI data. In Proceedings of the Brain Informatics: International Conference, BI 2017, Beijing, China, 16–18 November 2017; Proceedings. Springer: Cham, Switzerland, 2017; pp. 213–222. [Google Scholar]
- Trivedi, N.K.; Jain, S.; Agarwal, S. Identifying and Categorizing Alzheimer’s Disease with Lightweight Federated Learning Using Identically Distributed Images. In Proceedings of the 2024 11th International Conference on Reliability, Infocom Technologies and Optimization (Trends and Future Directions) (ICRITO), Noida, India, 14–15 March 2024; IEEE: New York, NY, YSA, 2024; pp. 1–5. [Google Scholar]
- Altalbe, A.; Javed, A.R. Privacy Preserved Brain Disorder Diagnosis Using Federated Learning. Comput. Syst. Sci. Eng. 2023, 47, 2187–2200. [Google Scholar] [CrossRef]
- Mandawkar, U.; Diwan, T. Alzheimer disease classification using tawny flamingo based deep convolutional neural networks via federated learning. Imaging Sci. J. 2022, 70, 459–472. [Google Scholar] [CrossRef]
- Castro, F.; Impedovo, D.; Pirlo, G. A Federated Learning System with Biometric Medical Image Authentication for Alzheimer’s Diagnosis. In Proceedings of the International Conference on Pattern Recognition Applications and Methods (ICPRAM), Rome, Italy, 24–26 February 2024; pp. 951–960. [Google Scholar]
- Biswas, M.; Chowdhury, S.U.; Nahar, N.; Hossain, M.S.; Andersson, K. A belief rule base expert system for staging non-small cell lung cancer under uncertainty. In Proceedings of the 2019 IEEE International Conference on Biomedical Engineering, Computer and Information Technology for Health (BECITHCON), Dhaka, Bangladesh, 28–30 November 2019; IEEE: New York, NY, YSA, 2019; pp. 47–52. [Google Scholar]
- Mandal, A.K.; Sarma, P.K.D. Usage of particle swarm optimization in digital images selection for monkeypox virus prediction and diagnosis. Malays. J. Comput. Sci. 2024, 37, 124–139. [Google Scholar]
- Alzheimer’s Disease Neuroimaging Initiative (ADNI). Available online: https://adni.loni.usc.edu. (accessed on 25 February 2024).
- Wimo, A.; Jonsson, L.; Gustavsson, A.; McDaid, D.; Ersek, K.; Georges, J.; Gulácsi, L.; Karpati, K.; Kenigsberg, P.; Valtonen, H. Utility score changes in Alzheimer’s disease. Value Health 2009, 12, 295–305. [Google Scholar]
- Raees, P.M.; Thomas, V. Automated detection of Alzheimer’s Disease using Deep Learning in MRI. J. Phys. Conf. Ser. 2021, 1921, 012024. [Google Scholar] [CrossRef]
- Buvaneswari, P.; Gayathri, R. Deep learning-based segmentation in classification of Alzheimer’s disease. Arab. J. Sci. Eng. 2021, 46, 5373–5383. [Google Scholar] [CrossRef]
- Saratxaga, C.L.; Moya, I.; Picón, A.; Acosta, M.; Moreno-Fernandez-de Leceta, A.; Garrote, E.; Bereciartua-Perez, A. MRI deep learning-based solution for Alzheimer’s disease prediction. J. Pers. Med. 2021, 11, 902. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Qing, Z.; Liu, R.; Zhang, X.; Lv, P.; Wang, M.; Wang, Y.; He, K.; Gao, Y.; Zhang, B. Deep learning-based classification and voxel-based visualization of frontotemporal dementia and Alzheimer’s disease. Front. Neurosci. 2021, 14, 626154. [Google Scholar] [CrossRef] [PubMed]





| Study | Data Type | Methodology | Accuracy | Key Findings | Limitations |
|---|---|---|---|---|---|
| Marwa et al. [19] | MRI Images | DNN | 99.68% | High accuracy for various stages | Limited generalizability |
| Ghazal et al. [20] | MRI Images | Transfer Learning (AlexNet) | 91.7% | Effective for stage classification | Potential overfitting |
| AlSaeed et al. [21] | MRI Images | CNN (ResNet50) | 85.7–99% | Superior performance in feature extraction | Not evaluated on diverse datasets |
| Helaly et al. [23] | MRI Images | CNN, Transfer Learning | 97% | VGG-19 model performed well in classification | Limited comparison with other DL models |
| Mohammed et al. [24] | MRI Images | CNN, Hybrid Models | 94.8% | Hybrid models improved diagnostic accuracy | Requires large datasets for training |
| Ji et al. [30] | MRI Images | Ensemble Learning | 97.65% | Early diagnosis of AD/mild cognitive impairment | Ensemble methods can be computationally expensive |
| Islam et al. [31] | MRI Images | CNN | Not Specified | Fast AD detection suitable for small datasets | Not detailed in feature extraction |
| Study | Data Type | Methodology | Accuracy | Key Findings | Limitations |
|---|---|---|---|---|---|
| Khalil et al. [5] | MRI Images, Simulation Data | Federated Learning (VHDL + FPGA) | 89% | Efficient hardware acceleration for FL with low power consumption | Limited to specific hardware settings |
| Mitrovska et al. [6] | Demographic and Diagnosis Data | Federated Averaging (FedAvg) + Secure Aggregation (SecAgg) | Not Specified | Enhanced privacy guarantees with FL; effective under diverse demographics | Potential complexity in real-world deployment |
| Trivedi et al. [32] | MRI Images | Federated Deep Learning (AlexNet) | 98.53% | High accuracy in AD classification with privacy-preserving setup | Requires IID datasets |
| Altalbe et al. [33] | Voice Disorder Data | Federated Learning (DNN) | 82.82% | Overfitting reduced through iterative training; good privacy preservation | Lower accuracy compared to other models |
| Mandawkar et al. [34] | Clinical Data | Tawny Flamingo Deep CNN + Federated Learning | 97.995% | High accuracy in AD detection; effective in distributed settings | Computationally intensive |
| Castro et al. [35] | RGB MRI Images | Federated Learning + Biometric Recognition | Comparable to Centralized ML | Privacy preservation in AD diagnosis with biometric authentication | Performance depends on biometric accuracy |
| Qian et al. [9] | sMRI Images | Federated Deep Forest (FeDeFo) | Not Specified | Personalized AD classification; effective handling of data discrepancies | Complexity in model personalization |
| Model Content | Details |
|---|---|
| 1st Convolution Layer 2D | Input Size 128 × 128 × 3, 16 filters of kernel size 3 × 3, he_normal initializer, L2 (0.01), LeakyReLU (alpha = 0.001) |
| 1st Max Pooling Layer 2D | Pool size 2 × 2 |
| Batch Normalization | Normalizes activations |
| 2nd Convolution Layer 2D | 32 filters of kernel size 3 × 3, he_normal initializer, L2 (0.01), LeakyReLU (alpha = 0.001) |
| 2nd Max Pooling Layer 2D | Pool size 2 × 2 |
| 3rd Convolution Layer 2D | 128 filters of kernel size 3 × 3, he_normal initializer, L2 (0.01), LeakyReLU (alpha = 0.001) |
| 3rd Max Pooling Layer 2D | Pool size 2 × 2 |
| Batch Normalization | Normalizes activations |
| Flattening Layer | Converts 2D matrix to 1D vector |
| Dense Layer | 128 units, he_normal initializer, L2 (0.01), LeakyReLU (alpha = 0.001) |
| Dropout Layer | Randomly deactivates 25% neurons |
| Dense Layer | 64 units, LeakyReLU (alpha = 0.001) |
| Dropout Layer | Randomly deactivates 25% neurons |
| Output Layer | 4 units, SoftMax activation |
| Optimization Function | Adam optimizer |
| Loss Function | Sparse Categorical Crossentropy |
| Metrics | Accuracy |
| Rule ID | Rule Weight | IF | THEN Group | Activation Weight | |
|---|---|---|---|---|---|
| CNN_Value | Age | ||||
| R1 | 1.0 | Low | Young-Old | CN | 0.36 |
| R2 | 1.0 | Low | Middle-Old | MCI | 0.40 |
| R3 | 1.0 | Low | Old-Old | MCI | 0.22 |
| R4 | 1.0 | Medium | Young-Old | MCI | 0.18 |
| R5 | 1.0 | Medium | Middle-Old | AD | 0.45 |
| R6 | 1.0 | Medium | Old-Old | AD | 0.30 |
| R7 | 1.0 | High | Young-Old | AD | 0.55 |
| R8 | 1.0 | High | Middle-Old | AD | 0.65 |
| R9 | 1.0 | High | Old-Old | AD | 0.80 |
| Hyperparameter | Value |
|---|---|
| Train-test split | 80% training, 20% testing |
| Scaling method | MinMaxScaler |
| PSO parameters: | Swarm size: 50 Maximum iterations: 100 Lower bounds: [0, 0, …, 0] (38 times) Upper bounds: [1, 1, …, 1] (38 times) |
| Number of clients | 3 |
| Iterations for federated learning | 5 |
| Rule ID | Rule Weight | CNN_Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.0 | 0.1656 | 0.7499 | 0.0845 |
| R2 | 0.7123 | 0.7404 | 0.2001 | 0.0595 |
| R3 | 0.0 | 0.4954 | 0.2110 | 0.2936 |
| R4 | 0.7882 | 0.0072 | 0.2619 | 0.7309 |
| R5 | 0.9821 | 0.6643 | 0.0000 | 0.3357 |
| R6 | 0.9650 | 0.5683 | 0.0000 | 0.4317 |
| R7 | 0.9971 | 0.0777 | 0.0212 | 0.9011 |
| R8 | 0.4432 | 0.3439 | 0.6561 | 0.0000 |
| R9 | 0.0542 | 0.0000 | 0.7270 | 0.2730 |
| Rule ID | Rule Weight | CNN_Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.7512 | 0.0026 | 0.6759 | 0.3215 |
| R2 | 0.5443 | 0.0000 | 0.5141 | 0.4859 |
| R3 | 0.0666 | 0.0002 | 0.6458 | 0.3540 |
| R4 | 0.5061 | 0.6815 | 0.3185 | 0.0000 |
| R5 | 0.8389 | 0.7446 | 0.2265 | 0.0289 |
| R6 | 0.3314 | 0.9831 | 0.0169 | 0.0000 |
| R7 | 1.0000 | 0.0059 | 0.3432 | 0.6510 |
| R8 | 0.0023 | 0.0014 | 0.4648 | 0.5338 |
| R9 | 0.1576 | 0.2998 | 0.1973 | 0.5029 |
| Rule ID | Rule Weight | CNN_Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.6234 | 0.5289 | 0.0000 | 0.4711 |
| R2 | 0.1875 | 0.5248 | 0.2184 | 0.2568 |
| R3 | 1.0000 | 1.0000 | 0.0000 | 0.0000 |
| R4 | 1.0000 | 0.2385 | 0.0177 | 0.7437 |
| R5 | 0.3468 | 0.4117 | 0.4524 | 0.1359 |
| R6 | 0.9086 | 0.0001 | 0.9976 | 0.0023 |
| R7 | 1.0000 | 0.0056 | 0.3551 | 0.6393 |
| R8 | 0.6326 | 0.0000 | 0.4097 | 0.5903 |
| R9 | 0.9736 | 0.5891 | 0.3160 | 0.0949 |
| Rule ID | Rule Weight | CNN_Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.2869 | 0.7223 | 0.2777 | 0.0000 |
| R2 | 0.6418 | 0.7559 | 0.1218 | 0.1223 |
| R3 | 0.4251 | 0.1436 | 0.3681 | 0.4883 |
| R4 | 0.9871 | 0.0046 | 0.3004 | 0.6950 |
| R5 | 0.4296 | 0.0000 | 0.4848 | 0.5152 |
| R6 | 0.0022 | 0.2094 | 0.0000 | 0.7906 |
| R7 | 0.3891 | 0.2848 | 0.3085 | 0.4067 |
| R8 | 0.4943 | 0.4355 | 0.0000 | 0.5645 |
| R9 | 0.4881 | 0.4510 | 0.3779 | 0.1711 |
| Rule ID | Rule Weight | CNN_Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.0000 | 0.4824 | 0.4045 | 0.1132 |
| R2 | 0.2866 | 0.0000 | 0.7747 | 0.2253 |
| R3 | 0.8641 | 0.0842 | 0.9158 | 0.0000 |
| R4 | 0.0007 | 0.7054 | 0.2771 | 0.0175 |
| R5 | 0.4554 | 0.3732 | 0.6268 | 0.0000 |
| R6 | 0.2630 | 0.6074 | 0.0000 | 0.3926 |
| R7 | 0.5805 | 0.8676 | 0.0346 | 0.0978 |
| R8 | 1.0000 | 0.0041 | 0.2227 | 0.7731 |
| R9 | 0.1764 | 0.0006 | 0.3066 | 0.6928 |
| Rule ID | Rule Weight | CNN_Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.0907 | 0.5785 | 0.3410 | 0.0805 |
| R2 | 0.6633 | 0.0039 | 0.2294 | 0.7666 |
| R3 | 0.3330 | 0.0841 | 0.0003 | 0.9156 |
| R4 | 0.0000 | 0.4099 | 0.3201 | 0.2700 |
| R5 | 0.4140 | 0.0090 | 0.2667 | 0.7243 |
| R6 | 0.6652 | 0.0003 | 0.6918 | 0.3079 |
| R7 | 0.9462 | 0.9070 | 0.0295 | 0.0634 |
| R8 | 0.9036 | 0.4595 | 0.0000 | 0.5405 |
| R9 | 0.0000 | 0.4054 | 0.1314 | 0.4632 |
| Rule ID | Rule Weight | CNN_Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.9997 | 0.0017 | 0.6385 | 0.3598 |
| R2 | 0.4324 | 0.4962 | 0.5038 | 0.0000 |
| R3 | 0.5750 | 0.7245 | 0.0000 | 0.2755 |
| R4 | 0.8920 | 0.1016 | 0.0050 | 0.8934 |
| R5 | 0.1937 | 0.7326 | 0.1164 | 0.1509 |
| R6 | 0.2851 | 0.0001 | 0.4976 | 0.5023 |
| R7 | 0.0632 | 0.8496 | 0.1204 | 0.0300 |
| R8 | 0.9109 | 0.7154 | 0.0000 | 0.2846 |
| R9 | 0.0000 | 0.6565 | 0.1659 | 0.1776 |
| Rule ID | Rule Weight | CNN_Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.0907 | 0.5785 | 0.3410 | 0.0805 |
| R2 | 0.6633 | 0.0039 | 0.2294 | 0.7666 |
| R3 | 0.3330 | 0.0842 | 0.0003 | 0.9156 |
| R4 | 0.0000 | 0.4099 | 0.3201 | 0.2700 |
| R5 | 0.4140 | 0.0090 | 0.2667 | 0.7243 |
| R6 | 0.6652 | 0.0003 | 0.6918 | 0.3079 |
| R7 | 0.9462 | 0.9070 | 0.0295 | 0.0634 |
| R8 | 0.9036 | 0.4595 | 0.0000 | 0.5405 |
| R9 | 0.0000 | 0.4054 | 0.1314 | 0.4632 |
| Rule ID | Rule Weight | CNN_Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.9959 | 0.0000 | 0.6876 | 0.3124 |
| R2 | 0.5408 | 0.5597 | 0.0000 | 0.4403 |
| R3 | 0.0000 | 0.3782 | 0.2889 | 0.3329 |
| R4 | 1.0000 | 0.0728 | 0.0007 | 0.9265 |
| R5 | 0.0863 | 0.7150 | 0.0405 | 0.2447 |
| R6 | 0.7342 | 0.1596 | 0.0000 | 0.8404 |
| R7 | 0.0796 | 0.0000 | 0.4875 | 0.5125 |
| R8 | 0.0001 | 0.5486 | 0.4514 | 0.0000 |
| R9 | 0.6853 | 0.0000 | 0.3189 | 0.6810 |
| Rule ID | Rule Weight | CNN Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.0956 | 0.3251 | 0.5028 | 0.1721 |
| R2 | 0.2457 | 0.6778 | 0.0000 | 0.3222 |
| R3 | 0.7022 | 0.0000 | 0.6496 | 0.3504 |
| R4 | 0.9938 | 0.7386 | 0.2160 | 0.0453 |
| R5 | 0.7375 | 0.0004 | 0.6380 | 0.3616 |
| R6 | 0.0011 | 0.0487 | 0.0742 | 0.8771 |
| R7 | 0.7026 | 0.0000 | 0.8207 | 0.1793 |
| R8 | 0.1263 | 0.0412 | 0.9588 | 0.0000 |
| R9 | 1.0000 | 0.7382 | 0.0000 | 0.2618 |
| Rule ID | Rule Weight | CNN Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.0000 | 0.3251 | 0.5028 | 0.1721 |
| R2 | 0.2457 | 0.6778 | 0.0000 | 0.3222 |
| R3 | 0.7022 | 0.0000 | 0.6496 | 0.3504 |
| R4 | 0.9938 | 0.7386 | 0.2160 | 0.0453 |
| R5 | 0.7375 | 0.0004 | 0.6380 | 0.3616 |
| R6 | 0.0011 | 0.0487 | 0.0742 | 0.8771 |
| R7 | 0.7026 | 0.0000 | 0.8207 | 0.1793 |
| R8 | 0.1263 | 0.0412 | 0.9588 | 0.0000 |
| R9 | 1.0000 | 0.7382 | 0.0000 | 0.2618 |
| Rule ID | Rule Weight | CNN Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.0000 | 0.3251 | 0.5028 | 0.1721 |
| R2 | 0.2457 | 0.6778 | 0.0000 | 0.3222 |
| R3 | 0.7022 | 0.0000 | 0.6496 | 0.3504 |
| R4 | 0.9938 | 0.7386 | 0.2160 | 0.0453 |
| R5 | 0.7375 | 0.0004 | 0.6380 | 0.3616 |
| R6 | 0.0011 | 0.0487 | 0.0742 | 0.8771 |
| R7 | 0.7026 | 0.0000 | 0.8207 | 0.1793 |
| R8 | 0.1263 | 0.0412 | 0.9588 | 0.0000 |
| R9 | 1.0000 | 0.7382 | 0.0000 | 0.2618 |
| Rule ID | Rule Weight | CNN Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.0000 | 0.2123 | 0.7077 | 0.0800 |
| R2 | 0.8953 | 0.0000 | 0.7188 | 0.2812 |
| R3 | 0.4688 | 0.4743 | 0.5055 | 0.0201 |
| R4 | 0.4004 | 0.3913 | 0.2215 | 0.3872 |
| R5 | 0.5339 | 0.0001 | 0.3351 | 0.6648 |
| R6 | 0.4359 | 0.8704 | 0.0903 | 0.0393 |
| R7 | 0.9764 | 0.6474 | 0.0510 | 0.3016 |
| R8 | 0.8233 | 0.0005 | 0.0981 | 0.9015 |
| R9 | 0.0301 | 0.4195 | 0.5728 | 0.0077 |
| Rule ID | Rule Weight | CNN Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.0000 | 0.2123 | 0.7077 | 0.0800 |
| R2 | 0.8953 | 0.0000 | 0.7188 | 0.2812 |
| R3 | 0.4688 | 0.4743 | 0.5055 | 0.0201 |
| R4 | 0.4004 | 0.3913 | 0.2215 | 0.3872 |
| R5 | 0.5339 | 0.0001 | 0.3351 | 0.6648 |
| R6 | 0.4359 | 0.8704 | 0.0903 | 0.0393 |
| R7 | 0.9764 | 0.6474 | 0.0510 | 0.3016 |
| R8 | 0.8233 | 0.0005 | 0.0981 | 0.9015 |
| R9 | 0.0301 | 0.4195 | 0.5728 | 0.0077 |
| Rule ID | Rule Weight | CNN Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.0000 | 0.2123 | 0.7077 | 0.0800 |
| R2 | 0.8953 | 0.0000 | 0.7188 | 0.2812 |
| R3 | 0.4688 | 0.4743 | 0.5055 | 0.0201 |
| R4 | 0.4004 | 0.3913 | 0.2215 | 0.3872 |
| R5 | 0.5339 | 0.0001 | 0.3351 | 0.6648 |
| R6 | 0.4359 | 0.8704 | 0.0903 | 0.0393 |
| R7 | 0.9764 | 0.6474 | 0.0510 | 0.3016 |
| R8 | 0.8233 | 0.0005 | 0.0981 | 0.9015 |
| R9 | 0.0301 | 0.4195 | 0.5728 | 0.0077 |
| Rule ID | Rule Weight | CNN Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.0489 | 0.0000 | 0.1579 | 0.8421 |
| R2 | 0.9898 | 0.7954 | 0.0000 | 0.2046 |
| R3 | 0.2992 | 0.2640 | 0.7360 | 0.0000 |
| R4 | 0.5001 | 0.6976 | 0.0000 | 0.3024 |
| R5 | 0.9721 | 0.4644 | 0.0000 | 0.5356 |
| R6 | 0.5106 | 0.0000 | 0.7867 | 0.2133 |
| R7 | 1.0000 | 0.0016 | 0.5032 | 0.4952 |
| R8 | 0.6459 | 0.7756 | 0.2244 | 0.0000 |
| R9 | 0.7654 | 0.6747 | 0.0183 | 0.3070 |
| Rule ID | Rule Weight | CNN Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.0489 | 0.0000 | 0.1579 | 0.8421 |
| R2 | 0.9898 | 0.7954 | 0.0000 | 0.2046 |
| R3 | 0.2992 | 0.2640 | 0.7360 | 0.0000 |
| R4 | 0.5001 | 0.6976 | 0.0000 | 0.3024 |
| R5 | 0.9721 | 0.4644 | 0.0000 | 0.5356 |
| R6 | 0.5106 | 0.0000 | 0.7867 | 0.2133 |
| R7 | 1.0000 | 0.0016 | 0.5032 | 0.4952 |
| R8 | 0.6459 | 0.7756 | 0.2244 | 0.0000 |
| R9 | 0.7654 | 0.6747 | 0.0183 | 0.3070 |
| Rule ID | Rule Weight | CNN Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.0489 | 0.0000 | 0.1579 | 0.8421 |
| R2 | 0.9898 | 0.7954 | 0.0000 | 0.2046 |
| R3 | 0.2992 | 0.2640 | 0.7360 | 0.0000 |
| R4 | 0.5001 | 0.6976 | 0.0000 | 0.3024 |
| R5 | 0.9721 | 0.4644 | 0.0000 | 0.5356 |
| R6 | 0.5106 | 0.0000 | 0.7867 | 0.2133 |
| R7 | 1.0000 | 0.0016 | 0.5032 | 0.4952 |
| R8 | 0.6459 | 0.7756 | 0.2244 | 0.0000 |
| R9 | 0.7654 | 0.6747 | 0.0183 | 0.3070 |
| Rule ID | Rule Weight | CNN Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.0000 | 0.6403 | 0.0000 | 0.3597 |
| R2 | 0.9376 | 0.2420 | 0.0000 | 0.7580 |
| R3 | 0.0000 | 0.0775 | 0.0506 | 0.8719 |
| R4 | 0.7595 | 0.1560 | 0.0007 | 0.8433 |
| R5 | 0.5355 | 0.8803 | 0.0056 | 0.1141 |
| R6 | 0.5329 | 0.0833 | 0.9167 | 0.0000 |
| R7 | 0.6066 | 0.0006 | 0.5928 | 0.4065 |
| R8 | 0.3410 | 0.5007 | 0.4993 | 0.0000 |
| R9 | 0.3354 | 0.0003 | 0.1079 | 0.8918 |
| Rule ID | Rule Weight | CNN Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.0000 | 0.0916 | 0.0789 | 0.8295 |
| R2 | 1.0000 | 0.5924 | 0.0000 | 0.4076 |
| R3 | 0.0064 | 0.0000 | 0.6615 | 0.3385 |
| R4 | 0.8879 | 0.0006 | 0.1202 | 0.8792 |
| R5 | 0.2335 | 0.6908 | 0.3092 | 0.0000 |
| R6 | 0.8953 | 0.0002 | 0.1252 | 0.8746 |
| R7 | 0.4955 | 0.0025 | 0.4704 | 0.5271 |
| R8 | 1.0000 | 0.6585 | 0.3393 | 0.0023 |
| R9 | 0.7271 | 0.6995 | 0.0000 | 0.3005 |
| Rule ID | Rule Weight | CNN Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.2760 | 0.6181 | 0.0000 | 0.3819 |
| R2 | 0.6066 | 0.0002 | 0.0000 | 0.9998 |
| R3 | 0.0000 | 0.1505 | 0.5868 | 0.2627 |
| R4 | 0.3360 | 0.0000 | 0.1487 | 0.8513 |
| R5 | 0.0648 | 0.2981 | 0.7019 | 0.0000 |
| R6 | 0.8948 | 0.0000 | 0.6912 | 0.3088 |
| R7 | 0.5753 | 0.4126 | 0.5874 | 0.0000 |
| R8 | 0.0295 | 0.5479 | 0.4335 | 0.0187 |
| R9 | 0.2985 | 0.5549 | 0.0000 | 0.4451 |
| Rule ID | Rule Weight | CNN Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.0000 | 0.2409 | 0.6108 | 0.1483 |
| R2 | 0.9916 | 0.0000 | 0.8724 | 0.1276 |
| R3 | 0.9982 | 0.0002 | 0.9226 | 0.0772 |
| R4 | 0.4423 | 0.0002 | 0.9988 | 0.0011 |
| R5 | 0.9752 | 0.8539 | 0.0000 | 0.1461 |
| R6 | 0.3286 | 0.4222 | 0.5778 | 0.0000 |
| R7 | 0.8676 | 0.9455 | 0.0442 | 0.0103 |
| R8 | 0.9292 | 0.0018 | 0.1299 | 0.8683 |
| R9 | 0.2586 | 0.7650 | 0.0000 | 0.2349 |
| Rule ID | Rule Weight | CNN Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.7287 | 0.0010 | 0.9990 | 0.0000 |
| R2 | 1.0000 | 0.0000 | 0.4366 | 0.5634 |
| R3 | 1.0000 | 0.0000 | 1.0000 | 0.0000 |
| R4 | 0.4041 | 0.0000 | 0.8518 | 0.1482 |
| R5 | 0.1697 | 0.0000 | 0.0000 | 1.0000 |
| R6 | 0.7189 | 0.3120 | 0.0000 | 0.6880 |
| R7 | 0.0050 | 0.4548 | 0.3923 | 0.1529 |
| R8 | 0.5496 | 0.0000 | 0.0269 | 0.9731 |
| R9 | 0.0409 | 0.6471 | 0.0000 | 0.3529 |
| Rule ID | Rule Weight | CNN Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.6438 | 0.4091 | 0.5909 | 0.0000 |
| R2 | 0.6435 | 0.0009 | 0.5823 | 0.4168 |
| R3 | 0.4093 | 0.0000 | 0.8276 | 0.1724 |
| R4 | 0.7355 | 0.0805 | 0.0000 | 0.9195 |
| R5 | 0.4807 | 0.0011 | 0.3556 | 0.6434 |
| R6 | 0.8471 | 0.6813 | 0.3187 | 0.0000 |
| R7 | 0.8134 | 0.5222 | 0.3630 | 0.1147 |
| R8 | 1.0000 | 0.0009 | 0.9573 | 0.0422 |
| R9 | 0.6295 | 1.0000 | 0.0000 | 0.0000 |
| Rule ID | Rule Weight | CNN Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.0259 | 0.0000 | 0.4371 | 0.5629 |
| R2 | 0.9548 | 0.0005 | 0.1798 | 0.8202 |
| R3 | 0.2521 | 0.0175 | 0.9825 | 0.0000 |
| R4 | 0.6134 | 0.8146 | 0.0064 | 0.1790 |
| R5 | 0.0410 | 0.0006 | 0.5337 | 0.4657 |
| R6 | 0.7805 | 0.0001 | 0.1645 | 0.8354 |
| R7 | 0.0000 | 0.5033 | 0.0631 | 0.4336 |
| R8 | 0.7242 | 0.1037 | 0.8963 | 0.0000 |
| R9 | 0.3425 | 0.4387 | 0.0000 | 0.5613 |
| Rule ID | Rule Weight | CNN Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.9841 | 0.7833 | 0.0378 | 0.1799 |
| R2 | 0.0186 | 0.2679 | 0.3714 | 0.3608 |
| R3 | 0.4292 | 0.1414 | 0.6634 | 0.1951 |
| R4 | 0.6449 | 0.0905 | 0.0000 | 0.9095 |
| R5 | 0.5903 | 0.0049 | 0.9838 | 0.0113 |
| R6 | 0.0000 | 0.0008 | 0.1789 | 0.8204 |
| R7 | 0.0808 | 0.6489 | 0.2400 | 0.1112 |
| R8 | 1.0000 | 0.0085 | 0.2246 | 0.7669 |
| R9 | 0.7350 | 0.1810 | 0.0010 | 0.8180 |
| Rule ID | Rule Weight | CNN Value | Age | Group |
|---|---|---|---|---|
| R1 | 0.5367 | 0.8086 | 0.0000 | 0.1914 |
| R2 | 0.4920 | 0.0005 | 0.2176 | 0.7818 |
| R3 | 0.2167 | 0.0002 | 0.1888 | 0.8111 |
| R4 | 0.3923 | 0.6196 | 0.3804 | 0.0000 |
| R5 | 0.9993 | 0.0051 | 0.2194 | 0.7755 |
| R6 | 0.0000 | 0.3821 | 0.2385 | 0.3794 |
| R7 | 0.3796 | 0.6376 | 0.2802 | 0.0822 |
| R8 | 0.2942 | 0.0012 | 0.2127 | 0.7861 |
| R9 | 0.0000 | 0.2058 | 0.0277 | 0.7665 |
| Rule ID | Rule Weight | CN | MCI | AD |
|---|---|---|---|---|
| R1 | 0.9612 | 0.0000 | 0.9689 | 0.0311 |
| R2 | 0.3388 | 0.0000 | 0.4785 | 0.5215 |
| R3 | 0.0850 | 0.8647 | 0.0747 | 0.0606 |
| R4 | 0.0806 | 0.4947 | 0.5053 | 0.0000 |
| R5 | 0.3647 | 0.0000 | 0.8442 | 0.1558 |
| R6 | 0.4242 | 0.0000 | 0.0772 | 0.9228 |
| R7 | 0.4484 | 0.7169 | 0.2821 | 0.0010 |
| R8 | 0.0000 | 0.0000 | 0.6721 | 0.3279 |
| R9 | 0.6411 | 0.0000 | 1.0000 | 0.0000 |
| Rule ID | Rule Weight | CN | MCI | AD |
|---|---|---|---|---|
| R1 | 0.9997 | 0.0758 | 0.0000 | 0.9242 |
| R2 | 0.2077 | 0.8508 | 0.1492 | 0.0000 |
| R3 | 0.3042 | 0.3163 | 0.6837 | 0.0000 |
| R4 | 0.2439 | 0.5159 | 0.0941 | 0.3899 |
| R5 | 0.5673 | 0.0007 | 0.6432 | 0.3561 |
| R6 | 0.8671 | 0.6333 | 0.2925 | 0.0742 |
| R7 | 0.9215 | 0.5557 | 0.4443 | 0.0000 |
| R8 | 0.5669 | 0.0033 | 0.0942 | 0.9025 |
| R9 | 0.7453 | 0.2629 | 0.7371 | 0.0000 |
| Rule ID | Rule Weight | CN | MCI | AD |
|---|---|---|---|---|
| R1 | 0.3629 | 0.4477 | 0.0000 | 0.5523 |
| R2 | 0.0000 | 0.2364 | 0.5648 | 0.1988 |
| R3 | 0.0000 | 0.2970 | 0.3287 | 0.3742 |
| R4 | 0.7316 | 0.4522 | 0.5478 | 0.0000 |
| R5 | 0.1970 | 0.5290 | 0.4710 | 0.0000 |
| R6 | 0.6037 | 0.6760 | 0.3241 | 0.0000 |
| R7 | 0.2779 | 0.0001 | 0.1816 | 0.8183 |
| R8 | 0.5331 | 0.9031 | 0.0449 | 0.0521 |
| R9 | 0.2306 | 0.6962 | 0.0000 | 0.3038 |
| Rule ID | Rule Weight | CN | MCI | AD |
|---|---|---|---|---|
| R1 | 0.0956 | 0.3251 | 0.5028 | 0.1721 |
| R2 | 0.2457 | 0.6778 | 0.0000 | 0.3222 |
| R3 | 0.7022 | 0.0000 | 0.6496 | 0.3504 |
| R4 | 0.9938 | 0.7386 | 0.2160 | 0.0453 |
| R5 | 0.7375 | 0.0004 | 0.6380 | 0.3616 |
| R6 | 0.0011 | 0.0487 | 0.0742 | 0.8771 |
| R7 | 0.7026 | 0.0000 | 0.8207 | 0.1793 |
| R8 | 0.1263 | 0.0412 | 0.9588 | 0.0000 |
| R9 | 1.0000 | 0.7382 | 0.0000 | 0.2618 |
| Rule ID | Rule Weight | CN | MCI | AD |
|---|---|---|---|---|
| R1 | 0.0000 | 0.2123 | 0.7077 | 0.0800 |
| R2 | 0.8953 | 0.0000 | 0.7188 | 0.2812 |
| R3 | 0.4688 | 0.4743 | 0.5055 | 0.0201 |
| R4 | 0.4004 | 0.3913 | 0.2215 | 0.3872 |
| R5 | 0.5339 | 0.0001 | 0.3351 | 0.6648 |
| R6 | 0.4359 | 0.8704 | 0.0903 | 0.0393 |
| R7 | 0.9764 | 0.6474 | 0.0510 | 0.3016 |
| R8 | 0.8233 | 0.0005 | 0.0981 | 0.9015 |
| R9 | 0.0301 | 0.4195 | 0.5728 | 0.0077 |
| Rule ID | Rule Weight | CN | MCI | AD |
|---|---|---|---|---|
| R1 | 0.0489 | 0.0000 | 0.1579 | 0.8421 |
| R2 | 0.9898 | 0.7954 | 0.0000 | 0.2046 |
| R3 | 0.2992 | 0.2640 | 0.7360 | 0.0000 |
| R4 | 0.5001 | 0.6976 | 0.0000 | 0.3024 |
| R5 | 0.9721 | 0.4644 | 0.0000 | 0.5356 |
| R6 | 0.5106 | 0.0000 | 0.7867 | 0.2133 |
| R7 | 1.0000 | 0.0016 | 0.5032 | 0.4952 |
| R8 | 0.6459 | 0.7756 | 0.2244 | 0.0000 |
| R9 | 0.7654 | 0.6747 | 0.0183 | 0.3070 |
| Rule ID | Rule Weight | CN | MCI | AD |
|---|---|---|---|---|
| R1 | 0.9762 | 0.0000 | 0.4092 | 0.5908 |
| R2 | 0.4676 | 0.0000 | 0.7109 | 0.2891 |
| R3 | 0.7043 | 0.0050 | 0.9950 | 0.0000 |
| R4 | 0.5023 | 0.1734 | 0.8266 | 0.0000 |
| R5 | 0.0000 | 0.0453 | 0.5596 | 0.3950 |
| R6 | 0.0000 | 0.0000 | 0.3649 | 0.6351 |
| R7 | 0.7269 | 0.0000 | 0.5659 | 0.4341 |
| R8 | 0.9804 | 0.0434 | 0.9566 | 0.0000 |
| R9 | 0.9975 | 0.7934 | 0.2053 | 0.0013 |
| Rule ID | Rule Weight | CN | MCI | AD |
|---|---|---|---|---|
| R1 | 0.2041 | 0.4060 | 0.4854 | 0.1087 |
| R2 | 0.7601 | 0.0000 | 0.8964 | 0.1036 |
| R3 | 0.6395 | 0.7190 | 0.2281 | 0.0529 |
| R4 | 1.0000 | 0.6089 | 0.3903 | 0.0008 |
| R5 | 0.0390 | 0.0000 | 0.0832 | 0.9168 |
| R6 | 0.6207 | 0.0000 | 0.8847 | 0.1153 |
| R7 | 0.3963 | 0.8816 | 0.0064 | 0.1120 |
| R8 | 0.3521 | 0.0014 | 0.2706 | 0.7280 |
| R9 | 0.0481 | 0.2013 | 0.6028 | 0.1959 |
| Rule ID | Rule Weight | CN | MCI | AD |
|---|---|---|---|---|
| R1 | 0.5367 | 0.0000 | 0.4371 | 0.5629 |
| R2 | 0.4920 | 0.0005 | 0.1798 | 0.8197 |
| R3 | 0.2167 | 0.0175 | 0.9825 | 0.0000 |
| R4 | 0.3923 | 0.8146 | 0.0064 | 0.1790 |
| R5 | 0.9993 | 0.0006 | 0.5347 | 0.4653 |
| R6 | 0.0000 | 0.0001 | 0.1645 | 0.8354 |
| R7 | 0.3796 | 0.5033 | 0.0631 | 0.4336 |
| R8 | 0.2942 | 0.1037 | 0.8963 | 0.0000 |
| R9 | 0.0000 | 0.4387 | 0.0000 | 0.5613 |
| Aggregator Client | Training | Testing | |||
|---|---|---|---|---|---|
| MSE | Accuracy | MSE | Accuracy | ||
| FedAvg Aggregator | Client 1 | 0.015088 | 0.40465 | 0.01072 | 0.48148 |
| Client 2 | 0.02309 | 0.35814 | 0.01808 | 0.37037 | |
| Client 3 | 0.01404 | 0.59813 | 0.01405 | 0.52830 | |
| Global | 0.00409 | 0.99845 | 0.00465 | 0.99999 | |
| FedProx Aggregator | Client 1 | 0.01391 | 0.59535 | 0.01040 | 0.61111 |
| Client 2 | 0.03767 | 0.41395 | 0.03701 | 0.38889 | |
| Client 3 | 0.02491 | 0.37383 | 0.02683 | 0.18868 | |
| Global | 0.00592 | 0.73602 | 0.00687 | 0.68944 | |
| Genetic Algorithm | Client 1 | 0.036076 | 0.31163 | 0.032451 | 0.29629 |
| Client 2 | 0.04106 | 0.35348 | 0.037835 | 0.24074 | |
| Client 3 | 0.03331 | 0.36448 | 0.03915 | 0.24528 | |
| Global | 0.00833 | 0.51086 | 0.00904 | 0.43478 | |
| Iteration | Client 1 Time (s) | Client 2 Time (s) | Client 3 Time (s) | Final PSO Time (s) |
|---|---|---|---|---|
| 1 | 253.88 | 375.92 | 296.97 | 742.59 |
| 2 | 251.42 | 249.82 | 252.57 | 746.66 |
| 3 | 279.37 | 252.04 | 251.23 | 734.71 |
| 4 | 259.25 | 251.35 | 249.78 | 739.96 |
| 5 | 249.15 | 249.71 | 250.31 | 734.90 |
| Total | 1293.07 | 1129.09 | 1300.86 | 3998.82 |
| Reference | Methodology | Dataset | Accuracy | Privacy-preserving | Uncertainty Issue |
|---|---|---|---|---|---|
| Umme et al. [1] | DementiaNet + Transfer Learning | MRI Dataset (6400 Images) | 97% | X | X |
| Raees et al. [40] | SVM + DNN | MCI dataset (111 people) | 90% | X | X |
| Buvaneswari et al. [41] | SegNet + ResNet-101 | ADNI dataset | 96% | X | X |
| Saratxaga et al. [42] | ResNet 18 + BrainNet | Collected From Kaggle | 80%, 90% | X | X |
| Hu et al. [43] | CNN | ADNI and NIFD dataset | 92% | X | X |
| Khalil et al. [5] | Hardware Acceleration (VHDL + FPGA) | Simulation Data | 89% | Achieved 87% sensitivity; Low power consumption (35–39 mW) | X |
| Mitrovska et al. [6] | FedAvg, Secure Aggregation (SecAgg) | Demographic Simulations | - | Highlighted privacy guarantees; Evaluated statistical variations | X |
| Trivedi et al. [32] | Federated Deep Learning + AlexNet | IID Dataset | 98.53% | Ensured data privacy; Tested with single/multiple clients | X |
| Altalbe et al. [33] | DNN within Federated Learning | Kay Elemetrics voice disorder data | 82.82% | Reduced overfitting; Preserved privacy and security | X |
| Mandawkar et al. [34] | Tawny Flamingo Deep CNN | Clinical Data | 98.252% (K-fold), 97.995% (training) | High accuracy; Tuned by Tawny Flamingo algorithm | X |
| Castro et al. [35] | Federated Learning + Biometric Authentication | OASIS, ADNI datasets | Comparable | Protected privacy; Prevented data poisoning | X |
| Qian et al. [9] | FeDeFo (Deep Forest + Federated Learning) | sMRI images | - | Personalized model while protecting data privacy | X |
| Proposed Research | Hybrid Deep and Federated Learning | ADNI and NIfTI files dataset | 99.9% | Ensured data privacy; Tested with multiple clients | Solved |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Basnin, N.; Mahmud, T.; Islam, R.U.; Andersson, K. An Evolutionary Federated Learning Approach to Diagnose Alzheimer’s Disease Under Uncertainty. Diagnostics 2025, 15, 80. https://doi.org/10.3390/diagnostics15010080
Basnin N, Mahmud T, Islam RU, Andersson K. An Evolutionary Federated Learning Approach to Diagnose Alzheimer’s Disease Under Uncertainty. Diagnostics. 2025; 15(1):80. https://doi.org/10.3390/diagnostics15010080
Chicago/Turabian StyleBasnin, Nanziba, Tanjim Mahmud, Raihan Ul Islam, and Karl Andersson. 2025. "An Evolutionary Federated Learning Approach to Diagnose Alzheimer’s Disease Under Uncertainty" Diagnostics 15, no. 1: 80. https://doi.org/10.3390/diagnostics15010080
APA StyleBasnin, N., Mahmud, T., Islam, R. U., & Andersson, K. (2025). An Evolutionary Federated Learning Approach to Diagnose Alzheimer’s Disease Under Uncertainty. Diagnostics, 15(1), 80. https://doi.org/10.3390/diagnostics15010080








