Information Geometry and Manifold Learning: A Novel Framework for Analyzing Alzheimer’s Disease MRI Data
Abstract
:1. Introduction
2. Related Works
3. Methodology
3.1. Information Geometry
3.2. Manifold Learning
4. Results
4.1. Dataset
4.2. Information Geometric Results
4.3. Manifold Learning Results
5. Discussions
5.1. Discussions on Information Geometric Results
5.2. Discussions on Manifold Learning Results
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nichols, E.; Szoeke, C.E.; Vollset, S.E.; Abbasi, N.; Abd-Allah, F.; Abdela, J.; Aichour, M.T.E.; Akinyemi, R.O.; Alahdab, F.; Asgedom, S.W.; et al. Global, regional, and national burden of Alzheimer’s disease and other dementias, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019, 18, 88–106. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Qin, W.; Zhu, M.; Jia, J. Model-based projection of dementia prevalence in China and worldwide: 2020–2050. J. Alzheimer’s Dis. 2021, 82, 1823–1831. [Google Scholar] [CrossRef] [PubMed]
- Livingston, G.; Huntley, J.; Liu, K.Y.; Costafreda, S.G.; Selbæk, G.; Alladi, S.; Ames, D.; Banerjee, S.; Burns, A.; Brayne, C.; et al. Dementia prevention, intervention, and care: 2024 report of the Lancet standing Commission. Lancet 2024, 404, 572–628. [Google Scholar] [CrossRef] [PubMed]
- Dubois, B.; von Arnim, C.A.; Burnie, N.; Bozeat, S.; Cummings, J. Biomarkers in Alzheimer’s disease: Role in early and differential diagnosis and recognition of atypical variants. Alzheimer’s Res. Ther. 2023, 15, 175. [Google Scholar] [CrossRef]
- Rafii, M.S.; Aisen, P.S. Detection and treatment of Alzheimer’s disease in its preclinical stage. Nat. Aging 2023, 3, 520–531. [Google Scholar] [CrossRef]
- Deng, Y.; Wang, H.; Gu, K.; Song, P. Alzheimer’s disease with frailty: Prevalence, screening, assessment, intervention strategies and challenges. Biosci. Trends 2023, 17, 283–292. [Google Scholar] [CrossRef]
- Vrahatis, A.G.; Skolariki, K.; Krokidis, M.G.; Lazaros, K.; Exarchos, T.P.; Vlamos, P. Revolutionizing the early detection of Alzheimer’s disease through non-invasive biomarkers: The role of artificial intelligence and deep learning. Sensors 2023, 23, 4184. [Google Scholar] [CrossRef]
- Trejo-Lopez, J.A.; Yachnis, A.T.; Prokop, S. Neuropathology of Alzheimer’s disease. Neurotherapeutics 2023, 19, 173–185. [Google Scholar] [CrossRef]
- Abushakra, S.; Porsteinsson, A.P.; Sabbagh, M.; Bracoud, L.; Schaerer, J.; Power, A.; Hey, J.A.; Scott, D.; Suhy, J.; Tolar, M.; et al. APOE ε4/ε4 homozygotes with early Alzheimer’s disease show accelerated hippocampal atrophy and cortical thinning that correlates with cognitive decline. Alzheimer’s Dement. Transl. Res. Clin. Interv. 2020, 6, e12117. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, D.; Zheng, H.; Cao, T.; Xia, K.; Su, M.; Meng, Q. The association between retina thinning and hippocampal atrophy in Alzheimer’s disease and mild cognitive impairment: A meta-analysis and systematic review. Front. Aging Neurosci. 2023, 15, 1232941. [Google Scholar] [CrossRef]
- Arya, A.D.; Verma, S.S.; Chakarabarti, P.; Chakrabarti, T.; Elngar, A.A.; Kamali, A.M.; Nami, M. A systematic review on machine learning and deep learning techniques in the effective diagnosis of Alzheimer’s disease. Brain Inform. 2023, 10, 17. [Google Scholar] [CrossRef] [PubMed]
- Dara, O.A.; Lopez-Guede, J.M.; Raheem, H.I.; Rahebi, J.; Zulueta, E.; Fernandez-Gamiz, U. Alzheimer’s disease diagnosis using machine learning: A survey. Appl. Sci. 2023, 13, 8298. [Google Scholar] [CrossRef]
- Houria, L.; Belkhamsa, N.; Cherfa, A.; Cherfa, Y. Multimodal magnetic resonance imaging for Alzheimer’s disease diagnosis using hybrid features extraction and ensemble support vector machines. Int. J. Imaging Syst. Technol. 2023, 33, 610–621. [Google Scholar] [CrossRef]
- Oliveira, M.J.; Ribeiro, P.; Rodrigues, P.M. Machine Learning-Driven GLCM Analysis of Structural MRI for Alzheimer’s Disease Diagnosis. Bioengineering 2024, 11, 1153. [Google Scholar] [CrossRef] [PubMed]
- Alkhatib, M.Q.; Al-Saad, M.; Aburaed, N.; Almansoori, S.; Zabalza, J.; Marshall, S.; Al-Ahmad, H. Tri-CNN: A three branch model for hyperspectral image classification. Remote Sens. 2023, 15, 316. [Google Scholar] [CrossRef]
- Carreras, J. Celiac Disease Deep Learning Image Classification Using Convolutional Neural Networks. J. Imaging 2024, 10, 200. [Google Scholar] [CrossRef]
- Mohammed, F.A.; Tune, K.K.; Assefa, B.G.; Jett, M.; Muhie, S. Medical Image Classifications Using Convolutional Neural Networks: A Survey of Current Methods and Statistical Modeling of the Literature. Mach. Learn. Knowl. Extr. 2024, 6, 699–735. [Google Scholar] [CrossRef]
- Gui, H.; Jiao, H.; Li, L.; Jiang, X.; Su, T.; Pang, Z. Breast Tumor Detection and Diagnosis Using an Improved Faster R-CNN in DCE-MRI. Bioengineering 2024, 11, 1217. [Google Scholar] [CrossRef]
- Lin, J.; Zhang, H.; Shang, H. Convolutional neural network incorporating multiple attention mechanisms for MRI classification of lumbar spinal stenosis. Bioengineering 2024, 11, 1021. [Google Scholar] [CrossRef]
- Stathopoulos, I.; Serio, L.; Karavasilis, E.; Kouri, M.A.; Velonakis, G.; Kelekis, N.; Efstathopoulos, E. Evaluating Brain Tumor Detection with Deep Learning Convolutional Neural Networks Across Multiple MRI Modalities. J. Imaging 2024, 10, 296. [Google Scholar] [CrossRef]
- Zahoor, M.M.; Khan, S.H.; Alahmadi, T.J.; Alsahfi, T.; Mazroa, A.S.A.; Sakr, H.A.; Alqahtani, S.; Albanyan, A.; Alshemaimri, B.K. Brain tumor MRI classification using a novel deep residual and regional CNN. Biomedicines 2024, 12, 1395. [Google Scholar] [CrossRef] [PubMed]
- Cherevko, Y.; Chepurna, O.; Kuleshova, Y. On Information Geometry Methods for Data Analysis. Geom. Integr. Quantization Pap. Lect. Ser. 2024, 29, 11–22. [Google Scholar] [CrossRef]
- Khan, G.A.; Hu, J.; Li, T.; Diallo, B.; Wang, H. Multi-view clustering for multiple manifold learning via concept factorization. Digit. Signal Process. 2023, 140, 104118. [Google Scholar] [CrossRef]
- Lee, M. The geometry of feature space in deep learning models: A holistic perspective and comprehensive review. Mathematics 2023, 11, 2375. [Google Scholar] [CrossRef]
- Meilă, M.; Zhang, H. Manifold learning: What, how, and why. Annu. Rev. Stat. Its Appl. 2024, 11, 393–417. [Google Scholar] [CrossRef]
- Song, W.; Zhang, X.; Yang, G.; Chen, Y.; Wang, L.; Xu, H. A Study on Dimensionality Reduction and Parameters for Hyperspectral Imagery Based on Manifold Learning. Sensors 2024, 24, 2089. [Google Scholar] [CrossRef]
- Tam, P.; Ros, S.; Song, I.; Kang, S.; Kim, S. A Survey of Intelligent End-to-End Networking Solutions: Integrating Graph Neural Networks and Deep Reinforcement Learning Approaches. Electronics 2024, 13, 994. [Google Scholar] [CrossRef]
- Zafeiropoulos, N.; Bitilis, P.; Tsekouras, G.E.; Kotis, K. Graph Neural Networks for Parkinson’s Disease Monitoring and Alerting. Sensors 2023, 23, 8936. [Google Scholar] [CrossRef]
- Seguin, C.; Sporns, O.; Zalesky, A. Brain network communication: Concepts, models and applications. Nat. Rev. Neurosci. 2023, 24, 557–574. [Google Scholar] [CrossRef]
- La Rosa, M.; Fiannaca, A.; La Paglia, L.; Urso, A. A graph neural network approach for the analysis of siRNA-target biological networks. Int. J. Mol. Sci. 2022, 23, 14211. [Google Scholar] [CrossRef]
- Miah, A.S.M.; Hasan, M.A.M.; Jang, S.W.; Lee, H.S.; Shin, J. Multi-stream general and graph-based deep neural networks for skeleton-based sign language recognition. Electronics 2023, 12, 2841. [Google Scholar] [CrossRef]
- Yu, Y.; Qian, W.; Zhang, L.; Gao, R. A graph-neural-network-based social network recommendation algorithm using high-order neighbor information. Sensors 2022, 22, 7122. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Huang, L.; Song, R.; Shen, Q.; Xu, H. A sequential graph neural network for short text classification. Algorithms 2021, 14, 352. [Google Scholar] [CrossRef]
- Abuhantash, F.; Abu Hantash, M.K.; AlShehhi, A. Comorbidity-based framework for Alzheimer’s disease classification using graph neural networks. Sci. Rep. 2024, 14, 21061. [Google Scholar] [CrossRef]
- Ravinder, M.; Saluja, G.; Allabun, S.; Alqahtani, M.S.; Abbas, M.; Othman, M.; Soufiene, B.O. Enhanced brain tumor classification using graph convolutional neural network architecture. Sci. Rep. 2023, 13, 14938. [Google Scholar] [CrossRef]
- Saeidi, M.; Karwowski, W.; Farahani, F.V.; Fiok, K.; Hancock, P.; Sawyer, B.D.; Christov-Moore, L.; Douglas, P.K. Decoding task-based fMRI data with graph neural networks, considering individual differences. Brain Sci. 2022, 12, 1094. [Google Scholar] [CrossRef]
- Sørensen, L.; Igel, C.; Pai, A.; Balas, I.; Anker, C.; Lillholm, M.; Nielsen, M.; Alzheimer’s Disease Neuroimaging Initiative. Differential diagnosis of mild cognitive impairment and Alzheimer’s disease using structural MRI cortical thickness, hippocampal shape, hippocampal texture, and volumetry. NeuroImage Clin. 2017, 13, 470–482. [Google Scholar] [CrossRef]
- Uysal, G.; Ozturk, M. Hippocampal atrophy based Alzheimer’s disease diagnosis via machine learning methods. J. Neurosci. Methods 2020, 337, 108669. [Google Scholar] [CrossRef]
- Lee, S.; Lee, H.; Kim, K.W. Magnetic resonance imaging texture predicts progression to dementia due to Alzheimer disease earlier than hippocampal volume. J. Psychiatry Neurosci. 2020, 45, 7–14. [Google Scholar] [CrossRef]
- Zhang, J.; Fan, Y.; Li, Q.; Thompson, P.M.; Ye, J.; Wang, Y. Empowering cortical thickness measures in clinical diagnosis of Alzheimer’s disease with spherical sparse coding. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, Australia, 18–21 April 2017; pp. 446–450. [Google Scholar]
- De Vos, F.; Schouten, T.M.; Hafkemeijer, A.; Dopper, E.G.; van Swieten, J.C.; de Rooij, M.; van der Grond, J.; Rombouts, S.A. Combining multiple anatomical MRI measures improves Alzheimer’s disease classification. Hum. Brain Mapp. 2016, 37, 1920–1929. [Google Scholar] [CrossRef]
- Wee, C.Y.; Yap, P.T.; Shen, D.; Initiative, A.D.N. Prediction of Alzheimer’s disease and mild cognitive impairment using cortical morphological patterns. Hum. Brain Mapp. 2013, 34, 3411–3425. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Qu, H.; Dong, X.; Dang, B.; Zang, H.; Gong, Y. Leveraging deep learning and xception architecture for high-accuracy mri classification in alzheimer diagnosis. arXiv 2024, arXiv:2403.16212. [Google Scholar]
- Fareed, M.M.S.; Zikria, S.; Ahmed, G.; Mui-Zzud-Din; Mahmood, S.; Aslam, M.; Jillani, S.F.; Moustafa, A.; Asad, M. ADD-Net: An effective deep learning model for early detection of Alzheimer disease in MRI scans. IEEE Access 2022, 10, 96930–96951. [Google Scholar] [CrossRef]
- AlSaeed, D.; Omar, S.F. Brain MRI analysis for Alzheimer’s disease diagnosis using CNN-based feature extraction and machine learning. Sensors 2022, 22, 2911. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Li, S.; Xiao, B.; Li, Y.; Wang, G.; Ma, X. Computer aided Alzheimer’s disease diagnosis by an unsupervised deep learning technology. Neurocomputing 2020, 392, 296–304. [Google Scholar] [CrossRef]
- Chakraborty, R.; Bouza, J.; Manton, J.; Vemuri, B.C. A deep neural network for manifold-valued data with applications to neuroimaging. In Proceedings of the Information Processing in Medical Imaging: 26th International Conference, IPMI 2019, Hong Kong, China, 2–7 June 2019; Proceedings 26. Springer: Berlin/Heidelberg, Germany, 2019; pp. 112–124. [Google Scholar]
- Mitchell-Heggs, R.; Prado, S.; Gava, G.P.; Go, M.A.; Schultz, S.R. Neural manifold analysis of brain circuit dynamics in health and disease. J. Comput. Neurosci. 2023, 51, 1–21. [Google Scholar] [CrossRef]
- Van der Haar, D.; Moustafa, A.; Warren, S.L.; Alashwal, H.; van Zyl, T. An Alzheimer’s disease category progression sub-grouping analysis using manifold learning on ADNI. Sci. Rep. 2023, 13, 10483. [Google Scholar] [CrossRef]
- Gunawardena, R.; Sarrigiannis, P.G.; Blackburn, D.J.; He, F. Kernel-based nonlinear manifold learning for EEG-based functional connectivity analysis and channel selection with application to Alzheimer’s disease. Neuroscience 2023, 523, 140–156. [Google Scholar] [CrossRef]
- Kim, S.; Kim, M.; Lee, J.E.; Park, B.y.; Park, H. Prognostic model for predicting Alzheimer’s disease conversion using functional connectome manifolds. Alzheimer’s Res. Ther. 2024, 16, 217. [Google Scholar] [CrossRef]
- Hua, J.C.; Kim, E.J.; He, F. Information Geometry Approach to Analyzing Simulated EEG Signals of Alzheimer’s Disease Patients and Healthy Control Subjects. In Proceedings of the 2023 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Istanbul, Turkey, 5–8 December 2023; pp. 2493–2500. [Google Scholar]
- Sarasua, I.; Lee, J.; Wachinger, C. Geometric deep learning on anatomical meshes for the prediction of Alzheimer’s disease. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; pp. 1356–1359. [Google Scholar]
- Huang, Z.; Mashour, G.A.; Hudetz, A.G. Functional geometry of the cortex encodes dimensions of consciousness. Nat. Commun. 2023, 14, 72. [Google Scholar] [CrossRef]
- Luppi, A.I.; Rosas, F.E.; Mediano, P.A.; Menon, D.K.; Stamatakis, E.A. Information decomposition and the informational architecture of the brain. Trends Cogn. Sci. 2024, 28, 352–368. [Google Scholar] [CrossRef] [PubMed]
- Almohammadi, A.; Wang, Y.K. Revealing brain connectivity: Graph embeddings for EEG representation learning and comparative analysis of structural and functional connectivity. Front. Neurosci. 2024, 17, 1288433. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.Y.; Zhu, J.D.; Tsai, S.J.; Yang, A.C. Exploring morphological similarity and randomness in Alzheimer’s disease using adjacent grey matter voxel-based structural analysis. Alzheimer’s Res. Ther. 2024, 16, 88. [Google Scholar] [CrossRef] [PubMed]
- Menardi, A.; Momi, D.; Vallesi, A.; Barabási, A.L.; Towlson, E.K.; Santarnecchi, E. Maximizing brain networks engagement via individualized connectome-wide target search. Brain Stimul. 2022, 15, 1418–1431. [Google Scholar] [CrossRef]
- Cao, J.; Yang, L.; Sarrigiannis, P.G.; Blackburn, D.; Zhao, Y. Dementia classification using a graph neural network on imaging of effective brain connectivity. Comput. Biol. Med. 2024, 168, 107701. [Google Scholar] [CrossRef]
- Gu, Y.; Peng, S.; Li, Y.; Gao, L.; Dong, Y. FC-HGNN: A heterogeneous graph neural network based on brain functional connectivity for mental disorder identification. Inf. Fusion 2025, 113, 102619. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X.; Chen, Y.; Yang, X. IFC-GNN: Combining interactions of functional connectivity with multimodal graph neural networks for ASD brain disorder analysis. Alex. Eng. J. 2024, 98, 44–55. [Google Scholar] [CrossRef]
- Zheng, K.; Yu, S.; Chen, B. Ci-gnn: A granger causality-inspired graph neural network for interpretable brain network-based psychiatric diagnosis. Neural Netw. 2024, 172, 106147. [Google Scholar] [CrossRef]
- Zhang, H.; Guilleminot, J.; Gomez, L.J. Stochastic modeling of geometrical uncertainties on complex domains, with application to additive manufacturing and brain interface geometries. Comput. Methods Appl. Mech. Eng. 2021, 385, 114014. [Google Scholar] [CrossRef]
- Chen, G.; Nash, T.A.; Cole, K.M.; Kohn, P.D.; Wei, S.M.; Gregory, M.D.; Eisenberg, D.P.; Cox, R.W.; Berman, K.F.; Kippenhan, J.S. Beyond linearity in neuroimaging: Capturing nonlinear relationships with application to longitudinal studies. NeuroImage 2021, 233, 117891. [Google Scholar] [CrossRef]
- Lei, B.; Cheng, N.; Frangi, A.F.; Tan, E.L.; Cao, J.; Yang, P.; Elazab, A.; Du, J.; Xu, Y.; Wang, T. Self-calibrated brain network estimation and joint non-convex multi-task learning for identification of early Alzheimer’s disease. Med Image Anal. 2020, 61, 101652. [Google Scholar] [CrossRef] [PubMed]
- Korhonen, O.; Zanin, M.; Papo, D. Principles and open questions in functional brain network reconstruction. Hum. Brain Mapp. 2021, 42, 3680–3711. [Google Scholar] [CrossRef] [PubMed]
- Pines, A.R.; Larsen, B.; Cui, Z.; Sydnor, V.J.; Bertolero, M.A.; Adebimpe, A.; Alexander-Bloch, A.F.; Davatzikos, C.; Fair, D.A.; Gur, R.C.; et al. Dissociable multi-scale patterns of development in personalized brain networks. Nat. Commun. 2022, 13, 2647. [Google Scholar] [CrossRef]
- Zhang, Y.; Tiňo, P.; Leonardis, A.; Tang, K. A survey on neural network interpretability. IEEE Trans. Emerg. Top. Comput. Intell. 2021, 5, 726–742. [Google Scholar] [CrossRef]
- Maqsood, S.; Damaševičius, R.; Maskeliūnas, R. Multi-modal brain tumor detection using deep neural network and multiclass SVM. Medicina 2022, 58, 1090. [Google Scholar] [CrossRef]
- Boehm, K.M.; Khosravi, P.; Vanguri, R.; Gao, J.; Shah, S.P. Harnessing multimodal data integration to advance precision oncology. Nat. Rev. Cancer 2022, 22, 114–126. [Google Scholar] [CrossRef]
- Smucny, J.; Shi, G.; Davidson, I. Deep learning in neuroimaging: Overcoming challenges with emerging approaches. Front. Psychiatry 2022, 13, 912600. [Google Scholar] [CrossRef]
- Abdi, H.; Williams, L.J. Principal component analysis. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Greenacre, M.; Groenen, P.J.; Hastie, T.; d’Enza, A.I.; Markos, A.; Tuzhilina, E. Principal component analysis. Nat. Rev. Methods Prim. 2022, 2, 100. [Google Scholar] [CrossRef]
- Borguet, S.; Léonard, O. The Fisher information matrix as a relevant tool for sensor selection in engine health monitoring. Int. J. Rotating Mach. 2008, 2008, 10. [Google Scholar] [CrossRef]
- Frieden, R.; Gatenby, R.A. Exploratory Data Analysis Using Fisher Information; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Amari, S.I. Information geometry. Jpn. J. Math. 2021, 16, 1–48. [Google Scholar] [CrossRef]
- Ay, N.; Jost, J.; Vân Lê, H.; Schwachhöfer, L. Information Geometry; Springer: Berlin/Heidelberg, Germany, 2017; Volume 64. [Google Scholar]
- De Lathauwer, L.; De Moor, B.; Vandewalle, J. Computation of the canonical decomposition by means of a simultaneous generalized Schur decomposition. SIAM J. Matrix Anal. Appl. 2004, 26, 295–327. [Google Scholar] [CrossRef]
- Bhatti, U.A.; Tang, H.; Wu, G.; Marjan, S.; Hussain, A. Deep learning with graph convolutional networks: An overview and latest applications in computational intelligence. Int. J. Intell. Syst. 2023, 2023, 8342104. [Google Scholar] [CrossRef]
- Jin, D.; Yu, Z.; Huo, C.; Wang, R.; Wang, X.; He, D.; Han, J. Universal graph convolutional networks. Adv. Neural Inf. Process. Syst. 2021, 34, 10654–10664. [Google Scholar]
- Vrahatis, A.G.; Lazaros, K.; Kotsiantis, S. Graph Attention Networks: A Comprehensive Review of Methods and Applications. Future Internet 2024, 16, 318. [Google Scholar] [CrossRef]
- Liu, Z.; Zhou, J. Graph attention networks. In Introduction to Graph Neural Networks; Springer: Berlin/Heidelberg, Germany, 2020; pp. 39–41. [Google Scholar]
- Bhatkar, S.; Gosavi, P.; Shelke, V.; Kenny, J. Link Prediction using GraphSAGE. In Proceedings of the 2023 International Conference on Advanced Computing Technologies and Applications (ICACTA), Mumbai, India, 6–7 October 2023; pp. 1–5. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
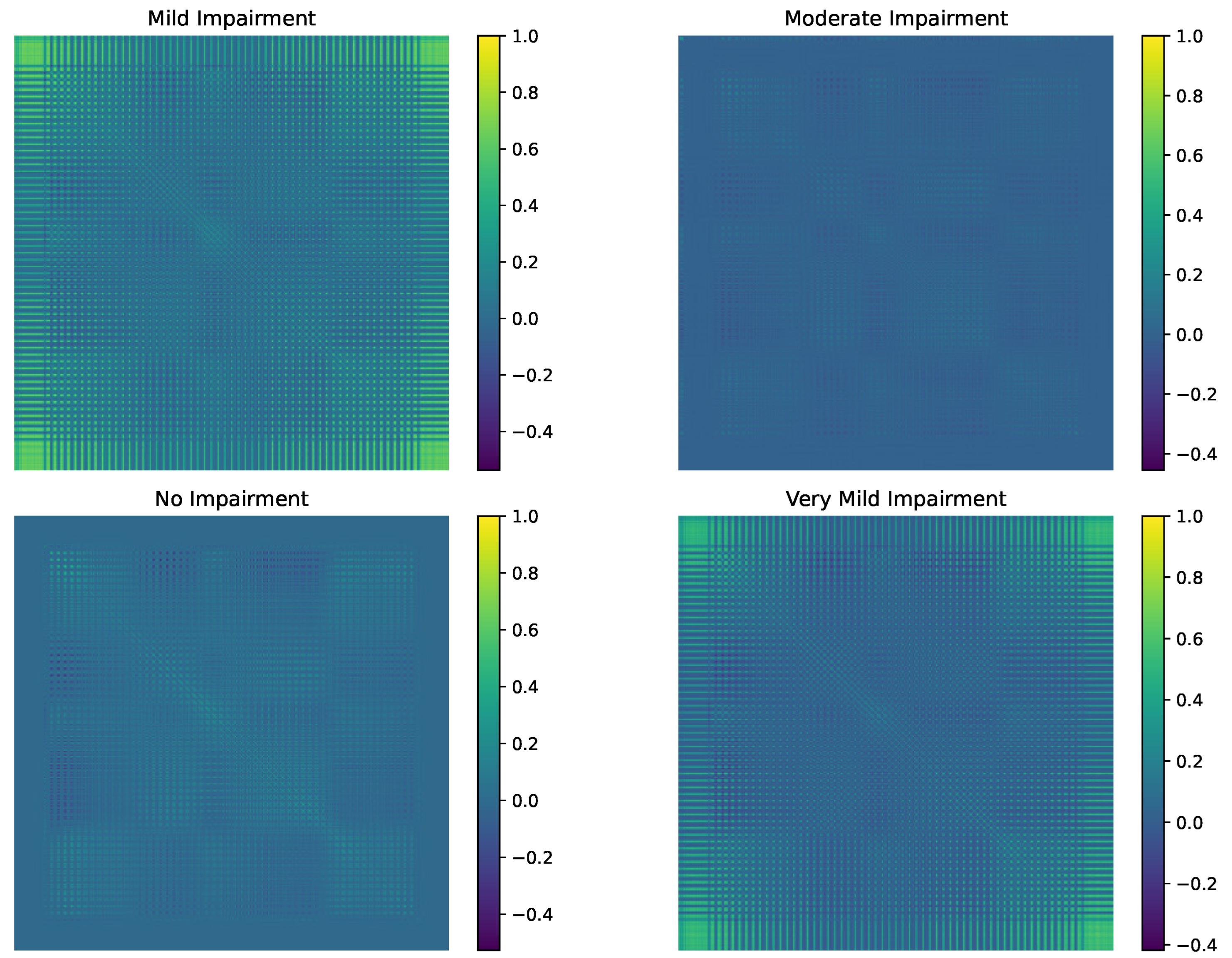
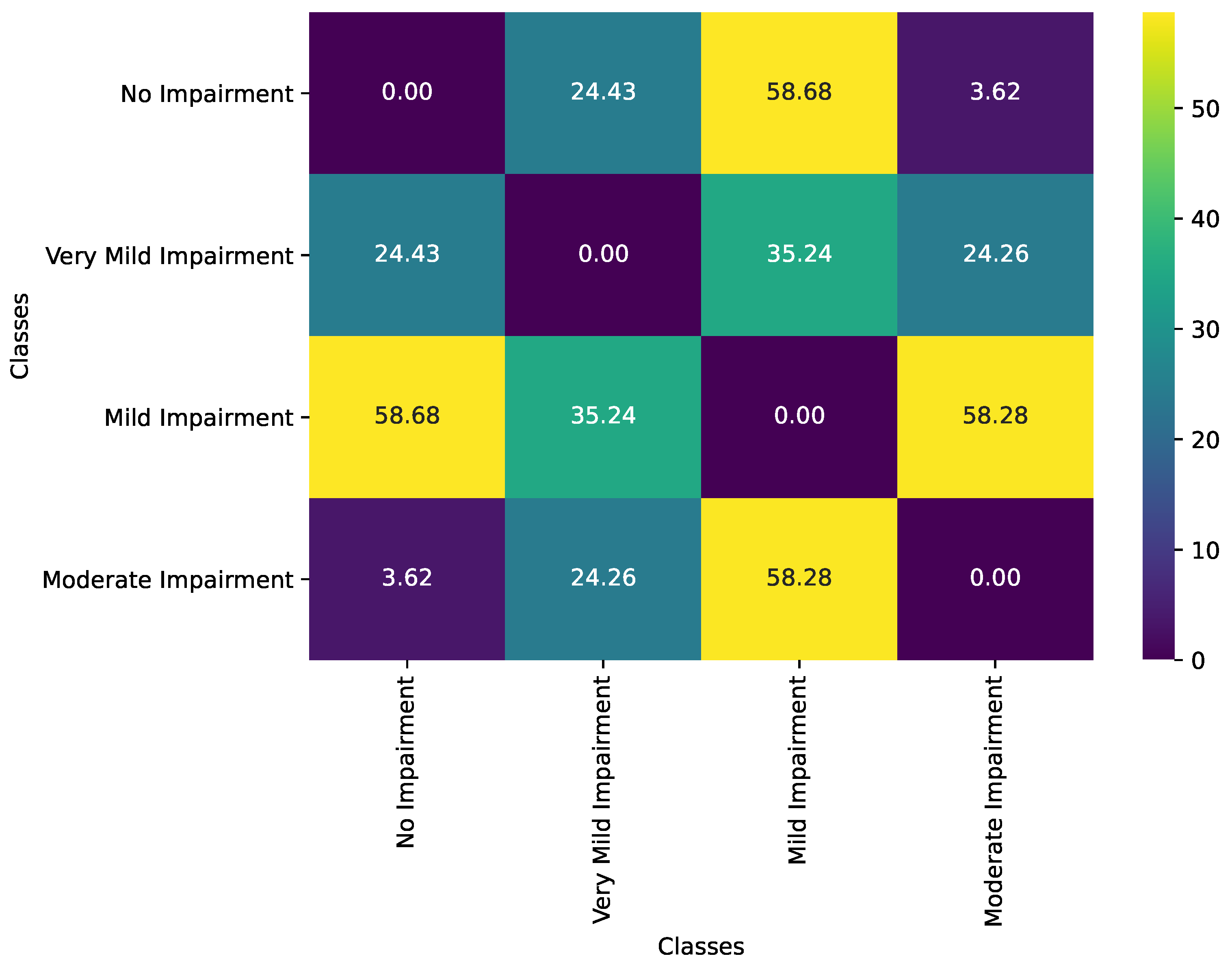
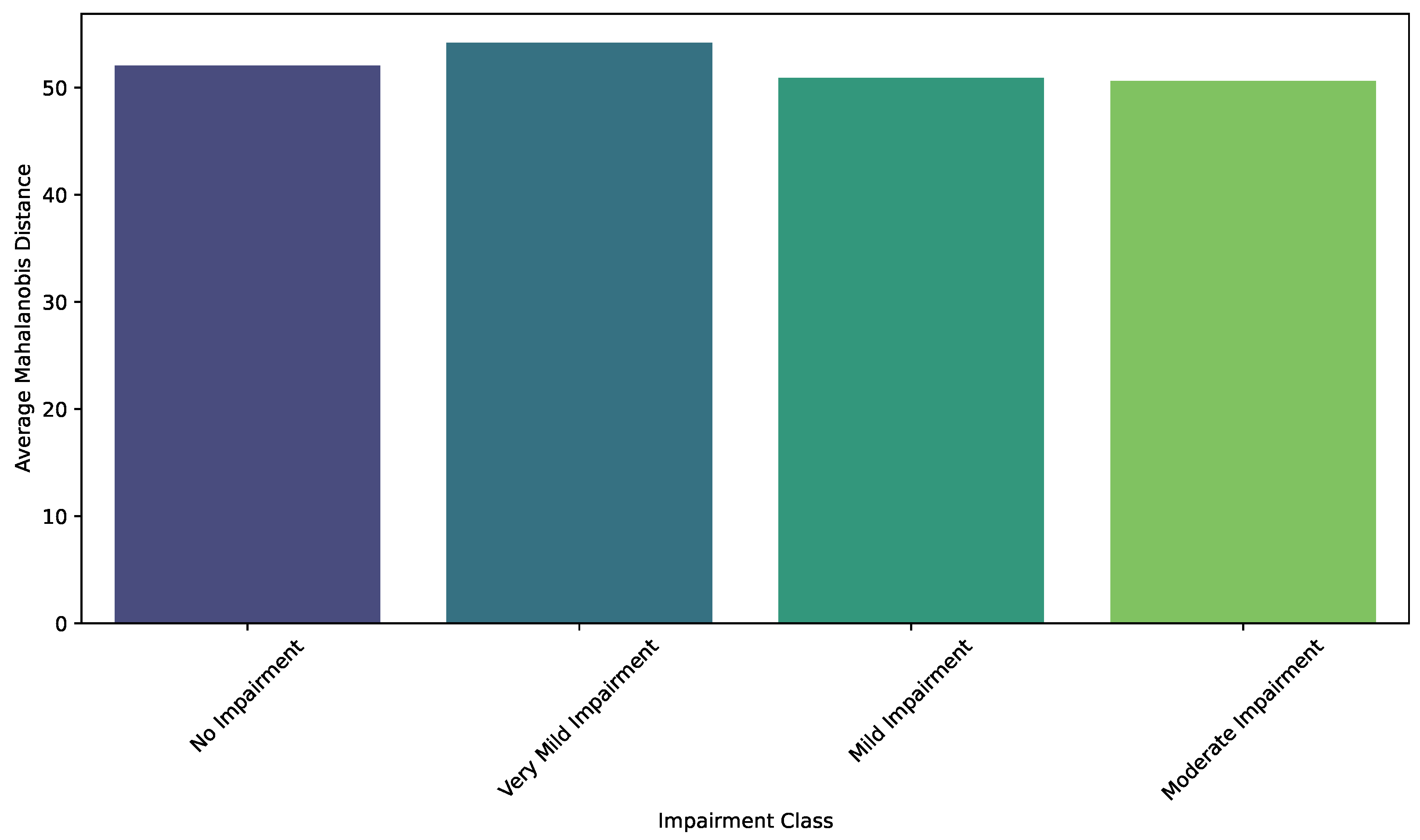



| Class Pair | Observed Geodesic Distance | p-Value |
|---|---|---|
| No Impairment vs. Very Mild Impairment | 24.43 | 0.1020 |
| No Impairment vs. Mild Impairment | 58.68 | 0.0000 |
| No Impairment vs. Moderate Impairment | 3.62 | 0.8380 |
| Very Mild Impairment vs. Mild Impairment | 35.24 | 0.0020 |
| Very Mild Impairment vs. Moderate Impairment | 24.26 | 0.1010 |
| Mild Impairment vs. Moderate Impairment | 58.28 | 0.0000 |
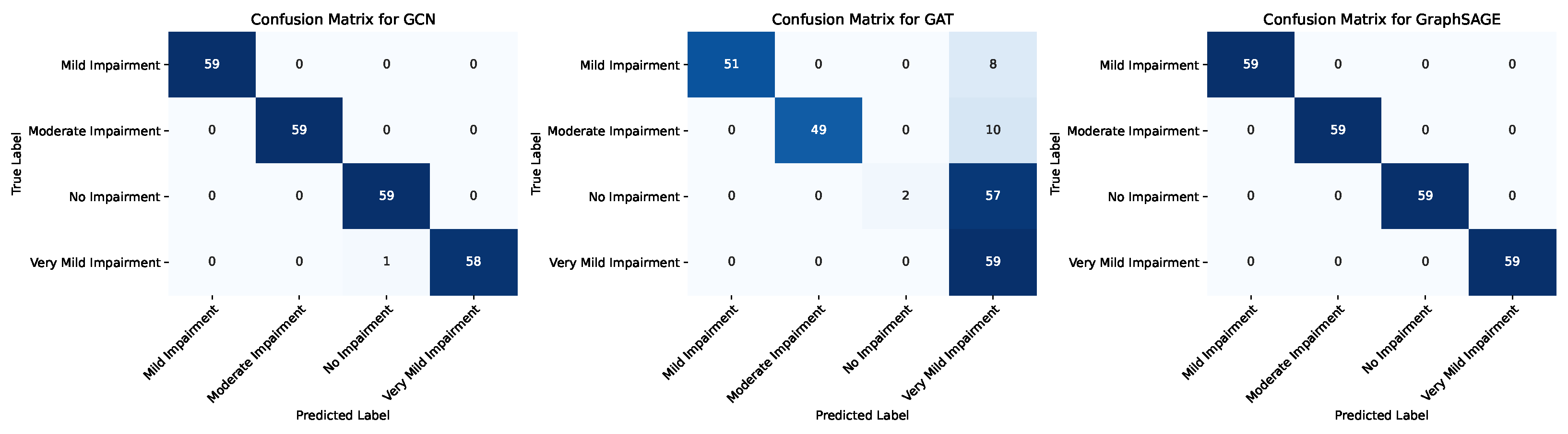
| Model | Class | Precision | Recall | F1-Score | Support |
|---|---|---|---|---|---|
| GCN | Mild Impairment | 1.0 | 1.0 | 1.0 | 39.0 |
| Moderate Impairment | 1.0 | 1.0 | 1.0 | 39.0 | |
| No Impairment | 1.0 | 1.0 | 1.0 | 40.0 | |
| Very Mild Impairment | 1.0 | 1.0 | 1.0 | 38.0 | |
| Accuracy | 1.0 | ||||
| Macro Avg | 1.0 | 1.0 | 1.0 | 156.0 | |
| Weighted avg | 1.0 | 1.0 | 1.0 | 156.0 | |
| GAT | Mild Impairment | 0.8421 | 0.4102 | 0.5517 | 39.0 |
| Moderate Impairment | 1.0 | 0.3077 | 0.4705 | 39.0 | |
| No Impairment | 0.4 | 1.0 | 0.5714 | 40.0 | |
| Very Mild Impairment | 1.0 | 0.6579 | 0.7936 | 38.0 | |
| Accuracy | 0.5961 | ||||
| Macro Avg | 0.8105 | 0.5939 | 0.5968 | 156.0 | |
| Weighted avg | 0.8066 | 0.5961 | 0.5954 | 156.0 | |
| GraphSAGE | Mild Impairment | 1.0 | 1.0 | 1.0 | 39.0 |
| Moderate Impairment | 1.0 | 1.0 | 1.0 | 39.0 | |
| No Impairment | 1.0 | 1.0 | 1.0 | 40.0 | |
| Very Mild Impairment | 1.0 | 1.0 | 1.0 | 38.0 | |
| Accuracy | 1.0 | ||||
| Macro Avg | 1.0 | 1.0 | 1.0 | 156.0 | |
| Weighted avg | 1.0 | 1.0 | 1.0 | 156.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akgüller, Ö.; Balcı, M.A.; Cioca, G. Information Geometry and Manifold Learning: A Novel Framework for Analyzing Alzheimer’s Disease MRI Data. Diagnostics 2025, 15, 153. https://doi.org/10.3390/diagnostics15020153
Akgüller Ö, Balcı MA, Cioca G. Information Geometry and Manifold Learning: A Novel Framework for Analyzing Alzheimer’s Disease MRI Data. Diagnostics. 2025; 15(2):153. https://doi.org/10.3390/diagnostics15020153
Chicago/Turabian StyleAkgüller, Ömer, Mehmet Ali Balcı, and Gabriela Cioca. 2025. "Information Geometry and Manifold Learning: A Novel Framework for Analyzing Alzheimer’s Disease MRI Data" Diagnostics 15, no. 2: 153. https://doi.org/10.3390/diagnostics15020153
APA StyleAkgüller, Ö., Balcı, M. A., & Cioca, G. (2025). Information Geometry and Manifold Learning: A Novel Framework for Analyzing Alzheimer’s Disease MRI Data. Diagnostics, 15(2), 153. https://doi.org/10.3390/diagnostics15020153





