Toll-like Receptor Gene Polymorphisms as Predictive Biomarkers for Response to Infliximab in Japanese Patients with Crohn’s Disease
Abstract
1. Introduction
- Th17/IL-17-signaling-mediated immune response in the pathogenesis of CD
- 2.
- Toll-like receptor (TLR) signaling shapes the Th17/IL-17 axis responses in CD
2. Materials and Methods
3. Results
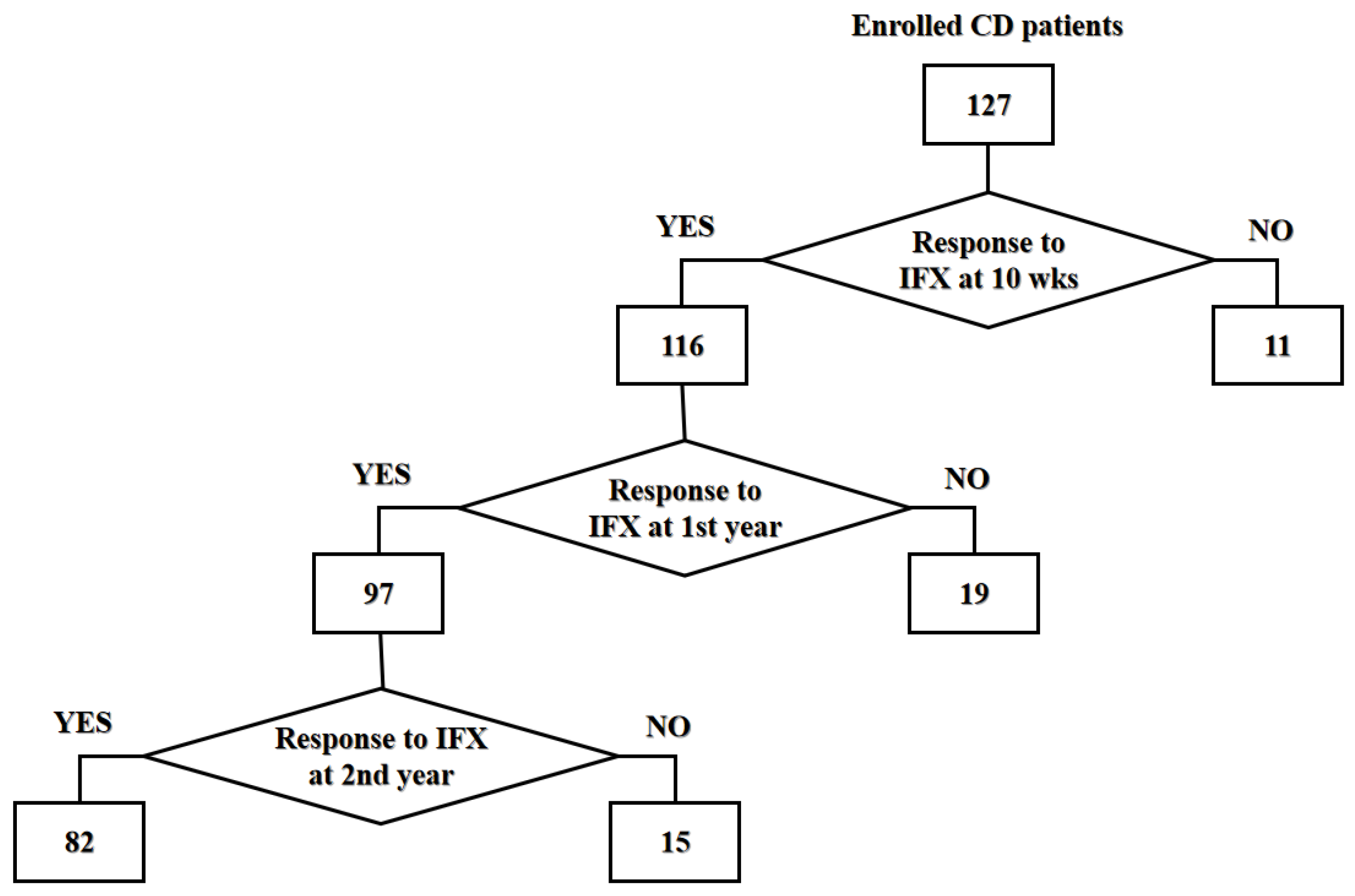
3.1. Clinical Characteristics of Study Population
3.2. Polymorphisms Associated with the Response to IFX at the 10-Week Treatment
3.3. Interaction of the Genetic Factors in Response to IFX at the 10-Week Treatment
3.4. Verification of Genetic Test to Predict the Response to IFX at the 10-Week Treatment
3.5. Polymorphisms Associated with the IFX Response at the 1-Year Treatment
3.6. Verification of Genetic Testing for Predicting the IFX Response at the 1-Year Treatment
3.7. Polymorphisms Associated with the IFX Response at the 2-Year Treatment
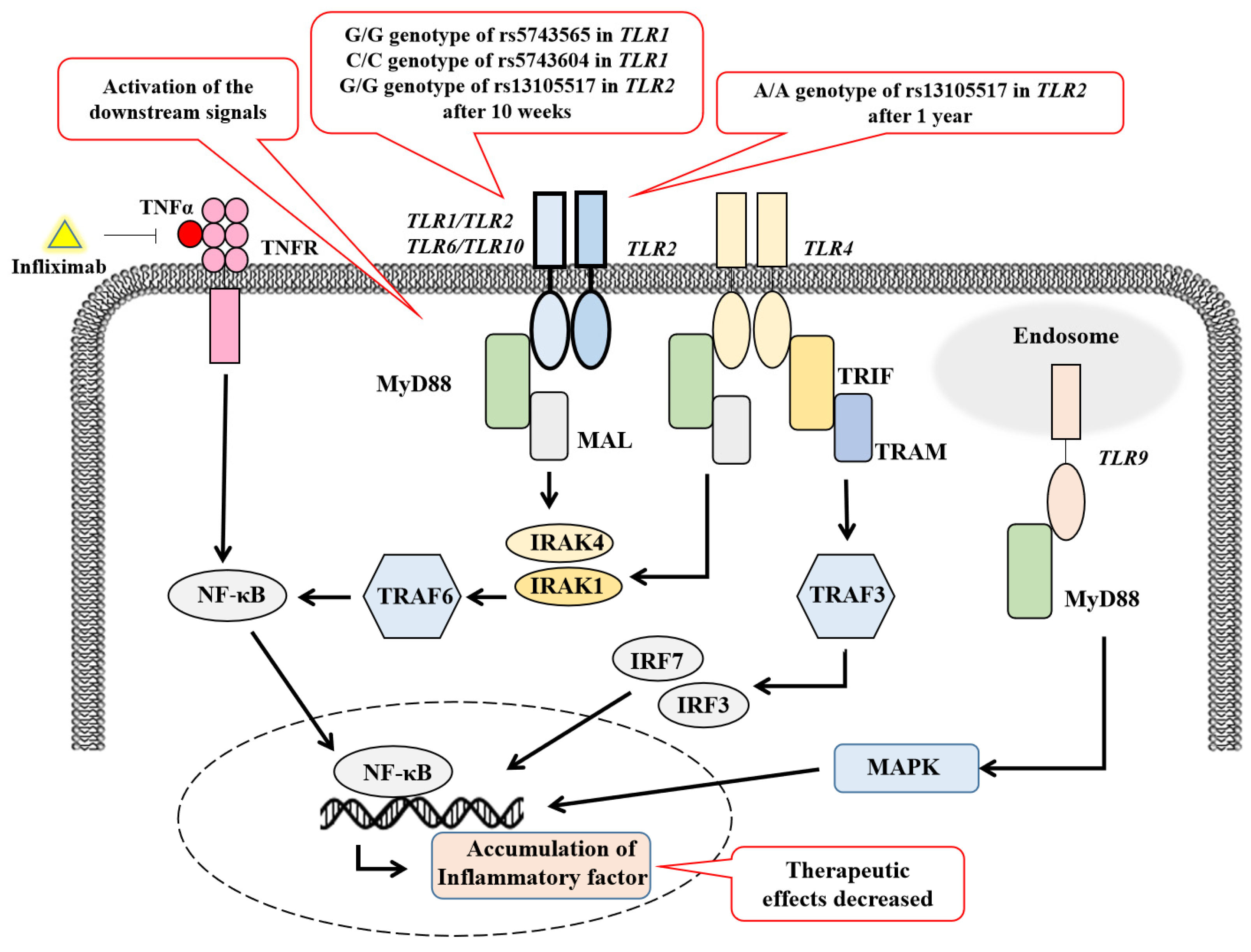
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Croft, M.; Salek-Ardakani, S.; Ware, C.F. Targeting the TNF and TNFR superfamilies in autoimmune disease and cancer. Nat. Rev. Drug Discov. 2024, 23, 939–961. [Google Scholar] [CrossRef] [PubMed]
- Leone, G.M.; Mangano, K.; Petralia, M.C.; Nicoletti, F.; Fagone, P. Past, Present and (Foreseeable) Future of Biological Anti-TNF Alpha Therapy. J. Clin. Med. 2023, 12, 1630. [Google Scholar] [CrossRef] [PubMed]
- Ben-Horin, S.; Chowers, Y. Review article: Loss of response to anti-TNF treatments in Crohn’s disease. Aliment. Pharmacol. Ther. 2011, 33, 987–995. [Google Scholar] [CrossRef]
- Navajas Hernández, P.; Mouhtar El Halabi, S.; González Parra, A.C.; Valdés Delgado, T.; Maldonado Pérez, B.; Castro Laria, L.; Charpentier, C.; Argüelles-Arias, F. Carriage of the HLA-DQA1⋆05 haplotype is associated with a higher risk of infratherapeutic drug concentration and higher immunogenicity in patients undergoing treatment with anti-TNF for inflammatory bowel disease. Therap. Adv. Gastroenterol. 2024, 17, 17562848241278145. [Google Scholar] [CrossRef]
- Srinivasan, A.; De Cruz, P.; Sam, M.; Toong, C.; van Langenberg, D.R. Dose intensification strategy influences infliximab pharmacokinetics but not clinical response after the same number of doses. J. Gastroenterol. Hepatol. 2023, 38, 724–732. [Google Scholar] [CrossRef]
- Qiu, P.; Ishimoto, T.; Fu, L.; Zhang, J.; Zhang, Z.; Liu, Y. The Gut Microbiota in Inflammatory Bowel Disease. Front. Cell. Infect. Microbiol. 2022, 12, 733992. [Google Scholar] [CrossRef] [PubMed]
- Saez, A.; Herrero-Fernandez, B.; Gomez-Bris, R.; Sánchez-Martinez, H.; Gonzalez-Granado, J.M. Pathophysiology of Inflammatory Bowel Disease: Innate Immune System. Int. J. Mol. Sci. 2023, 24, 1526. [Google Scholar] [CrossRef]
- Jarmakiewicz-Czaja, S.; Zielińska, M.; Sokal, A.; Filip, R. Genetic and Epigenetic Etiology of Inflammatory Bowel Disease: An Update. Genes 2022, 13, 2388. [Google Scholar] [CrossRef]
- Singh, N.; Bernstein, C.N. Environmental risk factors for inflammatory bowel disease. United Eur. Gastroenterol. J. 2022, 10, 1047–1053. [Google Scholar] [CrossRef]
- Abraham, C.; Cho, J.H. Inflammatory bowel disease. N. Engl. J. Med. 2009, 361, 2066–2078. [Google Scholar] [CrossRef]
- Liu, Z.J.; Yadav, P.K.; Su, J.L.; Wang, J.S.; Fei, K. Potential role of Th17 cells in the pathogenesis of inflammatory bowel disease. World J. Gastroenterol. 2009, 15, 5784–5788. [Google Scholar] [CrossRef]
- Blaschitz, C.; Raffatellu, M. Th17 cytokines and the gut mucosal barrier. J. Clin. Immunol. 2010, 30, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Hall, J.A.; Kroehling, L.; Wu, L.; Najar, T.; Nguyen, H.H.; Lin, W.Y.; Yeung, S.T.; Silva, H.M.; Li, D.; et al. Serum Amyloid A Proteins Induce Pathogenic Th17 Cells and Promote Inflammatory Disease. Cell 2020, 180, 79–91.e16. [Google Scholar] [CrossRef] [PubMed]
- Rafa, H.; Saoula, H.; Belkhelfa, M.; Medjeber, O.; Soufli, I.; Toumi, R.; de Launoit, Y.; Moralès, O.; Nakmouche, M.; Delhem, N.; et al. IL-23/IL-17A axis correlates with the nitric oxide pathway in inflammatory bowel disease: Immunomodulatory effect of retinoic acid. J. Interferon Cytokine Res. 2013, 33, 355–368. [Google Scholar] [CrossRef]
- Ghadimi, D.; Helwig, U.; Schrezenmeir, J.; Heller, K.J.; de Vrese, M. Epigenetic imprinting by commensal probiotics inhibits the IL-23/IL-17 axis in an in vitro model of the intestinal mucosal immune system. J. Leukoc. Biol. 2012, 92, 895–911. [Google Scholar] [CrossRef] [PubMed]
- Majumder, S.; Amatya, N.; Revu, S.; Jawale, C.V.; Wu, D.; Rittenhouse, N.; Menk, A.; Kupul, S.; Du, F.; Raphael, I.; et al. IL-17 metabolically reprograms activated fibroblastic reticular cells for proliferation and survival. Nat. Immunol. 2019, 20, 534–545. [Google Scholar] [CrossRef]
- Urabe, S.; Isomoto, H.; Ishida, T.; Maeda, K.; Inamine, T.; Kondo, S.; Higuchi, N.; Sato, K.; Uehara, R.; Yajima, H.; et al. Genetic Polymorphisms of IL-17F and TRAF3IP2 Could Be Predictive Factors of the Long-Term Effect of Infliximab against Crohn’s Disease. BioMed Res. Int. 2015, 2015, 416838. [Google Scholar] [CrossRef]
- Shen, Z.; Luo, W.; Tan, B.; Nie, K.; Deng, M.; Wu, S.; Xiao, M.; Wu, X.; Meng, X.; Tong, T.; et al. Roseburia intestinalis stimulates TLR5-dependent intestinal immunity against Crohn’s disease. EBioMedicine 2022, 85, 104285. [Google Scholar] [CrossRef]
- Liang, H.; Zhang, L.; Hoden, B.; Qu, B.; Derubeis, D.; Song, X.; Zhang, D. Delineating the Role of Toll-Like Receptors in Inflammatory Bowel Disease. Methods Mol. Biol. 2023, 2700, 221–228. [Google Scholar] [CrossRef]
- Abreu, M.T. Toll-like receptor signalling in the intestinal epithelium: How bacterial recognition shapes intestinal function. Nat. Rev. Immunol. 2010, 10, 131–144. [Google Scholar] [CrossRef]
- Hasan, U.; Chaffois, C.; Gaillard, C.; Saulnier, V.; Merck, E.; Tancredi, S.; Guiet, C.; Brière, F.; Vlach, J.; Lebecque, S.; et al. Human TLR10 is a functional receptor, expressed by B cells and plasmacytoid dendritic cells, which activates gene transcription through MyD88. J. Immunol. 2005, 174, 2942–2950. [Google Scholar] [CrossRef] [PubMed]
- Ah Kioon, M.D.; Laurent, P.; Chaudhary, V.; Du, Y.; Crow, M.K.; Barrat, F.J. Modulation of plasmacytoid dendritic cells response in inflammation and autoimmunity. Immunol. Rev. 2024, 323, 241–256. [Google Scholar] [CrossRef]
- Candelli, M.; Franza, L.; Pignataro, G.; Ojetti, V.; Covino, M.; Piccioni, A.; Gasbarrini, A.; Franceschi, F. Interaction between Lipopolysaccharide and Gut Microbiota in Inflammatory Bowel Diseases. Int. J. Mol. Sci. 2021, 22, 6242. [Google Scholar] [CrossRef]
- Kaur, A.; Kaushik, D.; Piplani, S.; Mehta, S.K.; Petrovsky, N.; Salunke, D.B. TLR2 Agonistic Small Molecules: Detailed Structure-Activity Relationship, Applications, and Future Prospects. J. Med. Chem. 2021, 64, 233–278. [Google Scholar] [CrossRef]
- Ito, K.; Kariya, M.; Yasui, K.; Takahashi, Y.; Takakura, Y. Development of Hydrophobic Interaction-based DNA Supramolecules as Efficient Delivery Carriers of CpG DNA to Immune cells. J. Pharm. Sci. 2022, 111, 1133–1141. [Google Scholar] [CrossRef]
- Marongiu, L.; Gornati, L.; Artuso, I.; Zanoni, I.; Granucci, F. Below the surface: The inner lives of TLR4 and TLR9. J. Leukoc. Biol. 2019, 106, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Hajjar, A.M.; O’Mahony, D.S.; Ozinsky, A.; Underhill, D.M.; Aderem, A.; Klebanoff, S.J.; Wilson, C.B. Cutting edge: Functional interactions between toll-like receptor (TLR) 2 and TLR1 or TLR6 in response to phenol-soluble modulin. J. Immunol. 2001, 166, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Coutinho-Wolino, K.S.; Almeida, P.P.; Mafra, D.; Stockler-Pinto, M.B. Bioactive compounds modulating Toll-like 4 receptor (TLR4)-mediated inflammation: Pathways involved and future perspectives. Nutr. Res. 2022, 107, 96–116. [Google Scholar] [CrossRef]
- Fore, F.; Budipranama, M.; Destiawan, R.A. TLR10 and Its Role in Immunity. Handb. Exp. Pharmacol. 2022, 276, 161–174. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, S.; Wang, Y. Diannexin alleviates myocardial ischemia-reperfusion injury by orchestrating cardiomyocyte oxidative damage, macrophage polarization and fibrotic process by TLR4-NF-kB-mediated inactivation of NLRP3 inflammasome. Int. Immunopharmacol. 2024, 130, 111668. [Google Scholar] [CrossRef]
- Zeng, F.; Li, Y.; Zhang, X.; Shen, L.; Zhao, X.; Beta, T.; Li, B.; Chen, R.; Huang, W. Immune regulation and inflammation inhibition of Arctium lappa L. polysaccharides by TLR4/NF-κB signaling pathway in cells. Int. J. Biol. Macromol. 2024, 254 Pt 2, 127700. [Google Scholar] [CrossRef] [PubMed]
- Capitani, M.; Al-Shaibi, A.A.; Pandey, S.; Gartner, L.; Taylor, H.; Hubrack, S.Z.; Agrebi, N.; Al-Mohannadi, M.J.; Al Kaabi, S.; Vogl, T.; et al. Biallelic TLR4 deficiency in humans. J. Allergy Clin. Immunol. 2023, 151, 783–790.e5. [Google Scholar] [CrossRef] [PubMed]
- Emele, R.E.; Salim, M.A.; Polat, F.; Diler, S.B. Investigation of TLR2 (-196 to -174del) and TLR9 (T-1486C) Gene Polymorphisms Association with Inflammatory Bowel Diseases. Asian Pac. J. Cancer Prev. 2024, 25, 2003–2010. [Google Scholar] [CrossRef]
- Azzam, N.; Nounou, H.; Alharbi, O.; Aljebreen, A.; Shalaby, M. CARD15/NOD2, CD14 and toll-like 4 receptor gene polymorphisms in Saudi patients with Crohn’s Disease. Int. J. Mol. Sci. 2012, 13, 4268–4280. [Google Scholar] [CrossRef]
- Kircheis, R.; Planz, O. Special Issue “The Role of Toll-Like Receptors (TLRs) in Infection and Inflammation 2.0”. Int. J. Mol. Sci. 2024, 25, 9709. [Google Scholar] [CrossRef] [PubMed]
- Bank, S.; Andersen, P.S.; Burisch, J.; Pedersen, N.; Roug, S.; Galsgaard, J.; Turino, S.Y.; Brodersen, J.B.; Rashid, S.; Rasmussen, B.K.; et al. Associations between functional polymorphisms in the NFκB signaling pathway and response to anti-TNF treatment in Danish patients with inflammatory bowel disease. Pharmacogenom. J. 2014, 14, 526–534. [Google Scholar] [CrossRef]
- Zhang, S.; Hu, Z.; Tanji, H.; Jiang, S.; Das, N.; Li, J.; Sakaniwa, K.; Jin, J.; Bian, Y.; Ohto, U.; et al. Small-molecule inhibition of TLR8 through stabilization of its resting state. Nat. Chem. Biol. 2018, 14, 58–64. [Google Scholar] [CrossRef]
- McKernan, D.P. Pattern recognition receptors as potential drug targets in inflammatory disorders. Adv. Protein Chem. Struct. Biol. 2020, 119, 65–109. [Google Scholar] [CrossRef]
- Limou, S.; Taverner, A.M.; Winkler, C.A. Ferret: A user-friendly Java tool to extract data from the 1000 Genomes Project. Bioinformatics 2016, 32, 2224–2226. [Google Scholar] [CrossRef][Green Version]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Ward, L.D.; Kellis, M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012, 40, D930–D934. [Google Scholar] [CrossRef] [PubMed]
- Gaiani, F.; Rotoli, B.M.; Ferrari, F.; Barilli, A.; Visigalli, R.; Carra, M.C.; de’Angelis, G.L.; de’Angelis, N.; Dall’Asta, V. Monocytes from infliximab-resistant patients with Crohn’s disease exhibit a disordered cytokine profile. Sci. Rep. 2020, 10, 12238. [Google Scholar] [CrossRef] [PubMed]
- Sazonovs, A.; Stevens, C.R.; Venkataraman, G.R.; Yuan, K.; Avila, B.; Abreu, M.T.; Ahmad, T.; Allez, M.; Ananthakrishnan, A.N.; Atzmon, G.; et al. Large-scale sequencing identifies multiple genes and rare variants associated with Crohn’s disease susceptibility. Nat. Genet. 2022, 54, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Hawn, T.R.; Misch, E.A.; Dunstan, S.J.; Thwaites, G.E.; Lan, N.T.; Quy, H.T.; Chau, T.T.; Rodrigues, S.; Nachman, A.; Janer, M.; et al. A common human TLR1 polymorphism regulates the innate immune response to lipopeptides. Eur. J. Immunol. 2007, 37, 2280–2289. [Google Scholar] [CrossRef]
- Arijs, I.; De Hertogh, G.; Lemaire, K.; Quintens, R.; Van Lommel, L.; Van Steen, K.; Leemans, P.; Cleynen, I.; Van Assche, G.; Vermeire, S.; et al. Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PLoS ONE 2009, 4, e7984. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]
| Characteristics | Groups | p-Value | |
|---|---|---|---|
| Responders | Non-Responders | ||
| 10 weeks (n = 127) | |||
| Number (%) | 116 (91.3) | 11 (8.7) | N/A |
| Age, mean ± SD (years) | 35.1 ± 12.3 | 35.2 ± 7.1 | 0.662 |
| Male/female (%) | 67/49 (57.8/42.2) | 10/1 (90.9/9.1) | 0.049 |
| 1 year (n = 116) | |||
| Number (%) | 97 (83.6) | 19 (16.4) | N/A |
| Age, mean ± SD (years) | 34.7 ± 12.8 | 37.3 ± 9.0 | 0.234 |
| Male/female (%) | 56/41 (57.7/42.3) | 11/8 (57.9/42.1) | 0.990 |
| 2 years (n = 97) | |||
| Number (%) | 82 (84.5) | 15 (15.5) | N/A |
| Age, mean ± SD (years) | 35.0 ± 13.3 | 32.8 ± 9.5 | 0.787 |
| Male/female (%) | 48/34 (58.5/41.5) | 8/7 (53.3/46.7) | 0.708 |
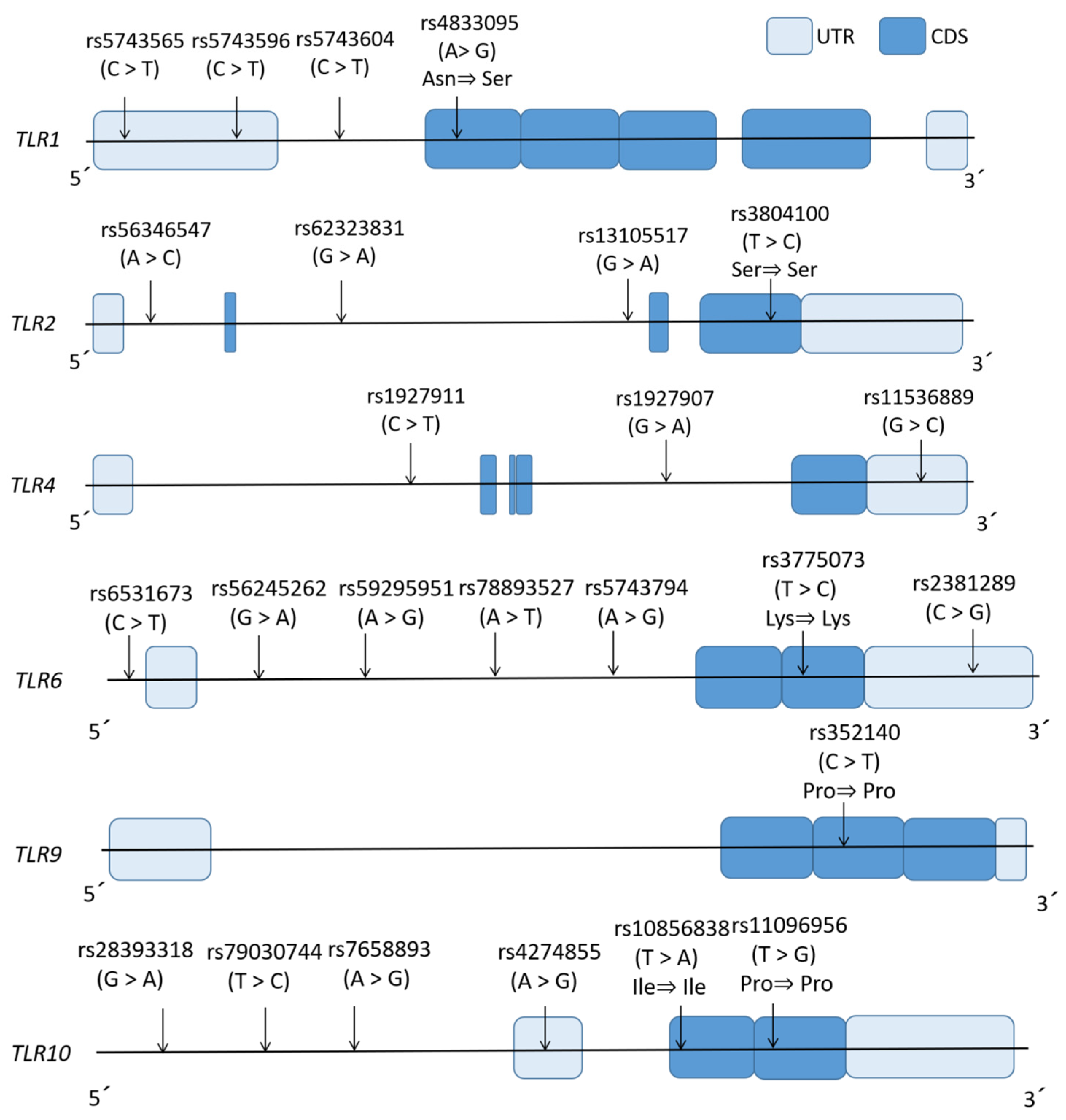
| Gene | Tag SNP | Major>Minor | dbSNP Function | Locus | Sequence of Primer (5’ to 3’) | Annealing Temperature (°C) | Cycle Number | Analytic Method (Restriction Enzyme) | |
|---|---|---|---|---|---|---|---|---|---|
| Forward | Reverse | ||||||||
| TLR1 | rs5743565 | A>G | 5’UTR | 4p14 | CCTCTGGGAATAACACCTCGT | GAAGGCCCCAGAGAGAAAAA | 58 | 30 | PCR-RFLP (HinF I) |
| rs5743596 | C>T | 5’UTR | AAATTTCCGGGTCTTTCAGC | ATCTGGGTTTTGGAGCCTTC | 55 | 50 | HRM with non-labeled probe | ||
| rs5743604 | C>T | Intronic | GAGCAGTCCCAATACCACCA | AGGAGAAGGCCTTGTGACAGT | 55 | 30 | PCR-RFLP (BmgB I) | ||
| rs4833095 | C>T | Missense | AAACCAGCTGGAGGATCCTAAT | CATTGTGTTCCCCACAAACA | 57 | 50 | HRM with non-labeled probe | ||
| TLR2 | rs56346547 | A>C | Intronic | GCGGACTTTCCCTTTTGCTT | GCTTCGCTGGTGTCCACATT | 57 | 30 | PCR-RFLP (BsmF I) | |
| rs62323831 | A>G | Intronic | 4q31.3 | GGCCAAATCTGGGGCTAGTT | CATATGCGAATCCCGACTCC | 58 | 50 | HRM with non-labeled probe | |
| rs13105517 | A>G | Intronic | 3p21.2 | ACTTCCTGGATTGCGGTGAT | AAACTCGAGGCAGACCAAGG | 60 | 30 | PCR-RFLP (Bts I) | |
| rs3804100 | C>T | Synonymous | TGCCTGGCCCTCTCTACAAA | GAGTTGCGGCAAATTCAAAG | 55 | 30 | PCR-RFLP (HpyCH4III) | ||
| TLR4 | rs1927911 | C>T | Intronic | 9q33.1 | TCATGGCCCAGATTTTGACA | AGCTGGCTTCTGCAAGGAAT | 55 | 30 | PCR-RFLP (Eco130 I) |
| rs1927907 | A>G | Intronic | GGCTGCCTGGTCTATCACAA | TTCAACCCTTGCTGCTTTCTC | 56 | 30 | PCR-RFLP (Hph I) | ||
| rs11536889 | C>G | 3’UTR | TTTGGGCTAGAGGCAGGAAG | TTTCCATTCCCTCCAGCAGT | 60 | 30 | PCR-RFLP (HpyCH4III) | ||
| TLR6 | rs6531673 | C>T | Intronic | 4p14 | GCATGATACTGCAGAAAGCAAGTACC | GCAACTCAAGTCATCCAGTGAA | 59 | 50 | HRM with non-labeled probe |
| rs56245262 | A>T | Intronic | ATCCACTTGGCCACTGAAAA | GGAGAATGGAGTGTGGCAGT | 56 | 30 | PCR-RFLP (Bcu I) | ||
| rs59295951 | A>G | Intronic | TCTTTTATCCTTCCCCACCA | GGTCACTGGTTGCAGCAGAT | 53 | 50 | HRM with non-labeled probe | ||
| rs78893527 | C>G | Intronic | CAACCCTGAATCTCCACACC | CTCTGGGGAATGCACACTTT | 53 | 50 | HRM with non-labeled probe | ||
| rs5743794 | G>A | Intronic | ACTGAGTTGCCTTTGCTCGT | GCCAGATAACTGACACCACCT | 55 | 50 | HRM with non-labeled probe | ||
| rs3775073 | A>G | Synonymous | GCTGGAATTCTTTGGAATCTGG | GCAAAGCTTCCAGTTTTACGAC | 55 | 50 | HRM with non-labeled probe | ||
| rs2381289 | T>C | 3’UTR | AGGAAGGCCAAGCAGATTTT | AGCGAGGGCTTCATTTTTCT | 53 | 50 | HRM with non-labeled probe | ||
| TLR9 | rs352140 | C>T | Synonymous | TTCCAGTTTGGGCAGGAAGT | GTCACGGAACAACCTGGTGA | 58 | 30 | PCR-RFLP (Bsh1236 I) | |
| TLR10 | rs28393318 | G>A | Intronic | 4p14 | TCCATCAGATCTGCCCCTAC | TGAGAGCGTGGGTTTCTTTT | 55 | 50 | HRM with non-labeled probe |
| rs79030744 | T>C | Intronic | AGGAGCTAAAGCCCAGAGGT | TCGGTCCTTAGGATGTCGTT | 55 | 50 | HRM with non-labeled probe | ||
| rs7658893 | A>G | Intronic | TGGTGGCAGTTATCAGGTCA | AACTGCCAGGGTCCTATCAA | 55 | 50 | HRM with non-labeled probe | ||
| rs4274855 | A>G | 5’UTR | CAAAGGCTCACAATGTCTGG | TCAGAGCATTGGCTGAGAAG | 57 | 50 | HRM with non-labeled probe | ||
| rs10856838 | T>A | Synonymous | GGGTTTTGAGCTCATCTTCATC | TTGGAGCAGTTGGTCATCAG | 55 | 50 | HRM with non-labeled probe | ||
| rs11096956 | T>G | Synonymous | CACAAATGCCACACATGCTT | TCAGATCCAAGTGTTCCAAGG | 52 | 50 | HRM with non-labeled probe | ||
| Gene | Tag SNP (Major>Minor) | Genotype | Groups | Inheritance Model * | Genotype Comparison | Gene | Tag SNP (Major>Minor) | Genotype | Groups | Inheritance Model * | Genotype Comparison | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Responders | Non-Responders | p-Value | OR | 95% CI | Responders | Non-Responders | p-Value | OR | 95% CI | ||||||||
| (n, %) | (n, %) | (n, %) | (n, %) | ||||||||||||||
| TLR1 | rs5743565 | MAF | 0.405 | 0.636 | Allele | 0.036 | 0.389 | 0.157–0.964 | TLR6 | rs6531673 | MAF | 0.315 | 0.318 | Allele | 0.973 | 0.984 | 0.385–2.516 |
| A>G | A/A | 39 (33.6) | 2 (18.2) | C>T | C/C | 52 (44.8) | 4 (36.4) | ||||||||||
| A/G | 60 (51.7) | 4 (36.4) | Dominant | 0.501 | 0.439 | 0.090–2.130 | C/T | 55 (47.4) | 7 (63.6) | Dominant | 0.754 | 0.703 | 0.195–2.534 | ||||
| G/G | 17 (14.7) | 5 (45.5) | Recessive | 0.023 | 0.206 | 0.057–0.751 | T/T | 9 (7.8) | 0 (N/A) | Recessive | 1.000 | 0.492 | 0.027–9.021 | ||||
| rs5743596 | MAF | 0.328 | 0.455 | Allele | 0.229 | 0.585 | 0.242–1.413 | rs56245262 | MAF | 0.263 | 0.318 | Allele | 0.448 | 1.434 | 0.563–3.651 | ||
| C>T | C/C | 51 (44.0) | 4 (36.4) | A>T | A/A | 39 (33.6) | 5 (45.5) | ||||||||||
| C/T | 54 (46.6) | 4 (36.4) | Dominant | 0.756 | 0.728 | 0.202–2.625 | A/T | 61 (52.6) | 5 (45.5) | Dominant | 0.512 | 1.645 | 0.472–5.731 | ||||
| T/T | 11 (9.5) | 3 (27.3) | Recessive | 0.104 | 0.279 | 0.065–1.209 | T/T | 16 (13.8) | 1 (9.1) | Recessive | 1.000 | 1.600 | 0.192–13.369 | ||||
| rs5743604 | MAF | 0.457 | 0.273 | Allele | 0.096 | 2.243 | 0.848–5.938 | rs59295951 | MAF | 0.280 | 0.318 | Allele | 0.705 | 0.834 | 0.325–2.139 | ||
| C>T | C/C | 33 (28.4) | 7 (63.6) | A>G | A/A | 58 (50.0) | 5 (45.5) | ||||||||||
| C/T | 60 (51.7) | 2 (18.2) | Dominant | 0.035 | 4.401 | 1.208–16.026 | A/G | 51 (44.0) | 5 (45.5) | Dominant | 1.000 | 0.833 | 0.241–2.884 | ||||
| T/T | 23 (19.8) | 2 (18.2) | Recessive | 1.000 | 1.113 | 0.225–5.507 | G/G | 7 (6.0) | 1 (9.1) | Recessive | 0.526 | 0.642 | 0.072–5.757 | ||||
| rs4833095 | MAF | 0.233 | 0.318 | Allele | 0.471 | 1.408 | 0.553–3.587 | rs78893527 | MAF | 0.164 | 0.182 | Allele | 0.768 | 0.881 | 0.283–2.750 | ||
| C>T | C/C | 43 (37.1) | 6 (54.5) | C>G | C/C | 81 (69.8) | 7 (63.6) | ||||||||||
| C/T | 54 (46.6) | 3 (27.3) | Dominant | 0.334 | 2.037 | 0.586–7.077 | C/G | 32 (27.6) | 4 (36.4) | Dominant | 0.736 | 0.756 | 0.208–2.750 | ||||
| T/T | 19 (16.4) | 2 (18.2) | Recessive | 1.000 | 0.881 | 0.176–4.405 | G/G | 3 (2.6) | 0 (N/A) | Recessive | 1.000 | 1.410 | 0.068–29.050 | ||||
| TLR2 | rs56346547 | MAF | 0.246 | 0.273 | Allele | 0.779 | 0.869 | 0.324–2.325 | rs5743794 | MAF | 0.371 | 0.409 | Allele | 0.722 | 0.851 | 0.349–2.073 | |
| A>C | A/A | 63 (54.3) | 6 (54.5) | G>A | G/G | 45 (38.8) | 4 (36.4) | ||||||||||
| A/C | 49 (42.2) | 4 (36.4) | Dominant | 1.000 | 1.009 | 0.292–3.495 | G/A | 56 (48.3) | 5 (45.5) | Dominant | 1.000 | 0.902 | 0.250–3.255 | ||||
| C/C | 4 (3.4) | 1 (9.1) | Recessive | 0.369 | 0.357 | 0.036–3.509 | A/A | 15 (12.9) | 2 (18.2) | Recessive | 0.642 | 0.668 | 0.132–3.396 | ||||
| rs62323831 | MAF | 0.319 | 0.545 | Allele | 0.032 | 0.390 | 0.161–0.944 | rs3775073 | MAF | 0.332 | 0.318 | Allele | 0.896 | 0.939 | 0.368–2.400 | ||
| G>A | G/G | 50 (43.1) | 2 (18.2) | A>G | A/A | 51 (44.0) | 5 (45.5) | ||||||||||
| G/A | 58 (50.0) | 6 (54.5) | Dominant | 0.198 | 0.293 | 0.061–1.418 | A/G | 53 (45.7) | 5 (45.5) | Dominant | 1.000 | 1.062 | 0.307–3.678 | ||||
| A/A | 8 (6.9) | 3 (27.3) | Recessive | 0.055 | 0.198 | 0.044–0.893 | G/G | 12 (10.3) | 1 (9.1) | Recessive | 1.000 | 1.154 | 0.136–9.814 | ||||
| rs13105517 | MAF | 0.397 | 0.136 | Allele | 0.020 | 4.161 | 1.197–14.472 | rs2381289 | MAF | 0.435 | 0.455 | Allele | 0.862 | 0.925 | 0.384–2.227 | ||
| G>A | G/G | 38 (32.8) | 8 (72.7) | T>C | T/T | 34 (29.3) | 3 (27.3) | ||||||||||
| G/A | 64 (55.2) | 3 (27.3) | Dominant | 0.017 | 5.473 | 1.374–21.786 | T/C | 63 (54.3) | 6 (54.5) | Dominant | 1.000 | 0.904 | 0.226–3.617 | ||||
| A/A | 14 (12.1) | 0 (N/A) | Recessive | 0.609 | 3.254 | 0.182–58.250 | C/C | 19 (16.4) | 2 (18.2) | Recessive | 1.000 | 0.881 | 0.176–4.405 | ||||
| rs3804100 | MAF | 0.349 | 0.500 | Allele | 0.159 | 0.536 | 0.223–1.291 | TLR10 | rs28393318 | MAF | 0.470 | 0.591 | Allele | 0.277 | 1.630 | 0.671–3.962 | |
| T>C | T/T | 48 (41.4) | 3 (27.3) | A>G | A/A | 31 (26.7) | 2 (18.2) | ||||||||||
| T/C | 55 (47.4) | 5 (45.5) | Dominant | 0.524 | 0.531 | 0.134–2.106 | A/G | 61 (52.6) | 5 (45.5) | Dominant | 0.727 | 0.609 | 0.125–2.977 | ||||
| C/C | 13 (11.2) | 3 (27.3) | Recessive | 0.144 | 0.337 | 0.079–1.430 | G/G | 24 (20.7) | 4 (36.4) | Recessive | 0.258 | 0.457 | 0.123–1.689 | ||||
| TLR4 | rs1927911 | MAF | 0.366 | 0.227 | Allele | 0.246 | 1.966 | 0.700–5.519 | rs79030744 | MAF | 0.259 | 0.227 | Allele | 1.000 | 1.186 | 0.419–3.355 | |
| C>T | C/C | 51 (44.0) | 7 (63.6) | T>C | T/T | 63 (54.3) | 7 (63.6) | ||||||||||
| C/T | 45 (38.8) | 3 (27.3) | Dominant | 0.343 | 2.230 | 0.619–8.039 | T/C | 46 (39.7) | 3 (27.3) | Dominant | 0.753 | 1.472 | 0.409–5.305 | ||||
| T/T | 20 (17.2) | 1 (9.1) | Recessive | 0.690 | 2.083 | 0.252–17.212 | C/C | 7 (6.0) | 1 (9.1) | Recessive | 0.526 | 0.642 | 0.072–5.757 | ||||
| rs1927907 | MAF | 0.366 | 0.364 | Allele | 0.980 | 1.012 | 0.408–2.511 | rs7658893 | MAF | 0.483 | 0.364 | Allele | 0.285 | 1.633 | 0.660–4.042 | ||
| G>A | G/G | 44 (37.9) | 4 (36.4) | A>G | A/A | 30 (25.9) | 3 (27.3) | ||||||||||
| G/A | 59 (50.9) | 6 (54.5) | Dominant | 1.000 | 0.935 | 0.259–3.378 | A/G | 60 (51.7) | 8 (72.7) | Dominant | 1.000 | 1.075 | 0.268–4.318 | ||||
| A/A | 13 (11.2) | 1 (9.1) | Recessive | 1.000 | 1.262 | 0.149–10.672 | G/G | 26 (22.4) | 0 (N/A) | Recessive | 0.119 | 6.735 | 0.384–118.200 | ||||
| rs11536889 | MAF | 0.310 | 0.455 | Allele | 0.167 | 0.540 | 0.223–1.307 | rs4274855 | MAF | 0.315 | 0.318 | Allele | 0.973 | 0.984 | 0.385–2.516 | ||
| G>C | G/G | 53 (45.7) | 2 (18.2) | C>T | C/C | 52 (44.8) | 4 (36.4) | ||||||||||
| G/C | 54 (46.6) | 8 (72.7) | Dominant | 0.112 | 0.264 | 0.055–1.276 | C/T | 55 (47.4) | 7 (63.6) | Dominant | 0.754 | 0.703 | 0.195–2.534 | ||||
| C/C | 9 (7.8) | 1 (9.1) | Recessive | 1.000 | 0.841 | 0.096–7.331 | T/T | 9 (7.8) | 0 (N/A) | Recessive | 1.000 | 2.033 | 0.111–37.270 | ||||
| TLR9 | rs352140 | MAF | 0.289 | 0.500 | Allele | 0.787 | 0.886 | 0.370–2.125 | rs10856838 | MAF | 0.159 | 0.136 | Allele | 0.774 | 1.441 | 0.408–5.089 | |
| C>T | C/C | 28 (24.1) | 3 (27.3) | T>A | T/T | 76 (65.5) | 8 (72.7) | ||||||||||
| C/T | 67 (57.8) | 5 (45.5) | Dominant | 0.729 | 1.179 | 0.293–4.748 | T/A | 37 (31.9) | 3 (27.3) | Dominant | 0.749 | 1.404 | 0.353–5.583 | ||||
| T/T | 21 (18.1) | 3 (27.3) | Recessive | 0.434 | 0.589 | 0.144–2.411 | A/A | 3 (2.6) | 0 (N/A) | Recessive | 1.000 | 0.709 | 0.034–14.610 | ||||
| rs11096956 | MAF | 0.371 | 0.409 | Allele | 0.722 | 0.851 | 0.349–2.073 | ||||||||||
| G>A | G/G | 45 (38.8) | 4 (36.4) | ||||||||||||||
| G/A | 56 (48.3) | 5 (45.5) | Dominant | 1.000 | 0.902 | 0.250–3.255 | |||||||||||
| A/A | 15 (12.9) | 2 (18.2) | Recessive | 0.642 | 0.668 | 0.132–3.396 | |||||||||||
| Factor | OR (95% CI) | p-Value * |
|---|---|---|
| A/A or A/G genotype of rs5743565 in TLR1 | 5.593 (1.407–22.239) | 0.015 |
| G/A or A/A genotype of rs13105517 in TLR2 | 6.124 (1.454–25.789) | 0.014 |
| Biomarker | TLR1 | TLR2 | Statistical Results | Genetic Test | ||||
|---|---|---|---|---|---|---|---|---|
| rs5743565 | rs13105517 | OR (95% CI) | p-Value * | Sensitivity | Specificity | PPV | NPV | |
| Marker 1 | A/A or A/G | – | 4.853 (1.331–17.690) | 0.023 | 85.3 | 45.5 | 94.3 | 22.7 |
| Marker 2 | – | G/A or A/A | 5.473 (1.374–21.790) | 0.017 | 67.2 | 72.7 | 96.3 | 17.4 |
| Marker 3 | A/A or A/G | G/A or A/A | 5.735 (1.186–27.730) | 0.024 | 56.0 | 81.8 | 97.0 | 15.0 |
| Gene | Tag SNP (Major>Minor) | Genotype | Groups | Inheritance Model * | Genotype Comparison | Gene | Tag SNP (Major>Minor) | Genotype | Groups | Inheritance Model * | Genotype Comparison | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Responders | Non-Responders | p-Value | OR | 95% CI | Responders | Non-Responders | p-Value | OR | 95% CI | ||||||||
| (n, %) | (n, %) | (n, %) | (n, %) | ||||||||||||||
| TLR1 | rs5743565 | MAF | 0.402 | 0.421 | Allele | 0.827 | 0.925 | 0.457–1.871 | TLR6 | rs6531673 | MAF | 0.330 | 0.237 | Allele | 0.259 | 1.586 | 0.709–3.550 |
| A>G | A/A | 33 (34.0) | 6 (31.6) | C>T | C/C | 41 (42.3) | 11 (57.9) | ||||||||||
| A/G | 50 (51.5) | 10 (52.6) | Dominant | 0.837 | 0.895 | 0.312–2.570 | C/T | 48 (49.5) | 7 (36.8) | Dominant | 0.210 | 1.878 | 0.694–5.084 | ||||
| G/G | 14 (14.4) | 3 (15.8) | Recessive | 1.000 | 0.900 | 0.232–3.494 | T/T | 8 (8.2) | 1 (5.3) | Recessive | 1.000 | 1.618 | 0.190–13.755 | ||||
| rs5743596 | MAF | 0.330 | 0.316 | Allele | 0.866 | 1.067 | 0.506–2.251 | rs56245262 | MAF | 0.418 | 0.316 | Allele | 0.242 | 1.553 | 0.740–3.259 | ||
| C>T | C/C | 42 (43.3) | 9 (47.4) | A>T | A/A | 30 (30.9) | 9 (47.4) | ||||||||||
| C/T | 46 (47.4) | 8 (42.1) | Dominant | 0.744 | 1.179 | 0.440–3.160 | A/T | 53 (54.6) | 8 (42.1) | Dominant | 0.165 | 2.010 | 0.741–5.453 | ||||
| T/T | 9 (9.3) | 2 (10.5) | Recessive | 1.000 | 0.869 | 0.172–4.384 | T/T | 14 (14.4) | 2 (10.5) | Recessive | 1.000 | 1.434 | 0.298–6.897 | ||||
| rs5743604 | MAF | 0.479 | 0.342 | Allele | 0.120 | 1.771 | 0.856–3.663 | rs59295951 | MAF | 0.263 | 0.368 | Allele | 0.185 | 0.611 | 0.294–1.272 | ||
| C>T | C/C | 26 (26.8) | 7 (36.8) | A>G | A/A | 50 (51.5) | 8 (42.1) | ||||||||||
| C/T | 49 (50.5) | 11 (57.9) | Dominant | 0.375 | 1.593 | 0.566–4.484 | A/G | 43 (44.3) | 8 (42.1) | Dominant | 0.452 | 0.684 | 0.253–1.847 | ||||
| T/T | 22 (22.7) | 1 (5.3) | Recessive | 0.116 | 5.280 | 0.667–41.841 | G/G | 4 (4.1) | 3 (15.8) | Recessive | 0.086 | 0.229 | 0.047–1.123 | ||||
| rs4833095 | MAF | 0.423 | 0.263 | Allele | 0.066 | 2.050 | 0.943–4.454 | rs78893527 | MAF | 0.160 | 0.184 | Allele | 0.710 | 0.842 | 0.340–2.083 | ||
| C>T | C/C | 33 (34.0) | 10 (52.6) | C>G | C/C | 69 (71.1) | 12 (63.2) | ||||||||||
| C/T | 46 (47.4) | 8 (42.1) | Dominant | 0.125 | 2.155 | 0.798–5.821 | C/G | 25 (25.8) | 7 (36.8) | Dominant | 0.489 | 0.696 | 0.248–1.949 | ||||
| T/T | 18 (18.6) | 1 (5.3) | Recessive | 0.193 | 4.102 | 0.514–32.787 | G/G | 3 (3.1) | 0 (N/A) | Recessive | 1.000 | 0.692 | 0.034–13.960 | ||||
| TLR2 | rs56346547 | MAF | 0.247 | 0.237 | Allele | 0.890 | 1.059 | 0.468–2.395 | rs5743794 | MAF | 0.381 | 0.316 | Allele | 0.444 | 1.336 | 0.636–2.808 | |
| A>C | A/A | 53 (54.6) | 10 (52.6) | G>A | G/G | 36 (37.1) | 9 (47.4) | ||||||||||
| A/C | 40 (41.2) | 9 (47.4) | Dominant | 0.872 | 0.922 | 0.344–2.471 | G/A | 48 (49.5) | 8 (42.1) | Dominant | 0.402 | 1.525 | 0.566–4.105 | ||||
| C/C | 4 (4.1) | 0 (N/A) | Recessive | 1.000 | 1.877 | 0.097–36.330 | A/A | 13 (13.4) | 2 (10.5) | Recessive | 1.000 | 1.315 | 0.272–6.369 | ||||
| rs62323831 | MAF | 0.335 | 0.237 | Allele | 0.235 | 1.624 | 0.726–3.632 | rs3775073 | MAF | 0.314 | 0.421 | Allele | 0.202 | 1.586 | 0.778–3.231 | ||
| G>A | G/G | 39 (40.2) | 11 (57.9) | A>G | A/A | 44 (45.4) | 7 (36.8) | ||||||||||
| G/A | 51 (52.6) | 7 (36.8) | Dominant | 0.155 | 2.045 | 0.754–5.543 | A/G | 45 (46.4) | 8 (42.1) | Dominant | 0.494 | 0.703 | 0.255–1.938 | ||||
| A/A | 7 (7.2) | 1 (5.3) | Recessive | 1.000 | 1.400 | 0.162–12.092 | G/G | 8 (8.2) | 4 (21.1) | Recessive | 0.108 | 0.337 | 0.090–1.261 | ||||
| rs13105517 | MAF | 0.361 | 0.579 | Allele | 0.012 | 0.411 | 0.202–0.833 | rs2381289 | MAF | 0.448 | 0.368 | Allele | 0.363 | 1.394 | 0.680–2.856 | ||
| G>A | G/G | 35 (36.1) | 3 (15.8) | T>C | T/T | 27 (27.8) | 7 (36.8) | ||||||||||
| G/A | 54 (55.7) | 10 (52.6) | Dominant | 0.111 | 0.332 | 0.090–1.220 | T/C | 53 (54.6) | 10 (52.6) | Dominant | 0.430 | 1.512 | 0.539–4.246 | ||||
| A/A | 8 (8.2) | 6 (31.6) | Recessive | 0.004 | 0.195 | 0.058–0.652 | C/C | 17 (17.5) | 2 (10.5) | Recessive | 0.735 | 1.806 | 0.381–8.562 | ||||
| rs3804100 | MAF | 0.376 | 0.211 | Allele | 0.050 | 2.262 | 0.984–5.200 | TLR10 | rs28393318 | MAF | 0.464 | 0.500 | Allele | 0.684 | 1.156 | 0.576–2.317 | |
| T>C | T/T | 37 (38.1) | 11 (57.9) | A>G | A/A | 28 (28.9) | 3 (15.8) | ||||||||||
| T/C | 47 (48.5) | 8 (42.1) | Dominant | 0.110 | 2.230 | 0.821–6.053 | A/G | 48 (49.5) | 13 (68.4) | Dominant | 0.395 | 0.462 | 0.125–1.711 | ||||
| C/C | 13 (13.4) | 0 (N/A) | Recessive | 0.123 | 6.231 | 0.355–109.500 | G/G | 21 (21.6) | 3 (15.8) | Recessive | 0.760 | 1.474 | 0.392–5.540 | ||||
| TLR4 | rs1927911 | MAF | 0.376 | 0.316 | Allele | 0.479 | 1.307 | 0.622–2.748 | rs79030744 | MAF | 0.278 | 0.158 | Allele | 0.121 | 2.057 | 0.814–5.198 | |
| C>T | C/C | 41 (42.3) | 10 (52.6) | T>C | T/T | 50 (51.5) | 13 (68.4) | ||||||||||
| C/T | 39 (40.2) | 6 (31.6) | Dominant | 0.405 | 1.518 | 0.566–4.070 | T/C | 40 (41.2) | 6 (31.6) | Dominant | 0.177 | 2.037 | 0.715–5.797 | ||||
| T/T | 17 (17.5) | 3 (15.8) | Recessive | 1.000 | 1.133 | 0.297–4.327 | C/C | 7 (7.2) | 0 (N/A) | Recessive | 0.597 | 3.232 | 0.177–59.030 | ||||
| rs1927907 | MAF | 0.392 | 0.237 | Allele | 0.070 | 2.075 | 0.931–4.625 | rs7658893 | MAF | 0.479 | 0.500 | Allele | 0.816 | 0.921 | 0.459–1.846 | ||
| G>A | G/G | 33 (34.0) | 11 (57.9) | A>G | A/A | 26 (26.8) | 4 (21.1) | ||||||||||
| G/A | 52 (53.6) | 7 (36.8) | Dominant | 0.050 | 2.667 | 0.978–7.267 | A/G | 49 (50.5) | 11 (57.9) | Dominant | 0.777 | 0.728 | 0.221–2.396 | ||||
| A/A | 12 (12.4) | 1 (5.3) | Recessive | 0.691 | 2.541 | 0.310–20.790 | G/G | 22 (22.7) | 4 (21.1) | Recessive | 1.000 | 1.100 | 0.331–3.656 | ||||
| rs11536889 | MAF | 0.294 | 0.395 | Allele | 0.219 | 0.638 | 0.310–1.311 | rs4274855 | MAF | 0.330 | 0.237 | Allele | 0.259 | 1.586 | 0.709–3.550 | ||
| G>C | G/G | 47 (48.5) | 6 (31.6) | C>T | C/C | 41 (42.3) | 11 (57.9) | ||||||||||
| G/C | 43 (44.3) | 11 (57.9) | Dominant | 0.177 | 0.491 | 0.172–1.398 | C/T | 48 (49.5) | 7 (36.8) | Dominant | 0.210 | 1.878 | 0.694–5.084 | ||||
| C/C | 7 (7.2) | 2 (10.5) | Recessive | 0.640 | 0.661 | 0.126–3.459 | T/T | 8 (8.2) | 1 (5.3) | Recessive | 1.000 | 1.618 | 0.190–13.755 | ||||
| TLR9 | rs352140 | MAF | 0.454 | 0.553 | Allele | 0.263 | 0.672 | 0.334–1.352 | rs10856838 | MAF | 0.196 | 0.132 | Allele | 0.494 | 1.608 | 0.588–4.394 | |
| C>T | C/C | 26 (26.8) | 2 (10.5) | T>A | T/T | 61 (62.9) | 15 (78.9) | ||||||||||
| C/T | 54 (55.7) | 13 (68.4) | Dominant | 0.155 | 0.321 | 0.069–1.487 | T/A | 34 (35.1) | 3 (15.8) | Dominant | 0.201 | 2.213 | 0.682–7.184 | ||||
| T/T | 17 (17.5) | 4 (21.1) | Recessive | 0.747 | 0.797 | 0.235–2.701 | A/A | 2 (2.1) | 1 (5.3) | Recessive | 0.418 | 0.379 | 0.033–4.403 | ||||
| rs11096956 | MAF | 0.381 | 0.316 | Allele | 0.444 | 1.336 | 0.636–2.808 | ||||||||||
| G>A | G/G | 36 (37.1) | 9 (47.4) | ||||||||||||||
| G/A | 48 (49.5) | 8 (42.1) | Dominant | 0.402 | 1.525 | 0.566–4.105 | |||||||||||
| A/A | 13 (13.4) | 2 (10.5) | Recessive | 1.000 | 1.315 | 0.272–6.369 | |||||||||||
| Biomarker | Statistical Results | Genetic Diagnosis | ||||
|---|---|---|---|---|---|---|
| OR (95% CI) | p-Value * | Sensitivity | Specificity | PPV | NPV | |
| G/G or G/A genotype of rs13105517 in TLR2 | 3.736 (1.029–13.57) | 0.004 | 91.8 | 31.6 | 87.3 | 42.9 |
| Gene | Tag SNP (Major>Minor) | Genotype | Groups (n,%) | Inheritance Model * | Genotype Comparison | Gene | Tag SNP (Major>Minor) | Genotype | Groups (n,%) | Inheritance Model * | Genotype Comparison | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Responders (n, %) | Non-Responders (n, %) | p-Value | OR | 95% CI | Responders (n, %) | Non-Responders (n, %) | p-Value | OR | 95% CI | |||||||||
| TLR1 | rs5743565 | MAF | 0.402 | 0.400 | Allele | 0.980 | 1.010 | 0.456–2.236 | TLR6 | rs6531673 | MAF | 0.348 | 0.233 | Allele | 0.221 | 1.750 | 0.708–4.327 | |
| A>G | A/A | 28 (34.1) | 5 (33.3) | C>T | C/C | 33 (40.2) | 8 (53.3) | |||||||||||
| A/G | 42 (51.2) | 8 (53.3) | Dominant | 1.000 | 0.964 | 0.300–3.096 | C/T | 41 (50.0) | 7 (46.7) | Dominant | 0.345 | 1.697 | 0.561–5.131 | |||||
| G/G | 12 (14.6) | 2 (13.3) | Recessive | 1.000 | 1.114 | 0.223–5.574 | T/T | 8 (9.8) | 0 (N/A) | Recessive | 0.351 | 0.283 | 0.015–5.164 | |||||
| rs5743596 | MAF | 0.341 | 0.267 | Allele | 0.423 | 1.426 | 0.597–3.407 | rs56245262 | MAF | 0.433 | 0.333 | Allele | 0.309 | 1.527 | 0.673–3.465 | |||
| C>T | C/C | 35 (42.7) | 7 (46.7) | A>T | A/A | 25 (30.5) | 5 (33.3) | |||||||||||
| C/T | 38 (46.3) | 8 (53.3) | Dominant | 0.775 | 1.175 | 0.389–3.547 | A/T | 43 (52.4) | 10 (66.7) | Dominant | 1.000 | 1.140 | 0.353–3.681 | |||||
| T/T | 9 (11.0) | 0 (N/A) | Recessive | 0.347 | 4.007 | 0.221–72.59 | T/T | 14 (17.1) | 0 (N/A) | Recessive | 0.117 | 6.562 | 0.371–116.100 | |||||
| rs5743604 | MAF | 0.470 | 0.533 | Allele | 0.520 | 0.774 | 0.355–1.689 | rs59295951 | MAF | 0.262 | 0.267 | Allele | 0.959 | 0.977 | 0.405–2.358 | |||
| C>T | C/C | 24 (29.3) | 2 (13.3) | A>G | A/A | 43 (52.4) | 7 (46.7) | |||||||||||
| C/T | 39 (47.6) | 10 (66.7) | Dominant | 0.341 | 0.372 | 0.078–1.774 | A/G | 35 (42.7) | 8 (53.3) | Dominant | 0.681 | 0.794 | 0.263–2.392 | |||||
| T/T | 19 (23.2) | 3 (20.0) | Recessive | 1.000 | 1.206 | 0.308–4.726 | G/G | 4 (4.9) | 0 (N/A) | Recessive | 1.000 | 1.777 | 0.091–34.740 | |||||
| rs4833095 | MAF | 0.421 | 0.433 | Allele | 0.898 | 0.950 | 0.433–2.084 | rs78893527 | MAF | 0.152 | 0.200 | Allele | 0.513 | 0.719 | 0.267–1.938 | |||
| C>T | C/C | 28 (34.1) | 5 (33.3) | C>G | C/C | 59 (72.0) | 10 (66.7) | |||||||||||
| C/T | 39 (47.6) | 7 (46.7) | Dominant | 1.000 | 0.964 | 0.300–3.096 | C/G | 21 (25.6) | 4 (26.7) | Dominant | 0.759 | 0.780 | 0.240–2.529 | |||||
| T/T | 15 (18.3) | 3 (20.0) | Recessive | 1.000 | 0.895 | 0.225–3.573 | G/G | 2 (2.4) | 1 (6.7) | Recessive | 0.399 | 0.350 | 0.030–4.124 | |||||
| TLR2 | rs56346547 | MAF | 0.268 | 0.133 | Allele | 0.166 | 2.383 | 0.787–7.215 | rs5743794 | MAF | 0.372 | 0.433 | Allele | 0.525 | 0.774 | 0.352–1.704 | ||
| A>C | A/A | 42 (51.2) | 11 (73.3) | G>A | G/G | 33 (40.2) | 3 (20.0) | |||||||||||
| A/C | 36 (43.9) | 4 (26.7) | Dominant | 0.160 | 2.619 | 0.770–8.905 | G/A | 37 (45.1) | 11 (73.3) | Dominant | 0.159 | 0.371 | 0.097–1.418 | |||||
| C/C | 4 (4.9) | 0 (N/A) | Recessive | 1.000 | 0.563 | 0.029–11.000 | A/A | 12 (14.6) | 1 (6.7) | Recessive | 0.685 | 2.400 | 0.288–19.960 | |||||
| rs62323831 | MAF | 0.311 | 0.467 | Allele | 0.097 | 0.516 | 0.234–1.136 | rs3775073 | MAF | 0.305 | 0.367 | Allele | 0.503 | 1.320 | 0.585–2.978 | |||
| G>A | G/G | 36 (43.9) | 3 (20.0) | A>G | A/A | 40 (48.8) | 4 (26.7) | |||||||||||
| G/A | 41 (50.0) | 10 (66.7) | Dominant | 0.095 | 0.319 | 0.084–1.218 | A/G | 34 (41.5) | 11 (73.3) | Dominant | 0.160 | 0.382 | 0.112–1.298 | |||||
| A/A | 5 (6.1) | 2 (13.3) | Recessive | 0.295 | 0.422 | 0.074–2.410 | G/G | 8 (9.8) | 0 (N/A) | Recessive | 0.351 | 3.537 | 0.194–64.600 | |||||
| rs13105517 | MAF | 0.354 | 0.400 | Allele | 0.627 | 0.821 | 0.370–1.822 | rs2381289 | MAF | 0.445 | 0.467 | Allele | 0.827 | 0.917 | 0.420–2.001 | |||
| G>A | G/G | 31 (37.8) | 4 (26.7) | T>C | T/T | 23 (28.0) | 4 (26.7) | |||||||||||
| G/A | 44 (53.7) | 10 (66.7) | Dominant | 0.562 | 0.598 | 0.175–2.043 | T/C | 45 (54.9) | 8 (53.3) | Dominant | 1.000 | 0.933 | 0.269–3.229 | |||||
| A/A | 7 (8.5) | 1 (6.7) | Recessive | 1.000 | 1.307 | 0.149–11.468 | C/C | 14 (17.1) | 3 (20.0) | Recessive | 0.723 | 0.824 | 0.205–3.306 | |||||
| rs3804100 | MAF | 0.366 | 0.433 | Allele | 0.483 | 0.754 | 0.343–1.661 | TLR10 | rs28393318 | MAF | 0.463 | 0.467 | Allele | 0.974 | 1.013 | 0.464–2.210 | ||
| T>C | T/T | 34 (41.5) | 3 (20.0) | A>G | A/A | 22 (26.8) | 6 (40.0) | |||||||||||
| T/C | 36 (43.9) | 11 (73.3) | Dominant | 0.153 | 0.353 | 0.092–1.347 | A/G | 44 (53.7) | 4 (26.7) | Dominant | 0.301 | 1.818 | 0.580–5.701 | |||||
| C/C | 12 (14.6) | 1 (6.7) | Recessive | 0.685 | 2.400 | 0.288–19.960 | G/G | 16 (19.5) | 5 (33.3) | Recessive | 0.305 | 0.485 | 0.145–1.617 | |||||
| TLR4 | rs1927911 | MAF | 0.378 | 0.367 | Allele | 0.906 | 1.050 | 0.469–2.353 | rs79030744 | MAF | 0.293 | 0.200 | Allele | 0.298 | 1.655 | 0.636–4.305 | ||
| C>T | C/C | 34 (41.5) | 7 (46.7) | T>C | T/T | 41 (50.0) | 9 (60.0) | |||||||||||
| C/T | 34 (41.5) | 5 (33.3) | Dominant | 0.708 | 1.235 | 0.409–3.731 | T/C | 34 (41.5) | 6 (40.0) | Dominant | 0.476 | 1.500 | 0.489–4.598 | |||||
| T/T | 14 (17.1) | 3 (20.0) | Recessive | 0.723 | 0.824 | 0.205–3.306 | C/C | 7 (8.5) | 0 (N/A) | Recessive | 0.591 | 3.079 | 0.167–56.820 | |||||
| rs1927907 | MAF | 0.409 | 0.300 | Allele | 0.263 | 1.612 | 0.695–3.736 | rs7658893 | MAF | 0.482 | 0.467 | Allele | 0.880 | 1.062 | 0.487–2.317 | |||
| G>A | G/G | 27 (32.9) | 6 (40.0%) | A>G | A/A | 21 (25.6) | 5 (33.3) | |||||||||||
| G/A | 43 (52.4) | 9 (60.0%) | Dominant | 0.595 | 1.358 | 0.438–4.209 | A/G | 43 (52.4) | 6 (40.0) | Dominant | 0.538 | 1.452 | 0.445–4.739 | |||||
| A/A | 12 (14.6) | 0 (N/A) | Recessive | 0.203 | 5.496 | 0.308–97.950 | G/G | 18 (22.0) | 4 (26.7) | Recessive | 0.740 | 0.773 | 0.220–2.722 | |||||
| rs11536889 | MAF | 0.293 | 0.300 | Allele | 0.936 | 0.966 | 0.413–2.259 | rs4274855 | MAF | 0.348 | 0.233 | Allele | 0.221 | 1.750 | 0.708–4.327 | |||
| G>C | G/G | 40 (48.8) | 7 (46.7) | C>T | C/C | 33 (40.2) | 8 (53.3) | |||||||||||
| G/C | 36 (43.9) | 7 (46.7) | Dominant | 0.880 | 0.919 | 0.305–2.769 | C/T | 41 (50.0) | 7 (46.7) | Dominant | 0.345 | 1.697 | 0.561–5.131 | |||||
| C/C | 6 (7.3) | 1 (6.7) | Recessive | 1.000 | 1.105 | 0.123–9.901 | T/T | 8 (9.8) | 0 (N/A) | Recessive | 0.351 | 3.537 | 0.194–64.600 | |||||
| TLR9 | rs352140 | MAF | 0.470 | 0.367 | Allele | 0.298 | 1.529 | 0.685–3.414 | rs10856838 | MAF | 0.201 | 0.167 | Allele | 0.805 | 1.260 | 0.448–3.540 | ||
| C>T | C/C | 21 (25.6) | 5 (33.3) | T>A | T/T | 51 (62.2) | 10 (66.7) | |||||||||||
| C/T | 45 (54.9) | 9 (60.0%) | Dominant | 0.538 | 1.452 | 0.445–4.739 | T/A | 29 (35.4) | 5 (33.3) | Dominant | 1.000 | 1.216 | 0.380–3.888 | |||||
| T/T | 16 (19.5) | 1 (6.7) | Recessive | 0.458 | 3.394 | 0.415–27.778 | A/A | 2 (2.4) | 0 (N/A) | Recessive | 1.000 | 0.963 | 0.044–21.060 | |||||
| rs11096956 | MAF | 0.372 | 0.433 | Allele | 0.525 | 0.774 | 0.352–1.704 | |||||||||||
| G>A | G/G | 33 (40.2) | 3 (20.0) | |||||||||||||||
| G/A | 37 (45.1) | 11 (73.3) | Dominant | 0.159 | 0.371 | 0.097–1.418 | ||||||||||||
| A/A | 12 (14.6) | 1 (6.7) | Recessive | 0.685 | 2.400 | 0.288–19.960 | ||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, J.; Kurumi, H.; Isomoto, H.; Ogihara, R.; Matsushima, K.; Machida, H.; Ishida, T.; Hirayama, T.; Yamaguchi, N.; Yoshida, Y.; et al. Toll-like Receptor Gene Polymorphisms as Predictive Biomarkers for Response to Infliximab in Japanese Patients with Crohn’s Disease. Diagnostics 2025, 15, 971. https://doi.org/10.3390/diagnostics15080971
Wei J, Kurumi H, Isomoto H, Ogihara R, Matsushima K, Machida H, Ishida T, Hirayama T, Yamaguchi N, Yoshida Y, et al. Toll-like Receptor Gene Polymorphisms as Predictive Biomarkers for Response to Infliximab in Japanese Patients with Crohn’s Disease. Diagnostics. 2025; 15(8):971. https://doi.org/10.3390/diagnostics15080971
Chicago/Turabian StyleWei, Jingjing, Hiroki Kurumi, Hajime Isomoto, Ryohei Ogihara, Kayoko Matsushima, Haruhisa Machida, Tetsuya Ishida, Tatsuro Hirayama, Naoyuki Yamaguchi, Yukina Yoshida, and et al. 2025. "Toll-like Receptor Gene Polymorphisms as Predictive Biomarkers for Response to Infliximab in Japanese Patients with Crohn’s Disease" Diagnostics 15, no. 8: 971. https://doi.org/10.3390/diagnostics15080971
APA StyleWei, J., Kurumi, H., Isomoto, H., Ogihara, R., Matsushima, K., Machida, H., Ishida, T., Hirayama, T., Yamaguchi, N., Yoshida, Y., & Tsukamoto, K. (2025). Toll-like Receptor Gene Polymorphisms as Predictive Biomarkers for Response to Infliximab in Japanese Patients with Crohn’s Disease. Diagnostics, 15(8), 971. https://doi.org/10.3390/diagnostics15080971







