Mapping Antimicrobial Resistance in Escherichia coli and Klebsiella pneumoniae from Complicated Urinary Tract Infections in Oman: Phenotypic and Genotypic Insights
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Period and Setting
2.2. Study Design
2.3. Inclusion and Exclusion Criteria
2.4. Sample Size
2.5. Bacterial Identification and Susceptibility Testing
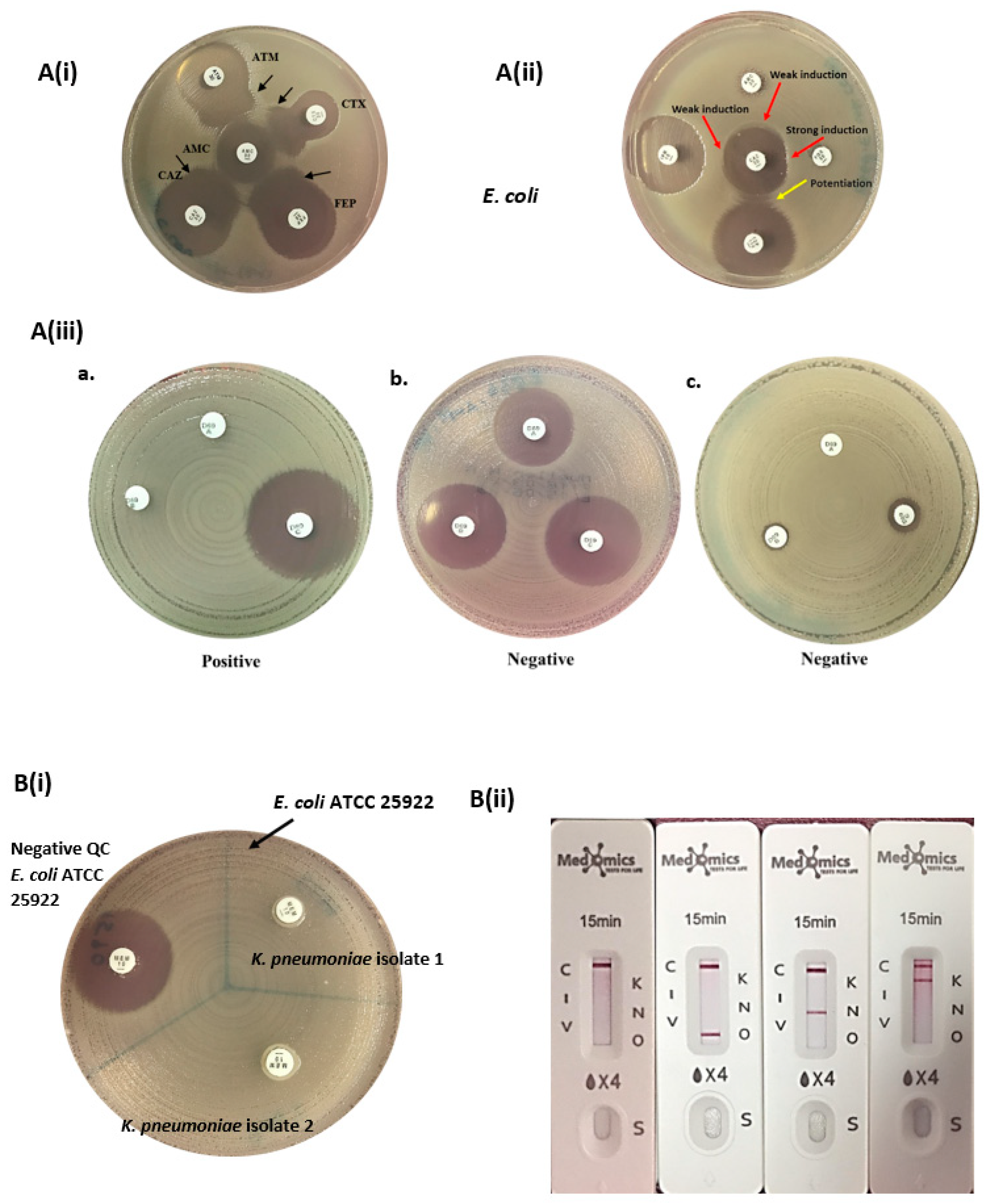
2.6. Manual Phenotypic Detection of ESBL, AmpC, and CPE
2.7. Whole Genome Sequencing (WGS)
2.8. Statistical Analyses
3. Results
3.1. Demographic Characteristics
3.2. Antimicrobial Susceptibility Profiles
3.3. Prevalence of ESBL, AmpC Beta-Lactamases, and Carbapenemases in cUTI
3.4. In Silico Identification of Resistance Genes from Genomic Sequences of AmpC-Producing E. coli
3.5. In Silico Identification of Resistance Genes from Genomic Sequences of Carbapenemase-Producing K. pneumoniae
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kot, B.; Grużewska, A.; Szweda, P.; Wicha, J.; Parulska, U. Antibiotic Resistance of Uropathogens Isolated from Patients Hospitalized in District Hospital in Central Poland in 2020. Antibiotics 2021, 10, 447. [Google Scholar] [CrossRef] [PubMed]
- López-Montesinos, I.; Horcajada, J.P. Oral and intravenous fosfomycin in complicated urinary tract infections. Rev. Esp. Quimioter. 2019, 32 (Suppl. S1), 37–44. [Google Scholar] [PubMed]
- Mamari, Y.A.L.; Sami, H.; Siddiqui, K.; Tahir, H.B.; Jabri, Z.A.L.; Muharrmi, Z.A.L.; Rizvi, S.G.; Rizvi, M. Trends of antimicrobial resistance in patients with complicated urinary tract infection: Suggested empirical therapy and lessons learned from a retrospective observational study in Oman. Urol. Ann. 2022, 14, 345–352. [Google Scholar] [CrossRef]
- Rizvi, M.; Malhotra, S.; Agarwal, J.; Siddiqui, A.H.; Devi, S.; Poojary, A.; Thakuria, B.; Princess, I.; Sami, H.; Gupta, A.; et al. Regional variations in antimicrobial susceptibility of community-acquired uropathogenic Escherichia coli in India: Findings of a multicentric study highlighting the importance of local antibiograms. IJID Reg. 2024, 11, 100370. [Google Scholar] [CrossRef] [PubMed]
- Wagenlehner, F.M.E.; Bjerklund Johansen, T.E.; Cai, T.; Koves, B.; Kranz, J.; Pilatz, A.; Tandogdu, Z. Epidemiology, definition and treatment of complicated urinary tract infections. Nat. Rev. Urol. 2020, 17, 586–600. [Google Scholar] [CrossRef]
- Walkty, A.; Karlowsky, J.A.; Lagace-Wiens, P.; Baxter, M.R.; Adam, H.J.; Zhanel, G.G. Antimicrobial resistance patterns of bacterial pathogens recovered from the urine of patients at Canadian hospitals from 2009 to 2020. JAC Antimicrob. Resist. 2022, 4, dlac122. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Zilberberg, M.D.; Nathanson, B.H.; Sulham, K.; Shorr, A.F. Antimicrobial Susceptibility and Cross-Resistance Patterns among Common Complicated Urinary Tract Infections in U.S. Hospitals, 2013 to 2018. Antimicrob. Agents Chemother. 2020, 64, e00346-20. [Google Scholar] [CrossRef]
- Ruppé, É.; Woerther, P.L.; Barbier, F. Mechanisms of antimicrobial resistance in Gram-negative bacilli. Ann. Intensive Care 2015, 5, 21. [Google Scholar] [CrossRef]
- Patel, R. MALDI-TOF MS for the diagnosis of infectious diseases. Clin. Chem. 2015, 61, 100–111. [Google Scholar] [CrossRef]
- Funke, G.; Funke-Kissling, P. Evaluation of the new VITEK 2 card for identification of clinically relevant gram-negative rods. J. Clin. Microbiol. 2004, 42, 4067–4071. [Google Scholar] [CrossRef] [PubMed]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 32nd ed.; CLSI: Wayne, PA, USA, 2022; CLSI Supplement M100; Available online: https://clsi.org/media/wi0pmpke/m100ed32_sample.pdf (accessed on 14 August 2024).
- Waheed, A.; Saleem, S.; Shahzad, N.; Akhtar, J.; Saeed, M.; Jameel, I.; Rasheed, F.; Jahan, S. Prevalence of Extended Spectrum [beta]-lactamase SHV and OXA Producing Gram Negative Bacteria at Tertiary Care Hospital of Lahore, Pakistan. Pak. J. Zool. 2019, 51, 2345. [Google Scholar] [CrossRef]
- Drieux, L.; Brossier, F.; Sougakoff, W.; Jarlier, V. Phenotypic detection of extended-spectrum beta-lactamase production in Enterobacteriaceae: Review and bench guide. Clin. Microbiol. Infect. 2008, 14 (Suppl. S1), 90–103. [Google Scholar] [CrossRef] [PubMed]
- Al Mamari, A.M.K.; Al Jabri, Z.; Sami, H.; Rizvi, S.G.A.; Chan, M.F.; Al Siyabi, T.; Al Muharrmi, Z.; Rizvi, M. Evaluation of six commercial and in-house phenotypic tests for detection of AmpC β-lactamases: Is routine detection possible? JAC Antimicrob. Resist. 2023, 5, dlad101. [Google Scholar] [CrossRef]
- Gupta, G.; Tak, V.; Mathur, P. Detection of AmpC β Lactamases in Gram-negative Bacteria. J. Lab. Physicians 2014, 6, 1–6. [Google Scholar] [CrossRef]
- Halstead, F.D.; Vanstone, G.L.; Balakrishnan, I. An evaluation of the Mast D69C AmpC Detection Disc Set for the detection of inducible and derepressed AmpC β-lactamases. J. Antimicrob. Chemother. 2012, 67, 2303–2304. [Google Scholar] [CrossRef][Green Version]
- CLSI. Analysis and Presentation of Cumulative Antimicrobial Susceptibility Test Data, 5th ed.; CLSI Guideline M39; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Li, X.; Chen, Y.; Gao, W.; Ye, H.; Shen, Z.; Wen, Z.; Wei, J. A 6-year study of complicated urinary tract infections in southern China: Prevalence, antibiotic resistance, clinical and economic outcomes. Ther. Clin. Risk Manag. 2017, 13, 1479–1487. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Palasik, B.N. Combating antimicrobial resistance with cefiderocol for complicated infections involving the urinary tract. Ther. Adv. Urol. 2022, 14, 17562872211065570. [Google Scholar] [CrossRef] [PubMed]
- Babiker, A.; Clarke, L.; Doi, Y.; Shields, R.K. Fosfomycin for treatment of multidrug-resistant pathogens causing urinary tract infection: A real-world perspective and review of the literature. Diagn. Microbiol. Infect. Dis. 2019, 95, 114856. [Google Scholar] [CrossRef]
- Mohamed, A.; Mohamud, M.; Mohamud, H. Epidemiology and Antimicrobial Susceptibility Pattern of Uropathogens in Patients with the Community- and Hospital-Acquired Urinary Tract Infections at a Tertiary Hospital in Somalia. Jundishapur J. Microbiol. 2020, 13, e107453. [Google Scholar] [CrossRef]
- Fernández-Martínez, M.; Ruiz del Castillo, B.; Lecea-Cuello, M.J.; Rodríguez-Baño, J.; Pascual, Á.; Martínez-Martínez, L. Prevalence of Aminoglycoside-Modifying Enzymes in Escherichia coli and Klebsiella pneumoniae Producing Extended Spectrum β-Lactamases Collected in Two Multicenter Studies in Spain. Microb. Drug Resist. 2018, 24, 367–376. [Google Scholar] [CrossRef]
- Abo-State, M.A.M.; Saleh, Y.E.S.; Ghareeb, H.M. Prevalence and sequence of aminoglycosides modifying enzymes genes among E. coli and Klebsiella species isolated from Egyptian hospitals. J. Radiat. Res. Appl. Sci. 2018, 11, 408–415. [Google Scholar]
- Arumugam, K.; Karande, G.S.; Patil, S.R. Prevalence of Carbapenemase Production Among Klebsiella and Escherichia coli Isolated from Urinary Tract Infections. Cureus 2024, 16, e70918. [Google Scholar] [CrossRef]
- Shash, R.Y.; Elshimy, A.A.; Soliman, M.Y.; Mosharafa, A.A. Molecular Characterization of Extended-Spectrum β-Lactamase Enterobacteriaceae Isolated from Egyptian Patients with Community- and Hospital-Acquired Urinary Tract Infection. Am. J. Trop. Med. Hyg. 2019, 100, 522–528. [Google Scholar] [CrossRef]
- Kaza, P.; Xavier, B.B.; Mahindroo, J.; Singh, N.; Baker, S.; Nguyen, T.N.T.; Mavuduru, R.S.; Mohan, B.; Taneja, N. Extensively Drug-Resistant Klebsiella pneumoniae Associated with Complicated Urinary Tract Infection in Northern India. Jpn. J. Infect. Dis. 2024, 77, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Chapelle, C.; Gaborit, B.; Dumont, R.; Dinh, A.; Vallée, M. Treatment of UTIs Due to Klebsiella pneumoniae Carbapenemase-Producers: How to Use New Antibiotic Drugs? A Narrative Review. Antibiotics 2021, 10, 1332. [Google Scholar] [CrossRef] [PubMed]
- Al Farsi, H.M.; Camporeale, A.; Ininbergs, K.; Al-Azri, S.; Al-Muharrmi, Z.; Al-Jardani, A.; Giske, C.G. Clinical and molecular characteristics of carbapenem non-susceptible Escherichia coli: A nationwide survey from Oman. PLoS ONE 2020, 15, e0239924. [Google Scholar] [CrossRef]
- Matsui, Y.; Hu, Y.; Rubin, J.; de Assis, R.S.; Suh, J.; Riley, L.W. Multilocus sequence typing of Escherichia coli isolates from urinary tract infection patients and from fecal samples of healthy subjects in a college community. Microbiologyopen 2020, 9, 1225–1233. [Google Scholar] [CrossRef] [PubMed]
- Doumith, M.; Day, M.; Ciesielczuk, H.; Hope, R.; Underwood, A.; Reynolds, R.; Wain, J.; Livermore, D.M.; Woodford, N. Rapid identification of major Escherichia coli sequence types causing urinary tract and bloodstream infections. J. Clin. Microbiol. 2015, 53, 160–166. [Google Scholar] [CrossRef]
- Mohamed, E.S.; Khairy, R.M.M.; Abdelrahim, S.S. Prevalence and molecular characteristics of ESBL and AmpC β -lactamase producing Enterobacteriaceae strains isolated from UTIs in Egypt. Antimicrob. Resist. Infect. Control 2020, 9, 198. [Google Scholar] [CrossRef]
- Jomehzadeh, N.; Ahmadi, K.; Rahmani, Z. Prevalence of plasmid-mediated AmpC β-lactamases among uropathogenic Escherichia coli isolates in southwestern Iran. Osong Public. Health Res. Perspect. 2021, 12, 390–395. [Google Scholar] [CrossRef] [PubMed]
- Mohamudha, P.R.; Harish, B.N.; Parija, S.C. Molecular description of plasmid-mediated AmpC β-lactamases among nosocomial isolates of Escherichia coli & Klebsiella pneumoniae from six different hospitals in India. Indian. J. Med. Res. 2012, 135, 114–119. [Google Scholar]
- Bala, R.; Singh, V.A.; Gupta, N.; Rakshit, P. Prevalence, multidrug-resistance and risk factors for AmpC β-lactamases producing Escherichia coli from hospitalized patients. J. Infect. Dev. Ctries. 2020, 14, 1466–1469. [Google Scholar] [CrossRef]
- Kamruzzaman, M.; Shoma, S.; Bari, S.M.N.; Ginn, A.N.; Wiklendt, A.M.; Partridge, S.R.; Faruque, S.M.; Iredell, J.R. Genetic diversity and antibiotic resistance in Escherichia coli from environmental surface water in Dhaka City, Bangladesh. Diagn. Microbiol. Infect. Dis. 2013, 76, 222–226. [Google Scholar] [CrossRef]
- Mata, C.; Miró, E.; Toleman, M.; Rivera, M.A.; Walsh, T.R.; Navarro, F. Association of blaDHA-1 and qnrB genes carried by broad-host-range plasmids among isolates of Enterobacteriaceae at a Spanish hospital. Clin. Microbiol. Infect. 2011, 17, 1514–1517. [Google Scholar] [CrossRef] [PubMed]
- Eftekhar, F.; Seyedpour, S.M. Prevalence of qnr and aac(6′)-Ib-cr Genes in Clinical Isolates of Klebsiella Pneumoniae from Imam Hussein Hospital in Tehran. Iran. J. Med. Sci. 2015, 40, 515–521. [Google Scholar]
- Ingti, B.; Laskar, M.A.; Choudhury, S.; Maurya, A.P.; Paul, D.; Talukdar, A.D.; Choudhury, M.D.; Dhar, D.; Chakravarty, A.; Bhattacharjee, A. Molecular and in silico analysis of a new plasmid-mediated AmpC β-lactamase (CMH-2) in clinical isolates of Klebsiella pneumoniae. Infect. Genet. Evol. 2017, 48, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Hennequin, C.; Ravet, V.; Robin, F. Plasmids carrying DHA-1 β-lactamases. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1197–1209. [Google Scholar] [CrossRef] [PubMed]
- Kao, C.Y.; Chen, J.W.; Liu, T.L.; Yan, J.J.; Wu, J.J. Comparative Genomics of Escherichia coli Sequence Type 219 Clones from the Same Patient: Evolution of the IncI1 blaCMY-Carrying Plasmid in Vivo. Front. Microbiol. 2018, 9, 1518. [Google Scholar] [CrossRef]
- Cascales, E.; Buchanan, S.K.; Duché, D.; Kleanthous, C.; Lloubès, R.; Postle, K.; Riley, M.; Slatin, S.; Cavard, D. Colicin biology. Microbiol. Mol. Biol. Rev. 2007, 71, 158–229. [Google Scholar] [CrossRef]
- AL-Quraini, M.; Rizvi, M.; AL-Jabri, Z.; Sami, H.; AL-Muzahmi, M.; AL-Muharrmi, Z.; Taneja, N.; Al-Busaidi, I.; Soman, R. Assessment of In-Vitro Synergy of Fosfomycin with Meropenem, Amikacin and Tigecycline in Whole Genome Sequenced Extended and Pan Drug Resistant Klebsiella Pneumoniae: Exploring A Colistin Sparing Protocol. Antibiotics 2022, 11, 153. [Google Scholar] [CrossRef]
- Al Fadhli, A.H.; Mouftah, S.F.; Jamal, W.Y.; Rotimi, V.O.; Ghazawi, A. Cracking the Code: Unveiling the Diversity of Carbapenem-Resistant Klebsiella pneumoniae Clones in the Arabian Peninsula through Genomic Surveillance. Antibiotics 2023, 12, 1081. [Google Scholar] [CrossRef]
- Nagaraj, G.; Shamanna, V.; Govindan, V.; Rose, S.; Sravani, D.; Akshata, K.P.; Shincy, M.R.; Venkatesha, V.T.; Abrudan, M.; Argimón, S.; et al. High-Resolution Genomic Profiling of Carbapenem-Resistant Klebsiella pneumoniae Isolates: A Multicentric Retrospective Indian Study. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2021, 73, 300–307. [Google Scholar] [CrossRef]
- Shukla, S.; Desai, S.; Bagchi, A.; Singh, P.; Joshi, M.; Joshi, C.; Patankar, J.; Maheshwari, G.; Rajni, E.; Shah, M.; et al. Diversity and Distribution of β-Lactamase Genes Circulating in Indian Isolates of Multidrug-Resistant Klebsiella pneumoniae. Antibiotics 2023, 12, 449. [Google Scholar] [CrossRef]
- Shankar, C.; Vasudevan, K.; Jacob, J.J.; Baker, S.; Isaac, B.J.; Neeravi, A.R.; Sethuvel, D.P.M.; George, B.; Veeraraghavan, B. Hybrid Plasmids Encoding Antimicrobial Resistance and Virulence Traits Among Hypervirulent Klebsiella pneumoniae ST2096 in India. Front. Cell Infect. Microbiol. 2022, 12, 875116. [Google Scholar] [CrossRef] [PubMed]
- Sundaresan, A.K.; Vincent, K.; Mohan, G.B.M.; Ramakrishnan, J. Association of sequence types, antimicrobial resistance and virulence genes in Indian isolates of Klebsiella pneumoniae: A comparative genomics study. J. Glob. Antimicrob. Resist. 2022, 30, 431–441. [Google Scholar] [CrossRef]
- Mancini, S.; Poirel, L.; Tritten, M.L.; Lienhard, R.; Bassi, C.; Nordmann, P. Emergence of an MDR Klebsiella pneumoniae ST231 producing OXA-232 and RmtF in Switzerland. J. Antimicrob. Chemother. 2018, 73, 821–823. [Google Scholar] [CrossRef]
- Di Pilato, V.; Henrici De Angelis, L.; Aiezza, N.; Baccani, I.; Niccolai, C.; Parisio, E.M.; Giordano, C.; Camarlinghi, G.; Barnini, S.; Forni, S.; et al. Resistome and virulome accretion in an NDM-1-producing ST147 sublineage of Klebsiella pneumoniae associated with an outbreak in Tuscany, Italy: A genotypic and phenotypic characterisation. Lancet Microbe 2022, 3, e224–e234. [Google Scholar] [CrossRef]
- Abid, F.B.; Tsui, C.K.M.; Doi, Y.; Deshmukh, A.; McElheny, C.L.; Bachman, W.C.; Fowler, E.L.; Albishawi, A.; Mushtaq, K.; Ibrahim, E.B.; et al. Molecular characterization of clinical carbapenem-resistant Enterobacterales from Qatar. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 1779–1785. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, L.; Jin, J.; Li, G.; Shao, H.; Song, Y.; Sun, Y.; Zhang, Y.; Cheng, J.; Li, L. Genomic Epidemiology and Characterization of Carbapenem-Resistant Klebsiella pneumoniae in ICU Inpatients in Henan Province, China: A Multicenter Cross-Sectional Study. Microbiol. Spectr. 2023, 11, e0419722. [Google Scholar] [CrossRef]
- Mathers, A.J.; Crook, D.; Vaughan, A.; Barry, K.E.; Vegesana, K.; Stoesser, N.; Parikh, H.I.; Sebra, R.; Kotay, S.; Walker, A.S.; et al. Klebsiella quasipneumoniae Provides a Window into Carbapenemase Gene Transfer, Plasmid Rearrangements, and Patient Interactions with the Hospital Environment. Antimicrob. Agents Chemother. 2019, 63, e02513-18. [Google Scholar] [CrossRef] [PubMed]
- Venkitapathi, S.; Wijesundara, Y.H.; Cornelius, S.A.; Herbert, F.C.; Gassensmith, J.J.; Zimmern, P.E.; De Nisco, N.J. Conserved FimK Truncation Coincides with Increased Expression of Type 3 Fimbriae and Cultured Bladder Epithelial Cell Association in Klebsiella quasipneumoniae. J. Bacteriol. 2022, 204, e0017222. [Google Scholar] [CrossRef]
- Shen, X.; Liu, L.; Yu, J.; Ai, W.; Cao, X.; Zhan, Q.; Guo, Y.; Wang, L.; Yu, F. High Prevalence of 16S rRNA Methyltransferase Genes in Carbapenem-Resistant Klebsiella pneumoniae Clinical Isolates Associated with Bloodstream Infections in 11 Chinese Teaching Hospitals. Infect. Drug Resist. 2020, 13, 2189–2197. [Google Scholar] [CrossRef]
- Saadatian Farivar, A.; Nowroozi, J.; Eslami, G.; Sabokbar, A. RAPD PCR Profile, Antibiotic Resistance, Prevalence of armA Gene, and Detection of KPC Enzyme in Klebsiella pneumoniae Isolates. Can. J. Infect. Dis. Med. Microbiol. 2018, 2018, 6183162. [Google Scholar] [CrossRef]
- Al Sheikh, Y.A.; Marie, M.A.M.; John, J.; Krishnappa, L.G.; Dabwab, K.H.M. Prevalence of 16S rRNA methylase genes among β-lactamase-producing Enterobacteriaceae clinical isolates in Saudi Arabia. Libyan J. Med. 2014, 9, 24432. [Google Scholar] [CrossRef] [PubMed]
- Dey, S.; Gaur, M.; Sahoo, R.K.; Das, A.; Jain, B.; Pati, S.; Subudhi, E. Genomic characterization of XDR Klebsiella pneumoniae ST147 co-resistant to carbapenem and colistin—The first report in India. J. Glob. Antimicrob. Resist. 2020, 22, 54–56. [Google Scholar] [CrossRef] [PubMed]
- Cerón, S.; Salem-Bango, Z.; Contreras, D.A.; Ranson, E.L.; Yang, S. Clinical and Genomic Characterization of Carbapenem-Resistant Klebsiella pneumoniae with Concurrent Production of NDM and OXA-48-like Carbapenemases in Southern California, 2016–2022. Microorganisms 2023, 11, 1717. [Google Scholar] [CrossRef] [PubMed]
- Boonyasiri, A.; Jauneikaite, E.; Brinkac, L.M.; Greco, C.; Lerdlamyong, K.; Tangkoskul, T.; Nguyen, K.; Thamlikitkul, V.; Fouts, D.E. Genomic and clinical characterisation of multidrug-resistant carbapenemase-producing ST231 and ST16 Klebsiella pneumoniae isolates colonising patients at Siriraj hospital, Bangkok, Thailand from 2015 to 2017. BMC Infect. Dis. 2021, 21, 142. [Google Scholar] [CrossRef]
- Piccirilli, A.; Cherubini, S.; Azzini, A.M.; Tacconelli, E.; Lo Cascio, G.; Maccacaro, L.; Bazaj, A.; Naso, L.; Amicosante, G.; Ltcf-Veneto Working Group; et al. Whole-Genome Sequencing (WGS) of Carbapenem-Resistant K. pneumoniae Isolated in Long-Term Care Facilities in the Northern Italian Region. Microorganisms 2021, 9, 1985. [Google Scholar] [CrossRef]
- Zeng, Y.; Li, W.; Zhao, M.; Li, J.; Liu, X.; Shi, L.; Yang, X.; Xia, H.; Yang, S.; Yang, L. The association between ambient temperature and antimicrobial resistance of Klebsiella pneumoniae in China: A difference-in-differences analysis. Front. Public. Health 2023, 11, 1158762. [Google Scholar] [CrossRef]
- Paskova, V.; Medvecky, M.; Skalova, A.; Chudejova, K.; Bitar, I.; Jakubu, V.; Bergerova, T.; Zemlickova, H.; Papagiannitsis, C.C.; Hrabak, J. Characterization of NDM-Encoding Plasmids from Enterobacteriaceae Recovered from Czech Hospitals. Front. Microbiol. 2018, 9, 1549. [Google Scholar] [CrossRef]
- Shen, S.; Han, R.; Yin, D.; Jiang, B.; Ding, L.; Guo, Y.; Wu, S.; Wang, C.; Zhang, H.; Hu, F. A Nationwide Genomic Study of Clinical Klebsiella pneumoniae Carrying blaOXA-232 and rmtF in China. Microbiol. Spectr. 2023, 11, e03863-22. [Google Scholar] [CrossRef]
- Spadar, A.; Phelan, J.; Elias, R.; Modesto, A.; Caneiras, C.; Marques, C.; Lito, L.; Pinto, M.; Cavaco-Silva, P.; Ferreira, H.; et al. Genomic epidemiological analysis of Klebsiella pneumoniae from Portuguese hospitals reveals insights into circulating antimicrobial resistance. Sci. Rep. 2022, 12, 13791. [Google Scholar] [CrossRef]
| Antibiotics | E. coli | K. pneumoniae | ||
|---|---|---|---|---|
| Number of Isolates Tested | Susceptibility% | Number of Isolates Tested | Susceptibility% | |
| Nitrofurantoin | 404 | 96% | 157 | 38% |
| Cotrimoxazole | 408 | 63% | 162 | 65% |
| Fosfomycin | 330 | 100% | 134 | 89% |
| Ampicillin | 408 | 29% | - | - |
| Cefazolin | 256 | 27% | 92 | 33% |
| Cefuroxime | 402 | 51% | 161 | 55% |
| Ceftriaxone | 407 | 54% | 161 | 61% |
| Ceftazidime | 406 | 57% | 162 | 60% |
| Cefepime | 407 | 56% | 163 | 63% |
| Amoxicillin-clavulanate | 152 | 66% | 77 | 65% |
| Ampicillin-sulbactam | 249 | 65% | 103 | 52% |
| Piperacillin-tazobactam | 408 | 95% | 163 | 72% |
| Gentamicin | 408 | 87% | 163 | 80% |
| Amikacin | 407 | 98% | 163 | 80% |
| Ciprofloxacin | 347 | 44% | 133 | 47% |
| Levofloxacin | 357 | 46% | 136 | 48% |
| Ertapenem | 371 | 98% | 146 | 81% |
| Imipenem | 408 | 98% | 163 | 83% |
| Meropenem | 406 | 99% | 163 | 84% |
| Minocycline | 270 | 87% | 98 | 61% |
| Phenotype | E. coli, n (%) | K. pneumoniae, n (%) | Total Prevalence Among E. coli (n = 406) and K. pneumoniae (n = 163) |
|---|---|---|---|
| ESBL | 164 (40.4%) | 33 (20.2%) | 197 (34.6%) |
| AmpC | 10 (2.5%) | 6 (3.7%) | 16 (2.8%) |
| ESBL + AmpC | 14 (3.4%) | 1 (0.6%) | 15 (2.6%) |
| CRE | 7 (1.7%) | 28 (17.2%) | 35 (6.2%) |
| Total | 195 | 68 | 263 |
| Isolate | Aminoglycoside | Macrolide | Quinolone | Folate Pathway Antagonist | Tetracycline | Beta-Lactam | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AmpC | ESBL | blaOXA | blaTEM | ||||||||||||||||||
| aac(3)-IIa | aadA5 | aph(3″)-Ib | aph(6)-Id | mph(A) | aac(6′)-Ib-cr | qnrB4 | qnrS1 | dfrA7 | dfrA14 | dfrA17 | sul1 | sul2 | tet(A) | Tet(B) | blaDHA-1 | blaCTX-M-15 | blaOXA-1 | blaTEM-1B | blaTEM-35 | ||
| 0410 | |||||||||||||||||||||
| 7659 | |||||||||||||||||||||
| 7826 | |||||||||||||||||||||
| 8268 | |||||||||||||||||||||
| 0495 | |||||||||||||||||||||
| 0539 | |||||||||||||||||||||
| 0402 | |||||||||||||||||||||
| 2536 | |||||||||||||||||||||
| 8100 | |||||||||||||||||||||
| 9044 | |||||||||||||||||||||
| 4976 | |||||||||||||||||||||
| Isolate | MLST | Aminoglycoside | Quinolone | Folate Pathway Antagonist | Tetracycline | Beta-Lactam | Fosfomycin | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aac(6′)-Ib | aadA1 | aadA2 | aph(3′)-Ia | aph(3′)-VI | armA | rmtF | aac(6′)-Ib-cr | qnrA | qnrB | qnrS | OqxA | OqxB | dfrA1 | dfrA5 | dfrA12 | dfrA14 | sul1 | sul2 | tet(A) | tet(D) | blaSHV-28 | blaSHV-106 | blaSHV-40 | blaSHV-56 | blaSHV-89 | blaSHV-11 | blaSHV-67 | blaTEM-1 | blaCTX-M-8 | blaCTX-M-15 | blaDHA-1 | blaOXA-1 | blaOXA-9 | blaOXA-48 | blaOXA-232 | blaNDM-5 | blaKPC-2 | blaOKP-B-3 | fosA5 | fosA6 | ||
| 4494 | ST-2096 | |||||||||||||||||||||||||||||||||||||||||
| 1395 | ST-2096 | |||||||||||||||||||||||||||||||||||||||||
| 4590 | ST-2096 | |||||||||||||||||||||||||||||||||||||||||
| 9002 | ST-2096 | |||||||||||||||||||||||||||||||||||||||||
| 4787 | ST-2096 | |||||||||||||||||||||||||||||||||||||||||
| 5038 | ST-2096 | |||||||||||||||||||||||||||||||||||||||||
| 0821 | ST-2096 | |||||||||||||||||||||||||||||||||||||||||
| 2541 | ST-2096 | |||||||||||||||||||||||||||||||||||||||||
| 2542 | ST-2096 | |||||||||||||||||||||||||||||||||||||||||
| 9590 | ST-2096 | |||||||||||||||||||||||||||||||||||||||||
| 8474 | ST-2096 | |||||||||||||||||||||||||||||||||||||||||
| 8637 | ST-2096 | |||||||||||||||||||||||||||||||||||||||||
| 1502 | ST-231 | |||||||||||||||||||||||||||||||||||||||||
| 0403 | ST-147 | |||||||||||||||||||||||||||||||||||||||||
| 2967 | ST-147 | |||||||||||||||||||||||||||||||||||||||||
| 0350 | ST-147 | |||||||||||||||||||||||||||||||||||||||||
| 4279 | ST-147 | |||||||||||||||||||||||||||||||||||||||||
| 6112 | ST-147 | |||||||||||||||||||||||||||||||||||||||||
| 7117 | ST-147 | |||||||||||||||||||||||||||||||||||||||||
| 0845 | ST-147 | |||||||||||||||||||||||||||||||||||||||||
| 2579 | ST-1770 | |||||||||||||||||||||||||||||||||||||||||
| 1776 | ST-111 | |||||||||||||||||||||||||||||||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
AL Shizawi, N.; AL Jabri, Z.; Khan, F.; Sami, H.; AL Siyabi, T.; AL Muharrmi, Z.; Sirasanagandla, S.R.; Rizvi, M. Mapping Antimicrobial Resistance in Escherichia coli and Klebsiella pneumoniae from Complicated Urinary Tract Infections in Oman: Phenotypic and Genotypic Insights. Diagnostics 2025, 15, 1062. https://doi.org/10.3390/diagnostics15091062
AL Shizawi N, AL Jabri Z, Khan F, Sami H, AL Siyabi T, AL Muharrmi Z, Sirasanagandla SR, Rizvi M. Mapping Antimicrobial Resistance in Escherichia coli and Klebsiella pneumoniae from Complicated Urinary Tract Infections in Oman: Phenotypic and Genotypic Insights. Diagnostics. 2025; 15(9):1062. https://doi.org/10.3390/diagnostics15091062
Chicago/Turabian StyleAL Shizawi, Nawal, Zaaima AL Jabri, Fatima Khan, Hiba Sami, Turkiya AL Siyabi, Zakariya AL Muharrmi, Srinivasa Rao Sirasanagandla, and Meher Rizvi. 2025. "Mapping Antimicrobial Resistance in Escherichia coli and Klebsiella pneumoniae from Complicated Urinary Tract Infections in Oman: Phenotypic and Genotypic Insights" Diagnostics 15, no. 9: 1062. https://doi.org/10.3390/diagnostics15091062
APA StyleAL Shizawi, N., AL Jabri, Z., Khan, F., Sami, H., AL Siyabi, T., AL Muharrmi, Z., Sirasanagandla, S. R., & Rizvi, M. (2025). Mapping Antimicrobial Resistance in Escherichia coli and Klebsiella pneumoniae from Complicated Urinary Tract Infections in Oman: Phenotypic and Genotypic Insights. Diagnostics, 15(9), 1062. https://doi.org/10.3390/diagnostics15091062







