Genome and Transcriptome Analyses Facilitate Genetic Control of Wohlfahrtia magnifica, a Myiasis-Causing Flesh Fly
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome Resources of W. magnifica
2.2. W. magnifica Sample Collection
2.3. RNA Isolation and Assessment
2.4. Illumina RNA-Seq Library Construction, Sequencing and Data Filtering
2.5. PacBio Iso-Seq Library Construction, Sequencing, and Data Processing
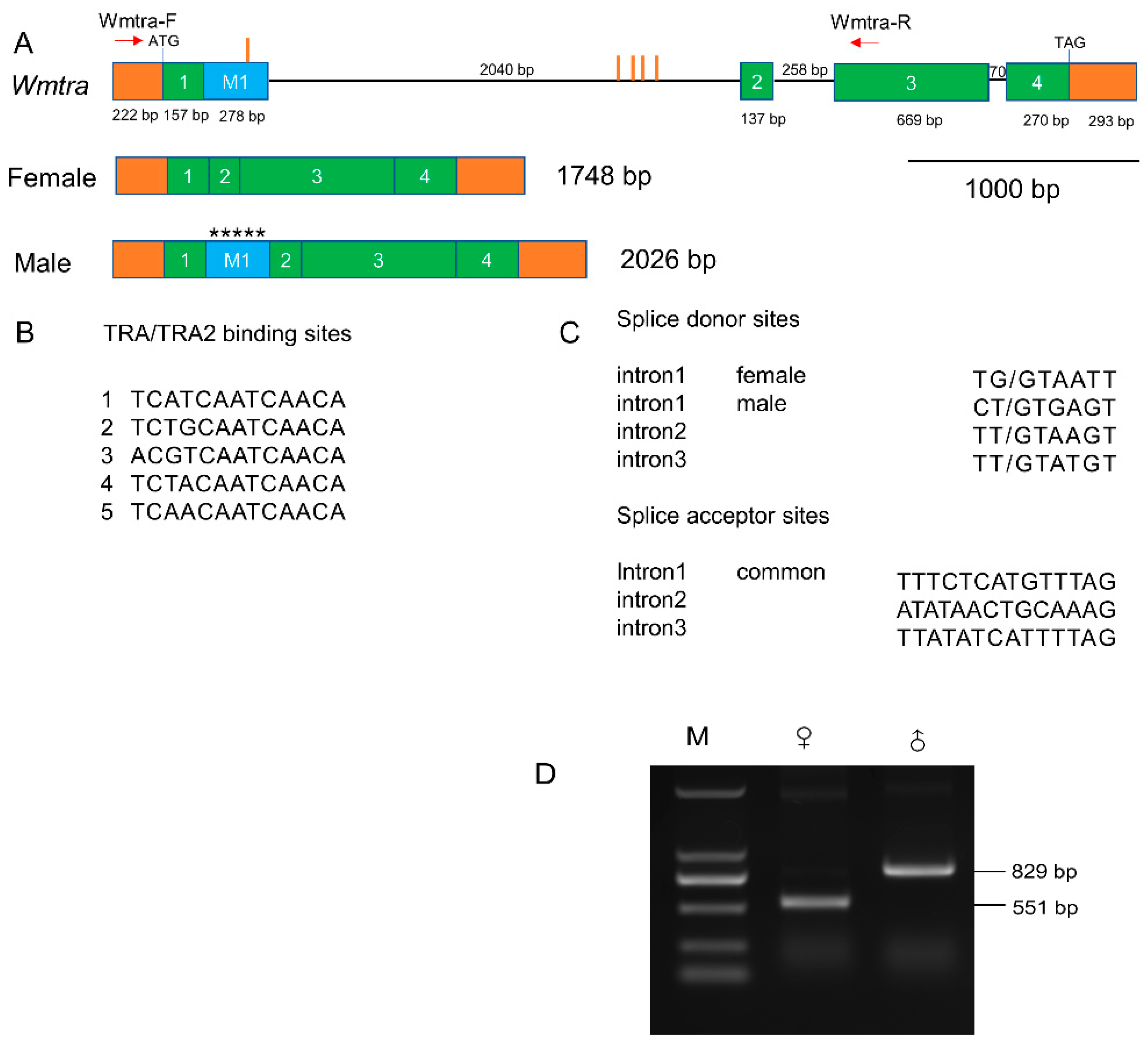
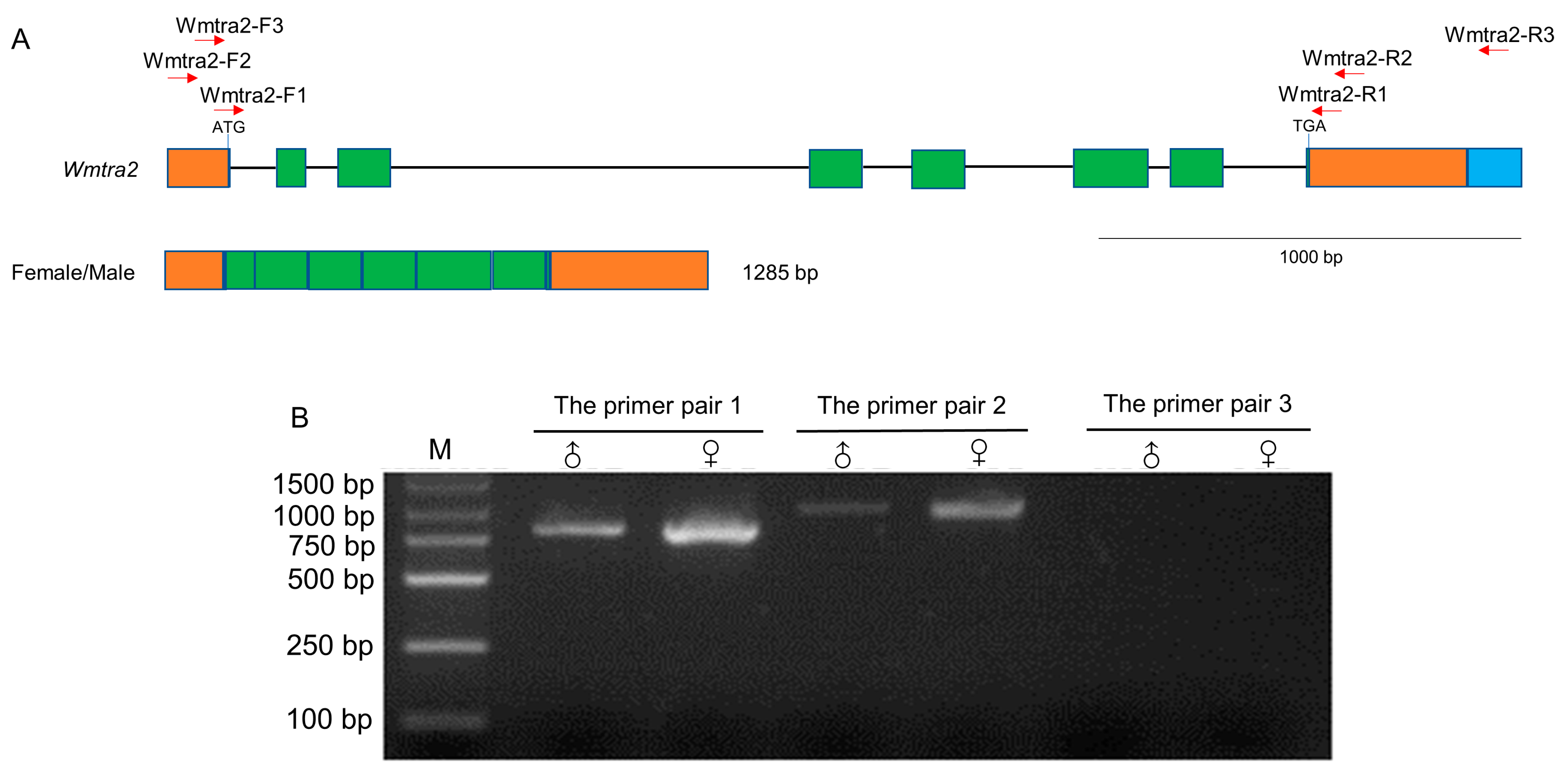
2.6. Isolation of the Wmtra and Wmtra2 Genes
2.7. Reverse Transcription Polymerase Chain Reaction (RT-PCR) Validation
2.8. Sequence Analysis
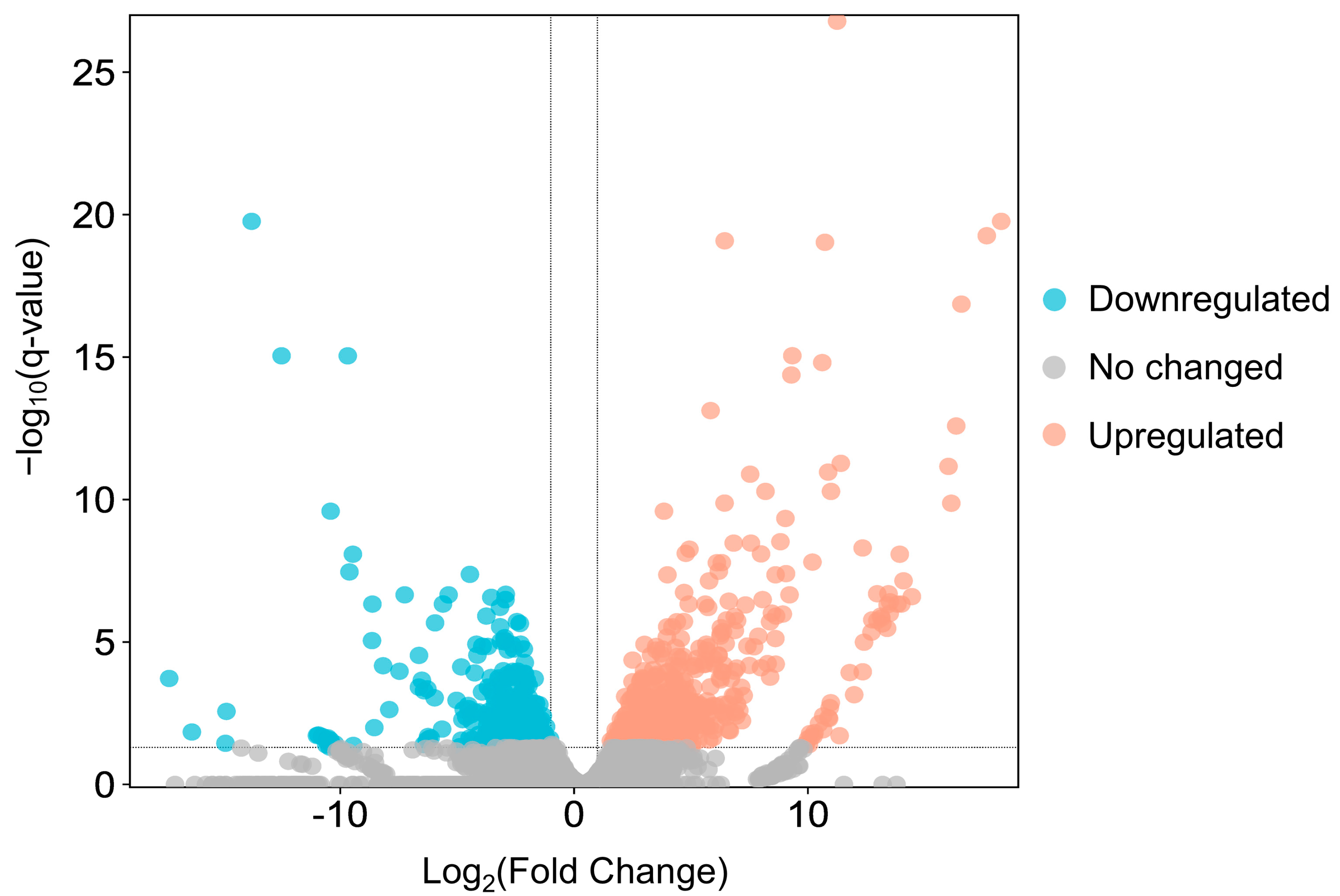
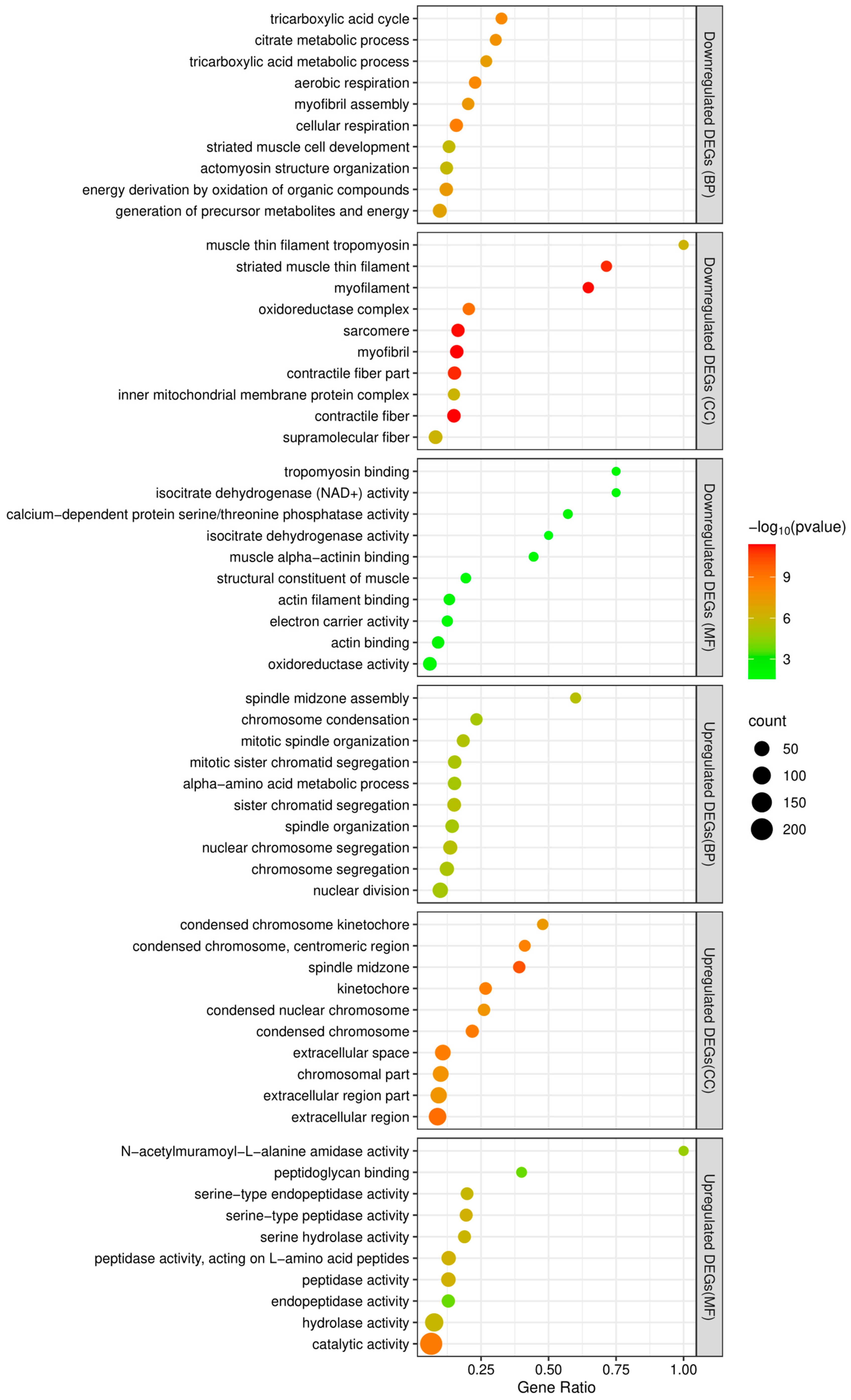
2.9. Identification of Differentially Expressed Genes (DEGs)
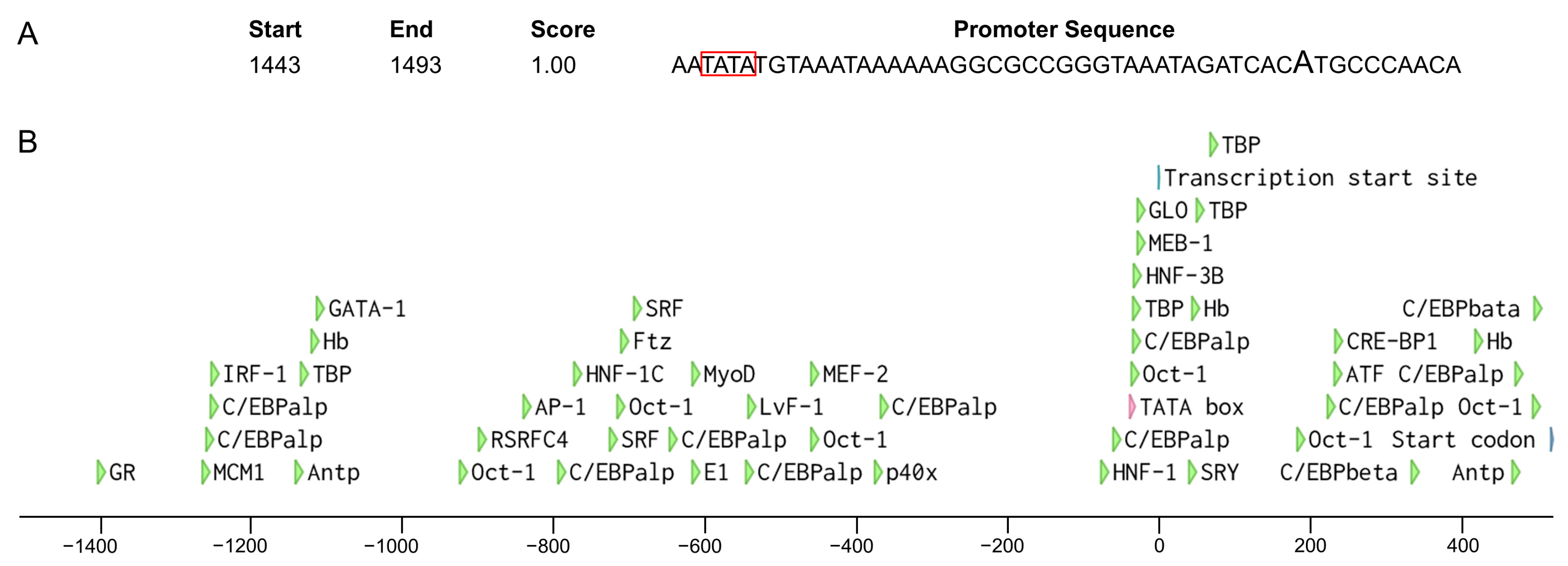
2.10. Promoter Analysis
2.11. Identification of Target Genes against W. magnifica Infestation
3. Results
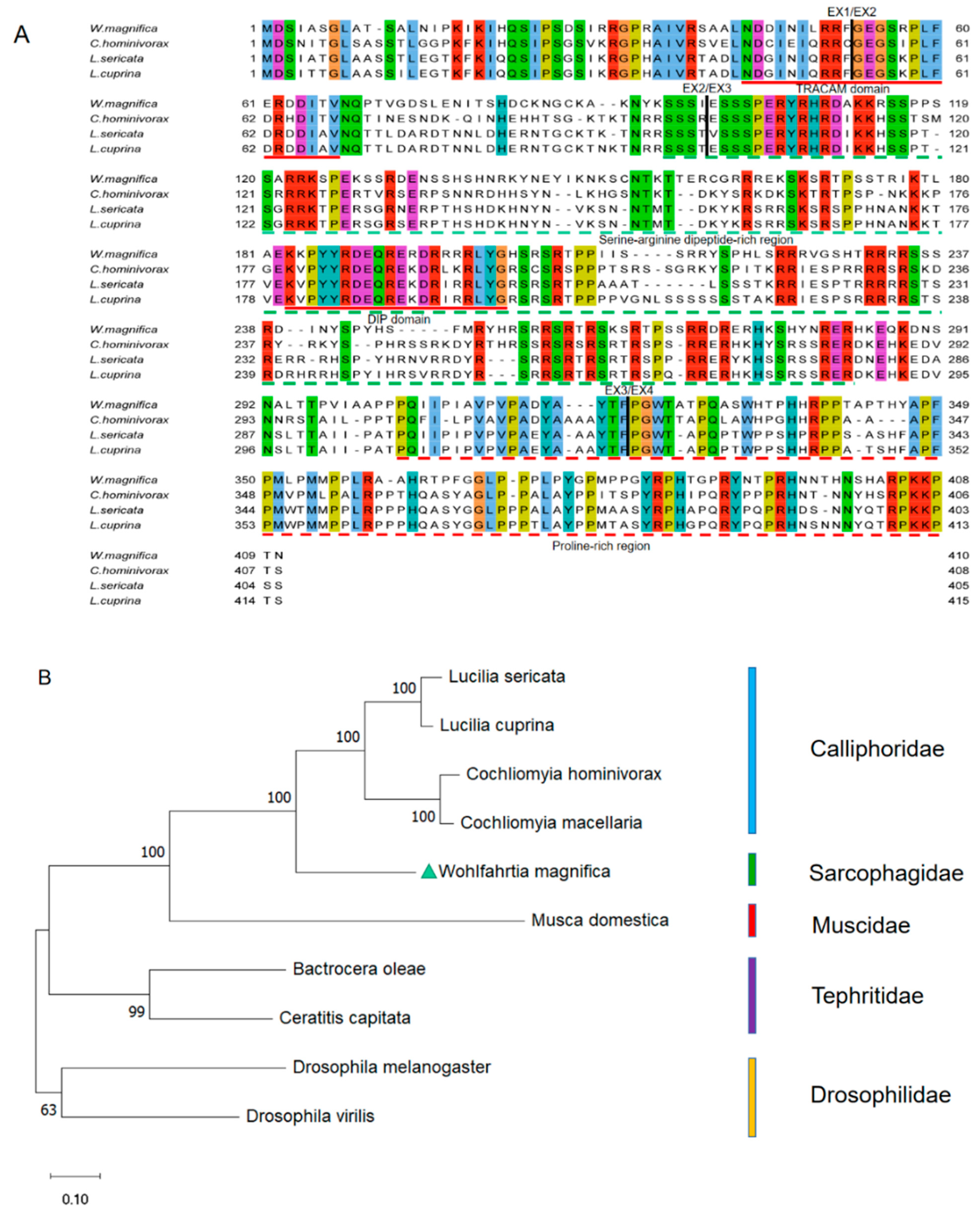
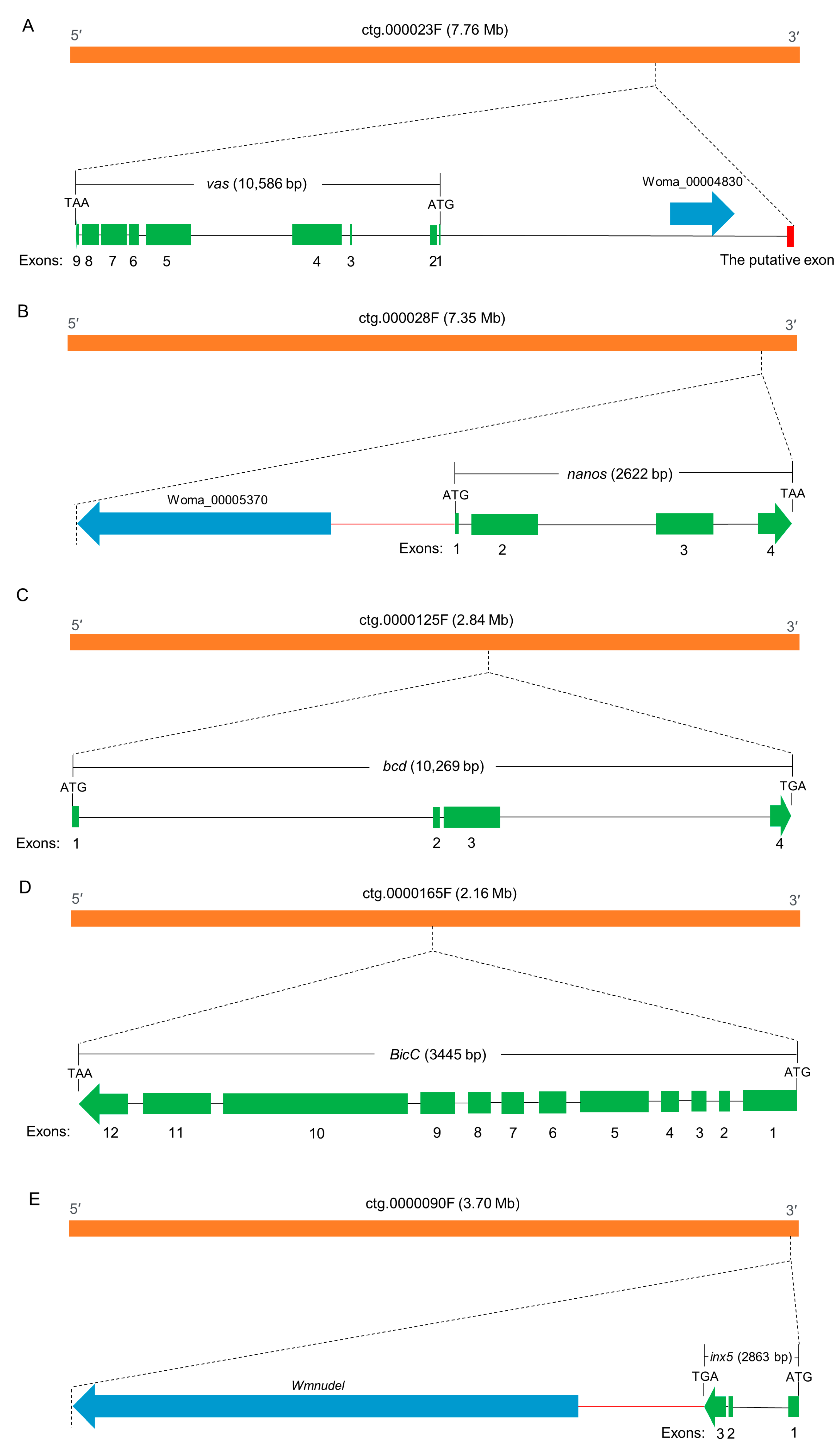
3.1. Isolation and Characterization of the Wmtra Gene
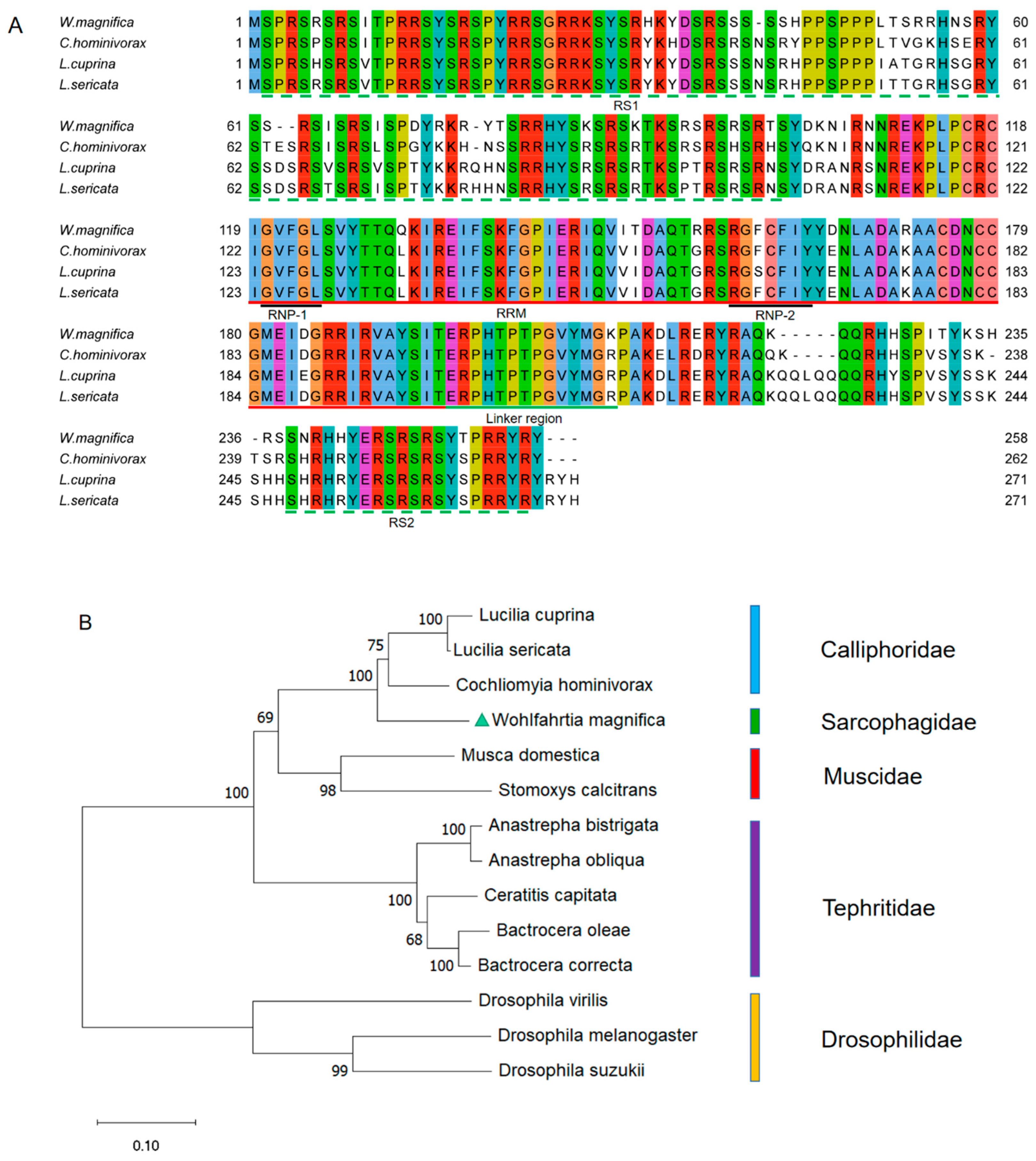
3.2. Isolation and Characterization of the Wmtra2 Gene
3.3. Gene Expression Analysis between Adult Females and Adult Males
3.4. Candidate Genes for Cas9-Based Homing Gene Drive
3.5. Potential Target Genes for Control Strategies against W. magnifica
4. Discussion
4.1. Acquisition of Full-Length Sequences of Transcripts
4.2. The Putative Sex Determination Mechanism of W. magnifica
4.3. Genetic Controls against W. magnifica Infestation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Farkas, R.; Képes, G.Y. Traumatic myiasis of horses caused by Wohlfahrtia magnifica. Acta Vet. Hung. 2001, 49, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Zhang, M.; Tang, L.; Ente, M.; Ma, X.; Chu, H.; Li, K.; Hu, D.; Zhang, D. First reports of nasal and traumatic myiasis infection in endangered Przewalski’s horses (Equus ferus przewalskii). Int. J. Parasitol. Parasites Wildl. 2019, 9, 21–24. [Google Scholar] [CrossRef] [PubMed]
- Dehghani, R.; Zarghi, I.; Sayyedi, H.R. Genital myiasis of a sheep by Wohlfahrtia magnifica, in Ghamsar, Kashan, Iran. Bangladesh J. Med. Sci. 2014, 13, 332–335. [Google Scholar] [CrossRef] [Green Version]
- Farkas, R.; Hall, M.J.R.; Daniel, M.; Börzsönyi, L. Efficacy of ivermectin and moxidectin injection against larvae of Wohlfahrtia magnifica (Diptera: Sarcophagidae) in sheep. Parasitol. Res. 1996, 82, 82–86. [Google Scholar] [CrossRef]
- Moshaverinia, A.; Moghaddas, E.; Maleki, M.; Borji, H. Gingival myiasis of camel (Camelus dromedarius) caused by Wohlfahrtia magnifica. Sci. Parasitol. 2013, 14, 85–87. [Google Scholar]
- Valentin, A.; Baumann, M.P.O.; Schein, E.; Bajanbileg, S. Genital myiasis (Wohlfahrtiosis) in camel herds of Mongolia. Vet. Parasitol. 1997, 73, 335–346. [Google Scholar] [CrossRef]
- Çiftçiŏglu, N.; Altintaş, K.; Haberal, M. A case of human orotracheal myiasis caused by Wohlfahrtia magnifica. Parasitol. Res. 1996, 83, 34–36. [Google Scholar] [CrossRef]
- Kokcam, I.; Saki, C.E. A case of cutaneous myiasis caused by Wohlfahrtia magnifica. J. Dermatol. 2005, 32, 459–463. [Google Scholar] [CrossRef]
- Farkas, R.; Hall, M.J.R.; Kelemen, F. Wound myiasis of sheep in Hungary. Vet. Parasitol. 1997, 69, 133–144. [Google Scholar] [CrossRef]
- Gaglio, G.; Brianti, E.; Abbene, S.; Giannetto, S. Genital myiasis by Wohlfahrtia magnifica (Diptera, Sarcophagidae) in Sicily (Italy). Parasitol. Res. 2011, 109, 1471–1474. [Google Scholar] [CrossRef]
- Giangaspero, A.; Traversa, D.; Trentini, R.; Scala, A.; Otranto, D. Traumatic myiasis by Wohlfahrtia magnifica in Italy. Vet. Parasitol. 2011, 175, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.J.R.; Testa, J.M.; Smith, L.; Adams, Z.J.O.; Khallaayoune, K.; Sotiraki, S.; Stefanakis, A.; Farkas, R.; Ready, P.D. Molecular genetic analysis of populations of Wohlfahrt’s wound myiasis fly, Wohlfahrtia magnifica, in outbreak populations from Greece and Morocco. Med. Vet. Entomol. 2009, 23, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Ruiz Martínez, I.; Leclercq, M. Data on distribution of screwworm fly Wohlfahrtia magnifica (Schiner) in southwestern Europe (Diptera: Sarcophagidae). Notes Fauniques Gembloux 1994, 28, 53–60. [Google Scholar]
- Sotiraki, S.; Farkas, R.; Hall, M.J.R. Fleshflies in the flesh: Epidemiology, population genetics and control of outbreaks of traumatic myiasis in the Mediterranean Basin. Vet. Parasitol. 2010, 174, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, M. Morphology of the Larva of Wohlfahrtia magnifica Schin. Found in a Wound on a Camel in Inner Mongolia. J. Chosen Nat. Hist. Soc. 1940, 7, 27–36. [Google Scholar]
- Farkas, R.; Hall, M.J.R.; Bouzagou, A.K.; Lhor, Y.; Khallaayoune, K. Traumatic myiasis in dogs caused by Wohlfahrtia magnifica and its importance in the epidemiology of wohlfahrtiosis of livestock. Med. Vet. Entomol. 2009, 23, 80–85. [Google Scholar] [CrossRef]
- Diakakis, N.; Papadopoulos, E.; Hall, M.J.R.; Desiris, A. Post-traumatic complication due to Wohlfahrtia magnifica larvae on a horse. Vet. Rec. 2006, 158, 170–172. [Google Scholar] [CrossRef]
- İpek, D.N.S.; Şaki, C.E.; Çay, M. The investigation of lipid peroxidation, anti-oxidant levels and some hematological parameters in sheep naturally infested with Wohlfahrtia magnifica larvae. Vet. Parasitol. 2012, 187, 112–118. [Google Scholar] [CrossRef]
- Martinez, R.I.; Cruz, S.M.D.; Rodriguez, R.; Lopez, D.M.; Parra, M.S.; Navio, F.A. Myiasis caused by wohlfahrt/a magnjfica in southern spain. Isr. J. Vet. Med. 1987, 43, 34–41. [Google Scholar]
- Remesar, S.; Otero, J.L.; Panadero, R.; Díez-Baños, P.; Díaz, P.; García-Díos, D.; Martínez-Calabuig, N.; Morrondo, M.P.; López, C. Traumatic myiasis by Wohlfahrtia magnifica in sheep flocks from southeastern Spain: Prevalence and risk factors. Med. Vet. Entomol. 2022, 36, 30–37. [Google Scholar] [CrossRef]
- Liu, J.; Hou, B.; Wuen, J.; Jiang, N.; Gao, T.; Hasi, S. Epidemiological Investigation on Genital Myiasis of Bactrian Camels in Parts of Inner Mongolia, China. J. Camel Pract. Res. 2022, 29, 229–235. [Google Scholar] [CrossRef]
- Sandeman, R.M.; Levot, G.W.; Heath, A.C.G.; James, P.J.; Greeff, J.C.; Scott, M.J.; Batterhamg, P.; Bowles, V.M. Control of the sheep blowfly in Australia and New Zealand–are we there yet? Int. J. Parasitol. 2014, 44, 879–891. [Google Scholar] [CrossRef] [PubMed]
- Bushland, R.C.; Lindquist, A.W.; Knipling, E.F. Eradication of screw-worms through release of sterilized males. Science 1955, 122, 287–288. [Google Scholar] [CrossRef]
- Knipling, E.F. Possibilities of insect control or eradication through the use of sexually sterile males. J. Econ. Entomol. 1955, 48, 459–462. [Google Scholar] [CrossRef]
- Wyss, J.H. Screwworm eradication in the Americas. Ann. N. Y. Acad. Sci. 2006, 916, 186–193. [Google Scholar] [CrossRef]
- Scott, M.J.; Concha, C.; Welch, J.B.; Phillips, P.L.; Skoda, S.R. Review of research advances in the screwworm eradication program over the past 25 years. Entomol. Exp. Appl. 2017, 164, 226–236. [Google Scholar] [CrossRef] [Green Version]
- Rendón, P.; McInnis, D.; Lance, D.; Stewart, J. Medfly (Diptera: Tephritidae) genetic sexing: Large-scale field comparison of males-only and bisexual sterile fly releases in Guatemala. J. Econ. Entomol. 2004, 97, 1547–1553. [Google Scholar] [CrossRef]
- Concha, C.; Yan, Y.; Arp, A.; Quilarque, E.; Sagel, A.; de León, A.P.; McMillan, W.O.; Skoda, S.; Scott, M.J. An early female lethal system of the New World screwworm, Cochliomyia hominivorax, for biotechnology-enhanced SIT. BMC Genet. 2020, 21, 143. [Google Scholar] [CrossRef]
- Yan, Y.; Scott, M.J. Building a transgenic sexing strain for genetic control of the Australian sheep blow fly Lucilia cuprina using two lethal effectors. BMC Genet. 2020, 21, 141. [Google Scholar] [CrossRef]
- Kyrou, K.; Hammond, A.M.; Galizi, R.; Kranjc, N.; Burt, A.; Beaghton, A.K.; Nolan, T.; Crisanti, A. A CRISPR-Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol. 2018, 36, 1062–1066. [Google Scholar] [CrossRef] [Green Version]
- Hammond, A.; Galizi, R.; Kyrou, K.; Simoni, A.; Siniscalchi, C.; Katsanos, D.; Gribble, M.; Baker, D.; Marois, E.; Russell, S.; et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 2016, 34, 78–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carballar-Lejarazú, R.; Ogaugwu, C.; Tushar, T.; Kelsey, A.; Pham, T.B.; Murphy, J.; Schmidt, H.; Lee, Y.; Lanzaro, G.C.; James, A.A. Next-generation gene drive for population modification of the malaria vector mosquito, Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2020, 117, 22805–22814. [Google Scholar] [CrossRef] [PubMed]
- Gantz, V.M.; Jasinskiene, N.; Tatarenkova, O.; Fazekas, A.; Macias, V.M.; Bier, E.; James, A.A. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl. Acad. Sci. USA 2015, 112, E6736–E6743. [Google Scholar] [CrossRef] [PubMed]
- Reid, W.; Williams, A.E.; Sanchez-Vargas, I.; Lin, J.; Juncu, R.; Olson, K.E.; Franz, A.W. Assessing single-locus CRISPR/Cas9-based gene drive variants in the mosquito Aedes aegypti via single-generation crosses and modeling. G3-Genes Genom Genet. 2022, 12, jkac280. [Google Scholar] [CrossRef] [PubMed]
- Carrami, E.M.; Eckermann, K.N.; Ahmed, H.M.; Sánchez, C.H.M.; Dippel, S.; Marshall, J.M.; Wimmer, E.A. Consequences of resistance evolution in a Cas9-based sex conversion-suppression gene drive for insect pest management. Proc. Natl. Acad. Sci. USA 2018, 115, 6189–6194. [Google Scholar] [CrossRef] [Green Version]
- Jia, Z.; Hasi, S.; Vogl, C.; Burger, P.A. Genomic insights into evolution and control of Wohlfahrtia magnifica, a widely distributed myiasis-causing fly of warm-blooded vertebrates. Mol. Ecol. Resour. 2022, 22, 2744–2757. [Google Scholar] [CrossRef]
- Bushnell, B. BBTools: A Suite of Fast, Multithreaded Bioinformatics Tools Designed for Analysis of DNA and RNA Sequence Data; Joint Genome Institute: Berkeley, CA, USA, 2018. [Google Scholar]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [Green Version]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Tamura, K.; Nei, M. MEGA: Molecular evolutionary genetics analysis software for microcomputers. Bioinformatics 1994, 10, 189–191. [Google Scholar] [CrossRef] [Green Version]
- Scott, M.J.; Benoit, J.B.; Davis, R.J.; Bailey, S.T.; Varga, V.; Martinson, E.O.; Hickner, P.V.; Syed, Z.; Cardoso, G.A.; Torres, T.T.; et al. Genomic analyses of a livestock pest, the New World screwworm, find potential targets for genetic control programs. Commun. Biol. 2020, 3, 1–14. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anstead, C.A.; Korhonen, P.K.; Young, N.D.; Hall, R.S.; Jex, A.R.; Murali, S.C.; Hughes, D.S.T.; LEE, S.F.; Perry, T.; Stroehlein, A.J.; et al. Lucilia cuprina genome unlocks parasitic fly biology to underpin future interventions. Nat. Commun. 2015, 6, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Vensko, S.P.; Belikoff, E.J.; Scott, M.J. Conservation and sex-specific splicing of the transformer gene in the Calliphorids Cochliomyia hominivorax, Cochliomyia macellaria and Lucilia sericata. PLoS ONE 2013, 8, e56303. [Google Scholar] [CrossRef] [Green Version]
- Concha, C.; Scott, M.J. Sexual development in Lucilia cuprina (Diptera, Calliphoridae) is controlled by the transformer gene. Genetics 2009, 182, 785–798. [Google Scholar] [CrossRef] [Green Version]
- Davis, R.J.; Belikoff, E.J.; Dickey, A.N.; Scholl, E.H.; Benoit, J.B.; Scott, M.J. Genome and transcriptome sequencing of the green bottle fly, Lucilia sericata, reveals underlying factors of sheep flystrike and maggot debridement therapy. Genomics 2021, 113, 3978–3988. [Google Scholar] [CrossRef]
- Hediger, M.; Henggeler, C.; Meier, N.; Perez, R.; Saccone, G.; Bopp, D. Molecular characterization of the key switch F provides a basis for understanding the rapid divergence of the sex-determining pathway in the housefly. Genetics 2010, 184, 155–170. [Google Scholar] [CrossRef] [Green Version]
- Verhulst, E.C.; van de Zande, L.; Beukeboom, L.W. Insect sex determination: It all evolves around transformer. Curr. Opin. Genet. Dev. 2010, 20, 376–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geuverink, E.; Beukeboom, L.W. Phylogenetic distribution and evolutionary dynamics of the sex determination genes doublesex and transformer in insects. Sex Dev. 2014, 8, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Manley, J.L.; Tacke, R. SR proteins and splicing control. Genes Dev. 1996, 10, 1569–1579. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, A.; Aoki, F.; Suzuki, M.G. Conserved domains in the transformer protein act complementary to regulate sex-specific splicing of its own pre-mRNA. Sex Dev. 2018, 12, 180–190. [Google Scholar] [CrossRef]
- Pane, A.; Salvemini, M.; Bovi, P.D.; Polito, C.; Saccone, G. The transformer gene in Ceratitis capitata provides a genetic basis for selecting and remembering the sexual fate. Development 2002, 129, 3715–3725. [Google Scholar] [CrossRef]
- Kandul, N.P.; Belikoff, E.J.; Liu, J.; Buchman, A.; Li, F.; Yamamoto, A.; Yang, T.; Shriner, I.; Scott, M.J.; Akbari, O.S. Genetically Encoded CRISPR Components Yield Efficient Gene Editing in the Invasive Pest Drosophila suzukii. CRISPR J. 2021, 4, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.H.; Huang, H.; Ward, C.M.; Su, J.T.; Schaeffer, L.V.; Guo, M.; Hay, B.A. A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science 2007, 316, 597–600. [Google Scholar] [CrossRef]
- Champer, J.; Yang, E.; Lee, E.; Liu, J.; Clark, A.G.; Messer, P.W. A CRISPR homing gene drive targeting a haplolethal gene removes resistance alleles and successfully spreads through a cage population. Proc. Natl. Acad. Sci. USA 2020, 117, 24377–24383. [Google Scholar] [CrossRef]
- Yadav, A.K.; Butler, C.; Yamamoto, A.; Patil, A.A.; Lloyd, A.L.; Scott, M.J. CRISPR/Cas9-based split homing gene drive targeting doublesex for population suppression of the global fruit pest Drosophila suzukii. Proc. Natl. Acad. Sci. USA 2023, 120, e2301525120. [Google Scholar] [CrossRef]
- Bottino-Rojas, V.; James, A.A. Use of Insect Promoters in Genetic Engineering to Control Mosquito-Borne Diseases. Biomolecules 2023, 13, 16. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, Z.; Hasi, S.; Zhan, D.; Hou, B.; Vogl, C.; Burger, P.A. Genome and Transcriptome Analyses Facilitate Genetic Control of Wohlfahrtia magnifica, a Myiasis-Causing Flesh Fly. Insects 2023, 14, 620. https://doi.org/10.3390/insects14070620
Jia Z, Hasi S, Zhan D, Hou B, Vogl C, Burger PA. Genome and Transcriptome Analyses Facilitate Genetic Control of Wohlfahrtia magnifica, a Myiasis-Causing Flesh Fly. Insects. 2023; 14(7):620. https://doi.org/10.3390/insects14070620
Chicago/Turabian StyleJia, Zhipeng, Surong Hasi, Deng Zhan, Bin Hou, Claus Vogl, and Pamela A. Burger. 2023. "Genome and Transcriptome Analyses Facilitate Genetic Control of Wohlfahrtia magnifica, a Myiasis-Causing Flesh Fly" Insects 14, no. 7: 620. https://doi.org/10.3390/insects14070620
APA StyleJia, Z., Hasi, S., Zhan, D., Hou, B., Vogl, C., & Burger, P. A. (2023). Genome and Transcriptome Analyses Facilitate Genetic Control of Wohlfahrtia magnifica, a Myiasis-Causing Flesh Fly. Insects, 14(7), 620. https://doi.org/10.3390/insects14070620






