A New, Non-Invasive Methodology for the Molecular Identification of Adult Sarcophagidae from Collections
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
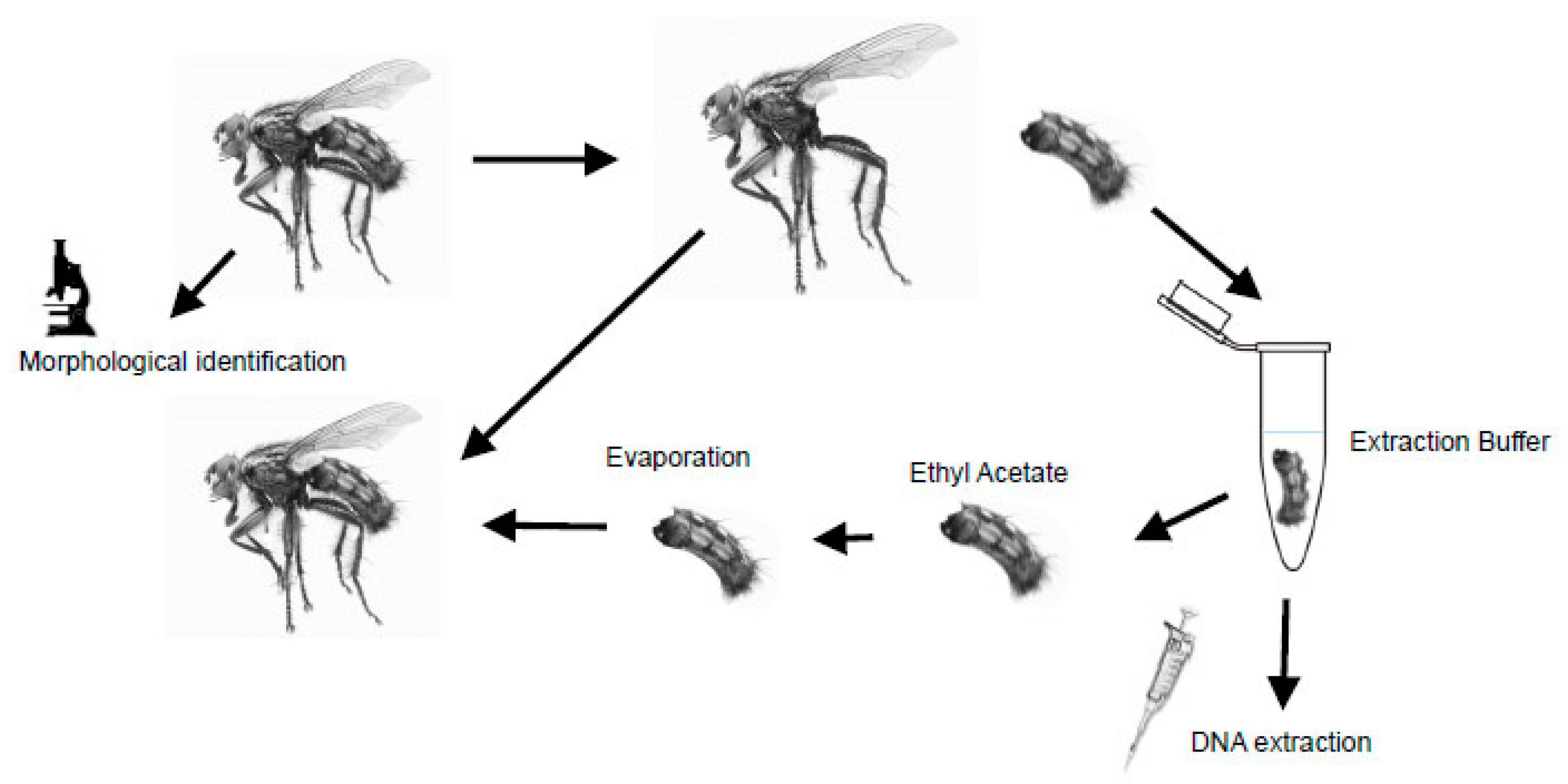
2.1. Specimen Preparation
2.2. DNA Extraction
- Twenty-five specimens were extracted using the QIAamp® DNA Mini kit. DNA was eluted in 200 μL sterile water; half of the abdomens (longitudinally dissected) of three additional specimens were tested with the same commercial kit.
- Fifteen specimens were extracted using the lab-made digestion buffer suggested by Gilbert et al. [22], followed by the QIAquick PCR Purification Kit®. DNA was eluted in 40 μL; half of the abdomens of three additional specimens (longitudinally dissected) were tested with the same commercial kit.
- Five specimens were extracted with the homemade digestion buffer proposed by Campos and Gilbert [29], followed by the QIAquick PCR Purification Kit®. DNA was eluted in 40 μL.
- Twelve specimens were extracted using the lab-made digestion buffer suggested by Santos et al. [30] and then treated as follows: eight abdomens processed using the QIAquick PCR Purification Kit®; two using the QIAamp® DNA Mini kit; and two with the QIAamp® DNA Investigator kit. DNA was eluted in 40 μL for the Purification Kit, 200 μL for the DNA Mini kit, and 100 μL for the Investigator kit.
- Twenty-four specimens were extracted using the QIAamp® DNA Investigator kit. Different elution volumes were used: DNA from 8 specimens was eluted in 100 μL, while for the rest of the samples the elution was performed using 50 μL.
2.3. DNA Amplification
2.4. Bioinformatics and Statistics
3. Results and Discussion
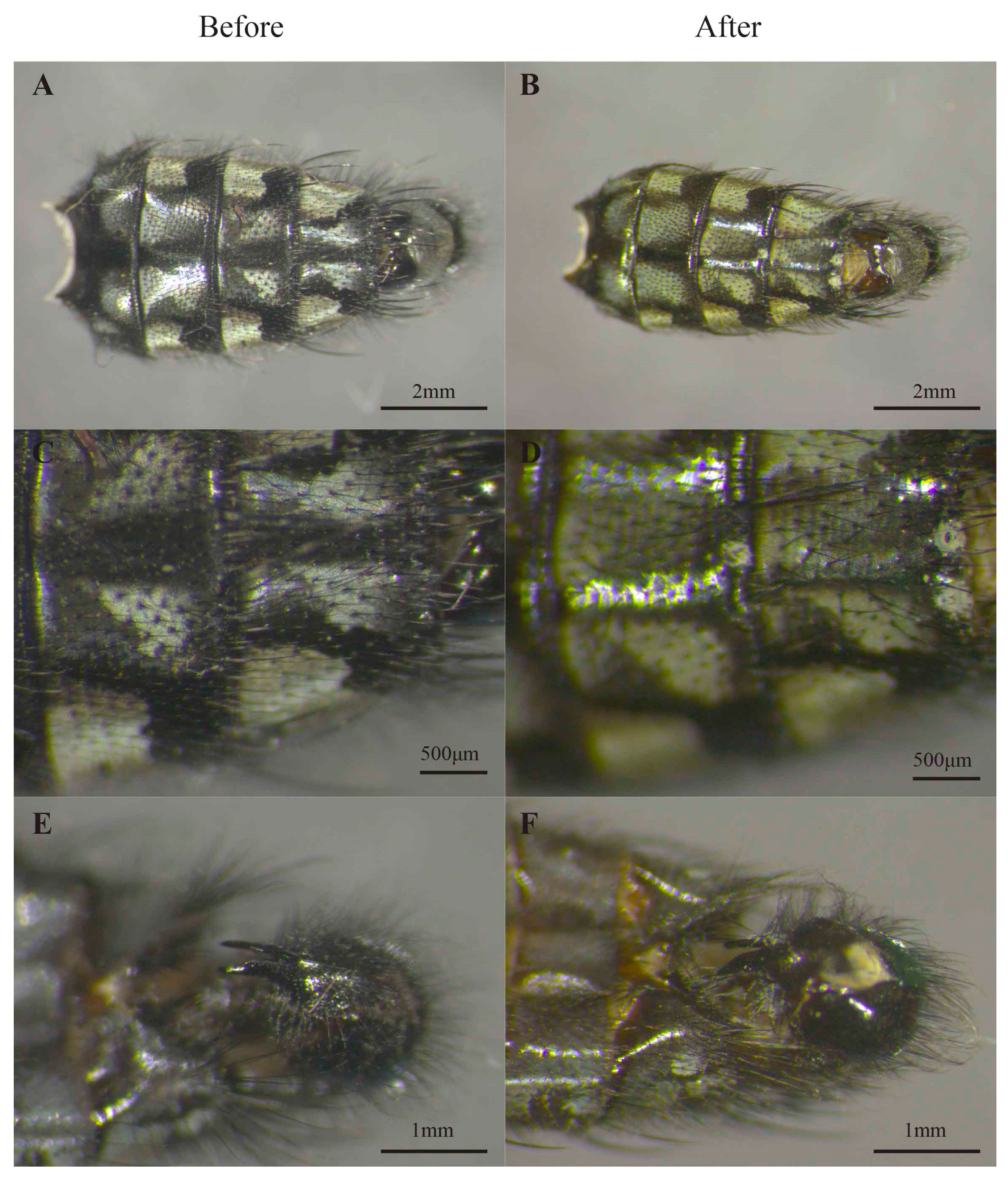
3.1. Morphological Identification
3.2. DNA Extraction
3.3. DNA Amplification
3.4. DNA Amplification and Sequence Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- King, G.A. Exaptation and synanthropic insects: A diachronic interplay between biology and culture. Environ. Archaeol. 2014, 19, 12–22. [Google Scholar] [CrossRef]
- Wells, J.D.; Pape, T.; Sperling, F.A. DNA-based identification and molecular systematics of forensically important Sarcophagidae (Diptera). J. Forensic Sci. 2001, 46, 1098–1102. [Google Scholar] [CrossRef]
- Amendt, J.; Richards, C.S.; Campobasso, C.P.; Zehner, R.; Hall, M.J. Forensic entomology: Applications and limitations. Forensic Sci. Med. Pathol. 2011, 7, 379–392. [Google Scholar] [CrossRef]
- Grzywacz, A.; Hall, M.J.; Pape, T.; Szpila, K. Muscidae (Diptera) of forensic importance-an identification key to third instar larvae of the Western Palaearctic region and a catalogue of the muscid carrion community. Int. J. Legal. Med. 2017, 131, 855–866. [Google Scholar] [CrossRef] [Green Version]
- Marshall, S.A. Flies: The Natural History & Diversity of Diptera; Firefly Books: Richmond Hill, ON, Canada, 2012; p. 616. [Google Scholar]
- Lo Pinto, S.; Giordani, G.; Tuccia, F.; Ventura, F.; Vanin, S. First records of Synthesiomyia nudiseta (Diptera: Muscidae) from forensic cases in Italy. For. Sci. Int. 2017, 276, e1–e7. [Google Scholar] [CrossRef] [PubMed]
- Courtney, G.W.; Pape, T.; Skevington, J.H.; Sinclair, B.J. Chapter 9. Biodiversity of Diptera. In Insect Biodiversity: Science and Society, 1st ed.; Foottit, R., Adler, P., Eds.; Blackwell Publishing: Hoboken, NJ, USA, 2009; pp. 185–222. [Google Scholar]
- Pape, T.; Blagoderov, V.; Mostovski, M.B. Order Diptera Linnaeus, 1758. Animal biodiversity: An outline of higher-level classification and survey of taxonomic richness. Zootaxa 2011, 3148, 222–229. [Google Scholar] [CrossRef] [Green Version]
- Szpila, K.; Richet, R.; Pape, T. Third instar larvae of flesh flies (Diptera: Sarcophagidae) of forensic importance—Critical review of characters and key for European species. Parasitol. Res. 2015, 114, 2279–2289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- GilArriortua, M.; Salona-Bordas, M.I.; Caine, L.M.; Pinheiro, F.; de Pancorbo, M.M. Technical Note: “Mitochondrial and nuclear DNA approaches for reliable identification of Lucilia (Diptera, Calliphoridae) species of forensic interest from Southern Europe”. Forensic Sci. Int. 2015, 257, 393–397. [Google Scholar] [CrossRef]
- Sonet, G.; Jordaens, K.; Braet, Y.; Bourguignon, L.; Dupont, E.; Backeljau, T.; De Meyer, M.; Desmyter, S. Utility of GenBank and the Barcode of Life Data Systems (BOLD) for the identification of forensically important Diptera from Belgium and France. Zookeys 2013, 365, 307–328. [Google Scholar] [CrossRef]
- Bortolini, S.; Giordani, G.; Tuccia, F.; Maistrello, L.; Vanin, S. Do longer sequences improve the accuracy of identification of forensically important Calliphoridae species? PeerJ 2018, 6, e5962. [Google Scholar] [CrossRef] [Green Version]
- Boehme, P.; Amendt, J.; Zehner, R. The use of COI barcodes for molecular identification of forensically important fly species in Germany. Parasitol. Res. 2012, 110, 2325–2332. [Google Scholar] [CrossRef]
- Adler, C.J.; Haak, W.; Donlon, D.; Cooper, A. Survival and recovery of DNA from ancient teeth and bones. J. Archaeol. Sci. 2011, 38, 956–964. [Google Scholar] [CrossRef]
- Stevens, J.; Wall, R. Genetic relationships between blowflies (Calliphoridae) of forensic importance. Forensic Sci. Int. 2001, 120, 116–123. [Google Scholar] [CrossRef]
- Tuccia, F.; Giordani, G.; Vanin, S. A general review of the most common COI primers for Calliphoridae identification in forensic entomology. Forensic Sci. Int. Genet. 2016, 24, e9–e11. [Google Scholar] [CrossRef]
- Wells, J.D.; Sperling, F.A. DNA-based identification of forensically important Chrysomyinae (Diptera: Calliphoridae). Forensic Sci. Int. 2001, 120, 110–115. [Google Scholar] [CrossRef]
- Zehner, R.; Amendt, J.; Schutt, S.; Sauer, J.; Krettek, R.; Povolny, D. Genetic identification of forensically important flesh flies (Diptera: Sarcophagidae). Int. J. Legal. Med. 2004, 118, 245–247. [Google Scholar] [CrossRef]
- Torres Tola, E.; Castillo Vega, P.; Calderón, A.B. Identificación molecular de Dípteros de importancia forense con el gen (COI Barcode), La Paz Bolivia. Medicina Legal de Costa Rica 2020, 37, 93–101. [Google Scholar]
- Schönberger, D.; Giordani, G.; Vanin, S.; Whitmore, D. A review of morphological characters for the identification of three common European species of Sarcophaga s.str. (Diptera: Sarcophagidae), with an emphasis on female terminalia. Zootaxa 2022, 5205, 463–480. [Google Scholar] [CrossRef]
- Tuccia, F.; Giordani, G.; Vanin, S. A combined protocol for identification of maggots of forensic interest. Sci. Justice 2016, 56, 264–268. [Google Scholar] [CrossRef]
- Gilbert, M.T.; Moore, W.; Melchior, L.; Worobey, M. DNA extraction from dry museum beetles without conferring external morphological damage. PLoS ONE 2007, 2, e272. [Google Scholar] [CrossRef] [Green Version]
- Panagiotakopulu, E. New records for ancient pests: Archaeoentomology in Egypt. J. Archaeol. Sci. 2001, 28, 1235–1246. [Google Scholar] [CrossRef] [Green Version]
- Panagiotakopulu, E.; Buckland, P.C. Forensic archaeoentomology—An insect fauna from a burial in York Minster. Forensic Sci. Int. 2012, 221, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Solomon, M. Archaeological records of storage pests: Sitophilus granarius (L.)(Coleoptera, Curculionidae) from an Egyptian pyramid tomb. J. Stored Prod. Res. 1965, 1, 105–107. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Elias, S.; Gilbert, M.T.; Haile, J.; Munch, K.; Kuzmina, S.; Froese, D.G.; Sher, A.; Holdaway, R.N.; Willerslev, E. Non-destructive sampling of ancient insect DNA. PLoS ONE 2009, 4, e5048. [Google Scholar] [CrossRef] [PubMed]
- Tin, M.M.; Economo, E.P.; Mikheyev, A.S. Sequencing degraded DNA from non-destructively sampled museum specimens for RAD-tagging and low-coverage shotgun phylogenetics. PLoS ONE 2014, 9, e96793. [Google Scholar] [CrossRef]
- Korlević, P.; McAlister, E.; Mayho, M.; Makunin, A.; Flicek, P.; Lawniczak, M.K. A Minimally Morphologically Destructive Approach for DNA Retrieval and Whole-Genome Shotgun Sequencing of Pinned Historic Dipteran Vector Species. Genome Biol. Evol. 2021, 13, evab226. [Google Scholar] [CrossRef]
- Campos, P.F.; Gilbert, T.M. DNA extraction from keratin and chitin. In Ancient DNA. Methods and Protocols; Shapiro, B., Hofreiter, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 43–49. [Google Scholar]
- Santos, D.; Ribeiro, G.C.; Cabral, A.D.; Sperança, M.A. A non-destructive enzymatic method to extract DNA from arthropod specimens: Implications for morphological and molecular studies. PLoS ONE 2018, 13, e0192200. [Google Scholar] [CrossRef] [Green Version]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Stubbs, A.E.; Falk, S.J. British Hoverflies: An Illustrated Identification Guide; British Entomological & Natural History Society; CAB Direct: Glasgow, UK, 1983; p. 253. [Google Scholar]
- Papp, L.; Darvas, B.E. Contributions to a Manual of Palaeartic Diptera, Vol. 3 Higher Brachycera; Science Herald: Budapest, Hungary, 1998; pp. 617–648. [Google Scholar]
- Whitmore, D.; Dupont, S.; Falk, S. Key to Adult Flesh Flies (Diptera: Sarcophagidae) of the British Isles; OSF: Peoria, IL, USA, 2020. [Google Scholar] [CrossRef]
- Buenaventura, E.; Szpila, K.; Cassel, B.; Wiegmann, B.M.; Pape, T. Anchored hybrid enrichment challenges the traditional classification of flesh flies (Diptera: Sarcophagidae). System. Entomol. 2020, 45, 281–301. [Google Scholar] [CrossRef] [Green Version]
- Sharma, M.; Singh, D.; Sharma, A.K. Mitochondrial DNA based identification of forensically important Indian flesh flies (Diptera: Sarcophagidae). Forensic Sci. Int. 2015, 247, 1–6. [Google Scholar] [CrossRef]
- Jordaens, K.; Sonet, G.; Richet, R.; Dupont, E.; Braet, Y.; Desmyter, S. Identification of forensically important Sarcophaga species (Diptera: Sarcophagidae) using the mitochondrial COI gene. Int. J. Legal. Med. 2013, 127, 491–504. [Google Scholar] [CrossRef]
- Phillips, A.J.; Simon, C. Simple, efficient, and nondestructive DNA extraction protocol for arthropods. Ann. Entomol. Soc. Am. 1995, 88, 281–283. [Google Scholar] [CrossRef]
- Rowley, D.L.; Coddington, J.A.; Gates, M.W.; Norrbom, A.L.; Ochoa, R.A.; Vandenberg, N.J.; Greenstone, M.H. Vouchering DNA-barcoded specimens: Test of a nondestructive extraction protocol for terrestrial arthropods. Mol. Ecol. Notes 2007, 7, 915–924. [Google Scholar] [CrossRef] [Green Version]

| Oligo Name | Sequence (5′ -> 3′) | Tm (°C) |
|---|---|---|
| Sar111_FW | TCGCAACAATGGTTATTCTCT | 54.0 |
| Sar111_RV | TCARTTTCCAAAYCCTCCAAT | 54.2 |
| Sar211_FW | GTAATTGTTACAGCYCATGC | 54.2 |
| Sar211_RV | TTCCAGCTCCRTTTTCTACT | 54.0 |
| Sar311_FW | CYCGAATRAAYAATATAAGTTTTTG | 56.4 |
| Sar311_RV | CCTAAAATTGAAGAAATTCCWGCTA | 60.3 |
| Sar411_FW | CTAATATTGCYCATGGRGGAGC | 58.5 |
| Sar411_RV | CGRTCAGTTAATARTATRGTRATWGC | 50.8 |
| Sar511_FW | GGWATTACHTTTGAYCGAAT | 54.7 |
| Sar511_RV | GAYTCTTGRCTAATAATGTGAG | 52.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giordani, G.; Whitmore, D.; Vanin, S. A New, Non-Invasive Methodology for the Molecular Identification of Adult Sarcophagidae from Collections. Insects 2023, 14, 635. https://doi.org/10.3390/insects14070635
Giordani G, Whitmore D, Vanin S. A New, Non-Invasive Methodology for the Molecular Identification of Adult Sarcophagidae from Collections. Insects. 2023; 14(7):635. https://doi.org/10.3390/insects14070635
Chicago/Turabian StyleGiordani, Giorgia, Daniel Whitmore, and Stefano Vanin. 2023. "A New, Non-Invasive Methodology for the Molecular Identification of Adult Sarcophagidae from Collections" Insects 14, no. 7: 635. https://doi.org/10.3390/insects14070635
APA StyleGiordani, G., Whitmore, D., & Vanin, S. (2023). A New, Non-Invasive Methodology for the Molecular Identification of Adult Sarcophagidae from Collections. Insects, 14(7), 635. https://doi.org/10.3390/insects14070635






