Transcriptomics Provide Insights into the Photoperiodic Regulation of Reproductive Diapause in the Green Lacewing, Chrysoperla nipponensis (Okamoto) (Neuroptera: Chrysopidae)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Insects
2.2. RNA Extraction
2.3. Illumina RNA-seq Library Construction and Sequencing
2.4. Illumina Data Analysis
2.5. KEGG Enrichment Analysis of Differentially Expressed Genes
2.6. Real-Time Quantitative PCR
2.7. RNA Interference
2.8. Statistical Analysis
3. Results
3.1. Transcriptome Analysis of Larva and Female Adults Reared under a Short Photoperiod and a Long Photoperiod
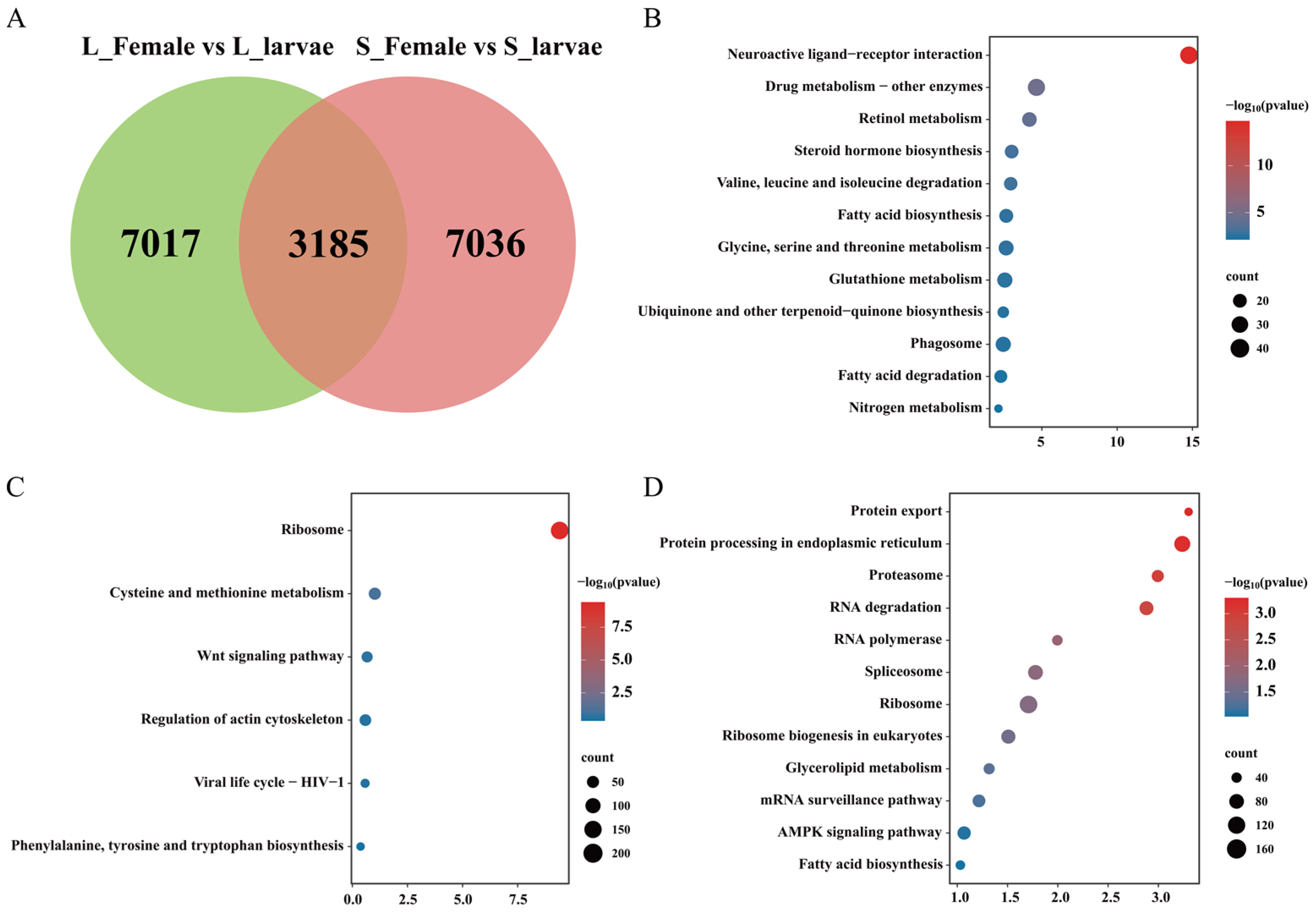
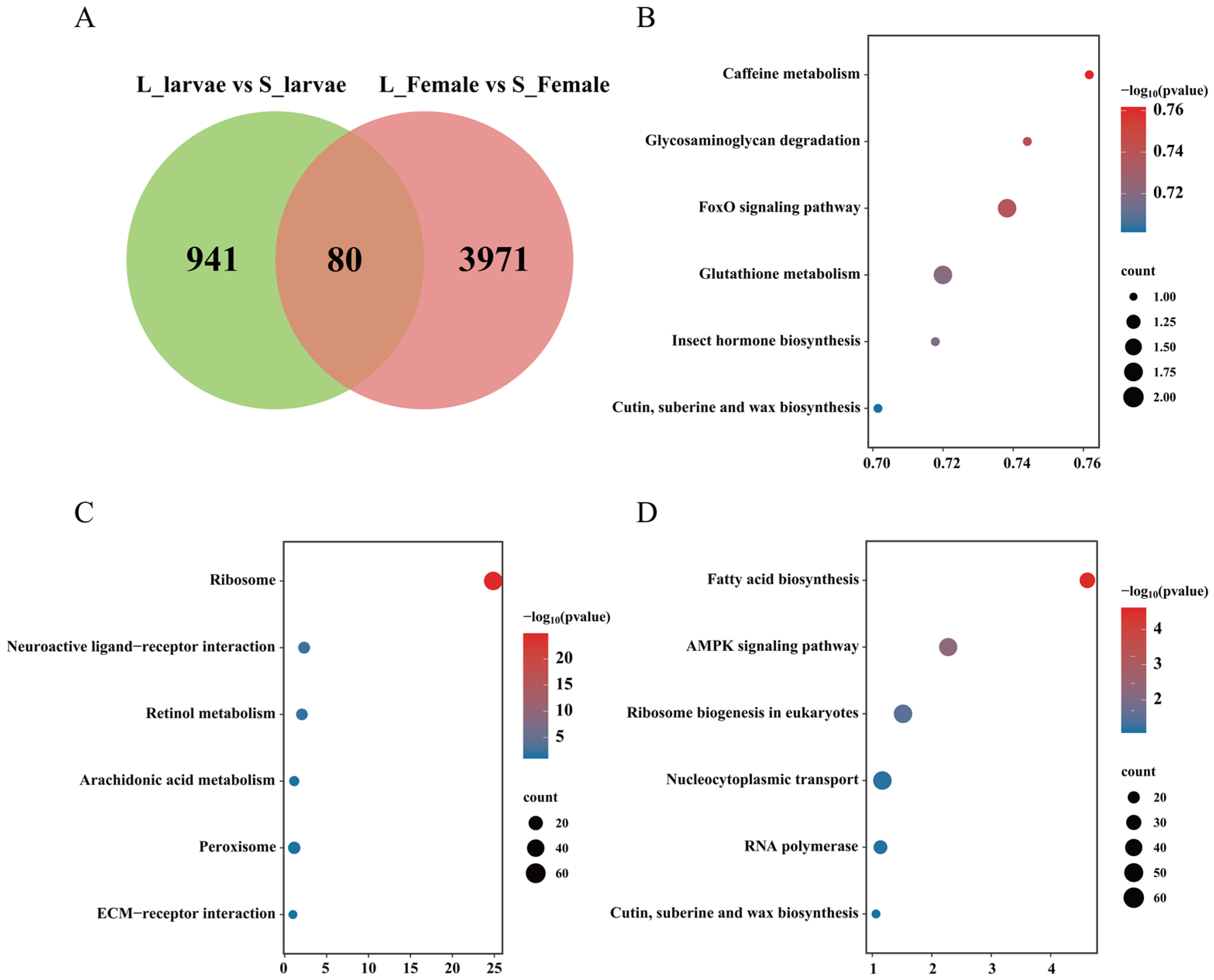
3.2. Transcriptome Undergoes Changes during the Developmental Process
3.3. Transcriptome Undergoes Changes in Different Photoperiods
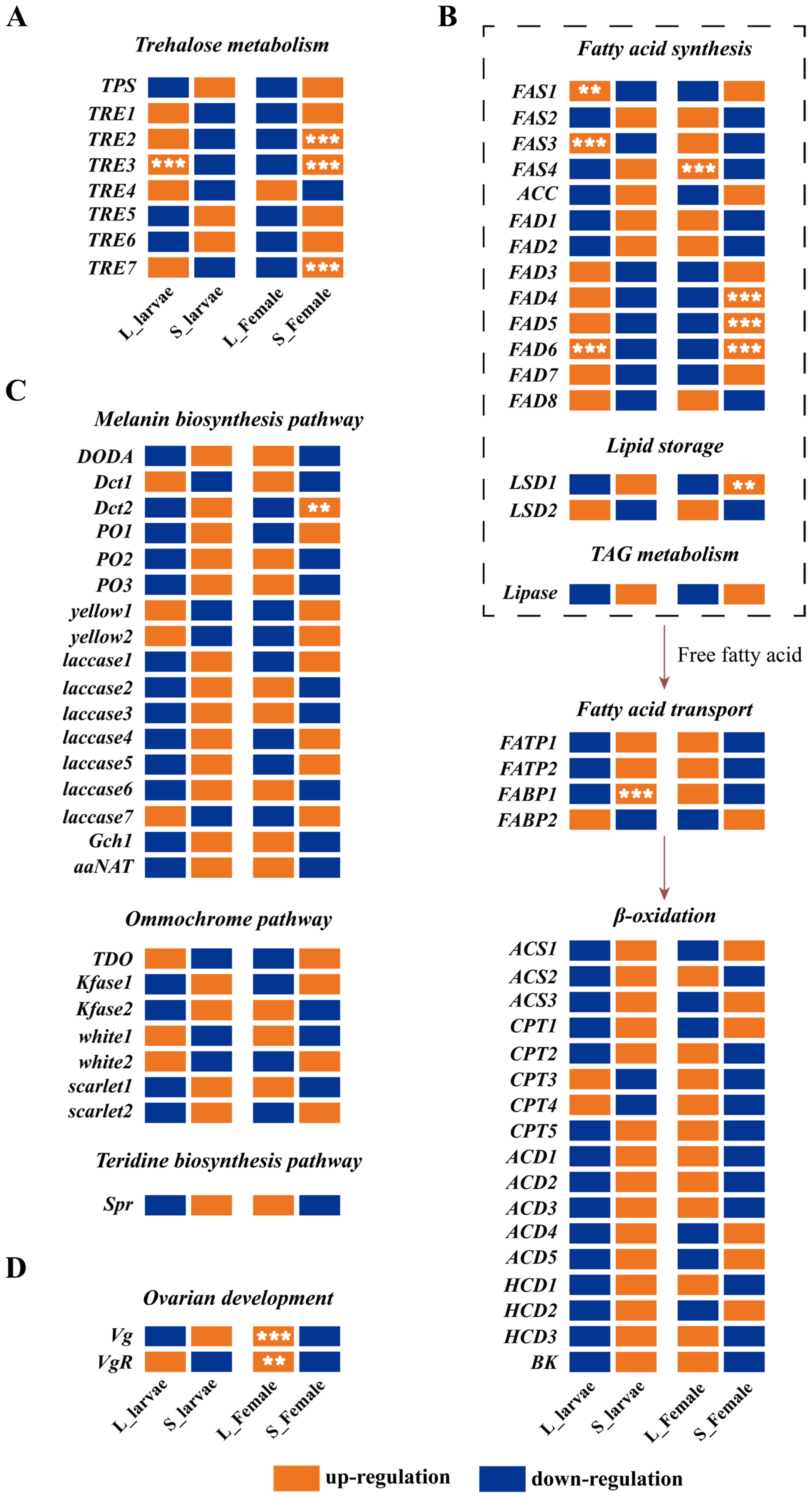
3.4. Expression Analysis of Multiple Metabolic Pathways
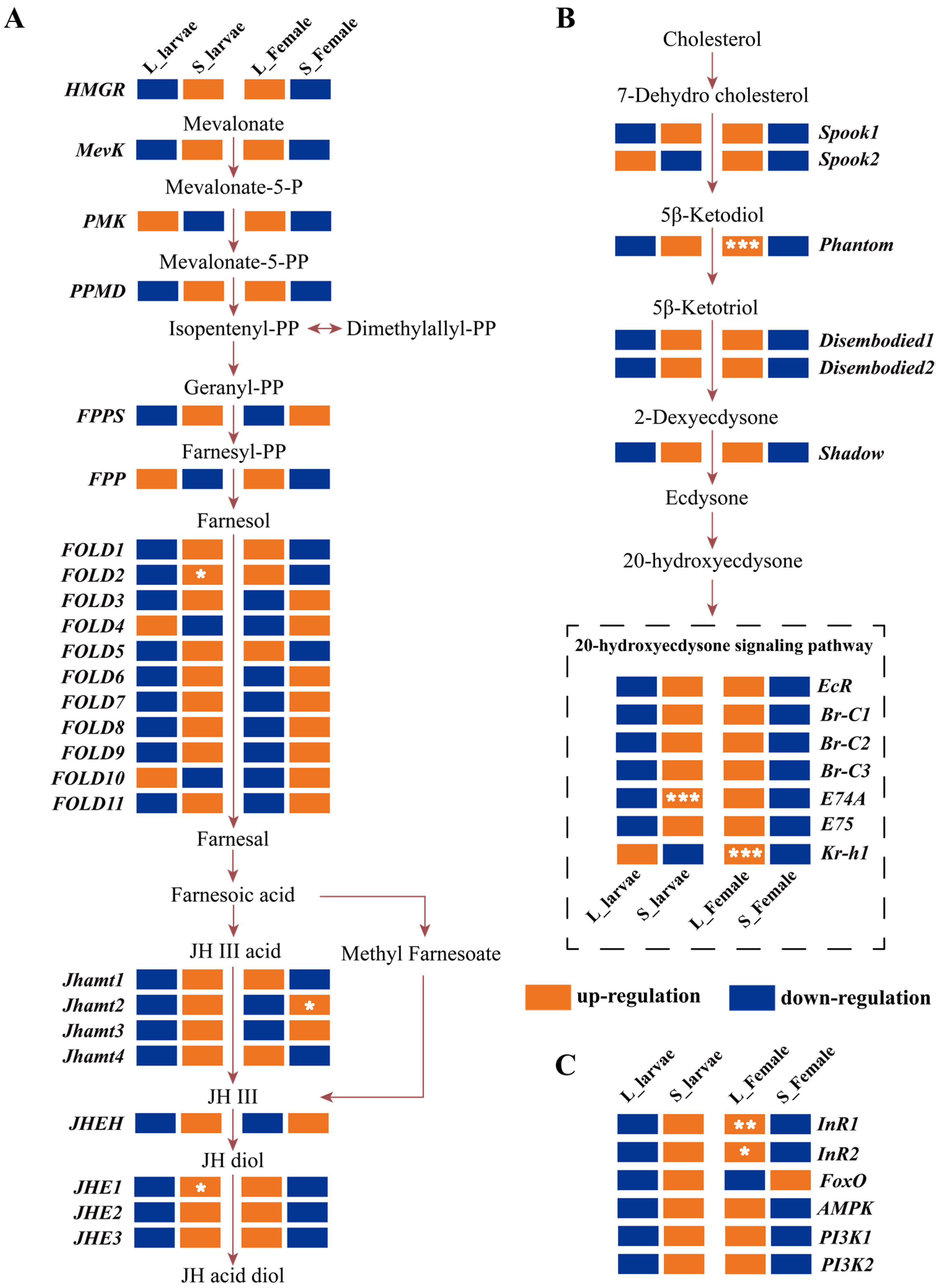
3.5. Hormone Gene Expression Profile in the Green Lacewing
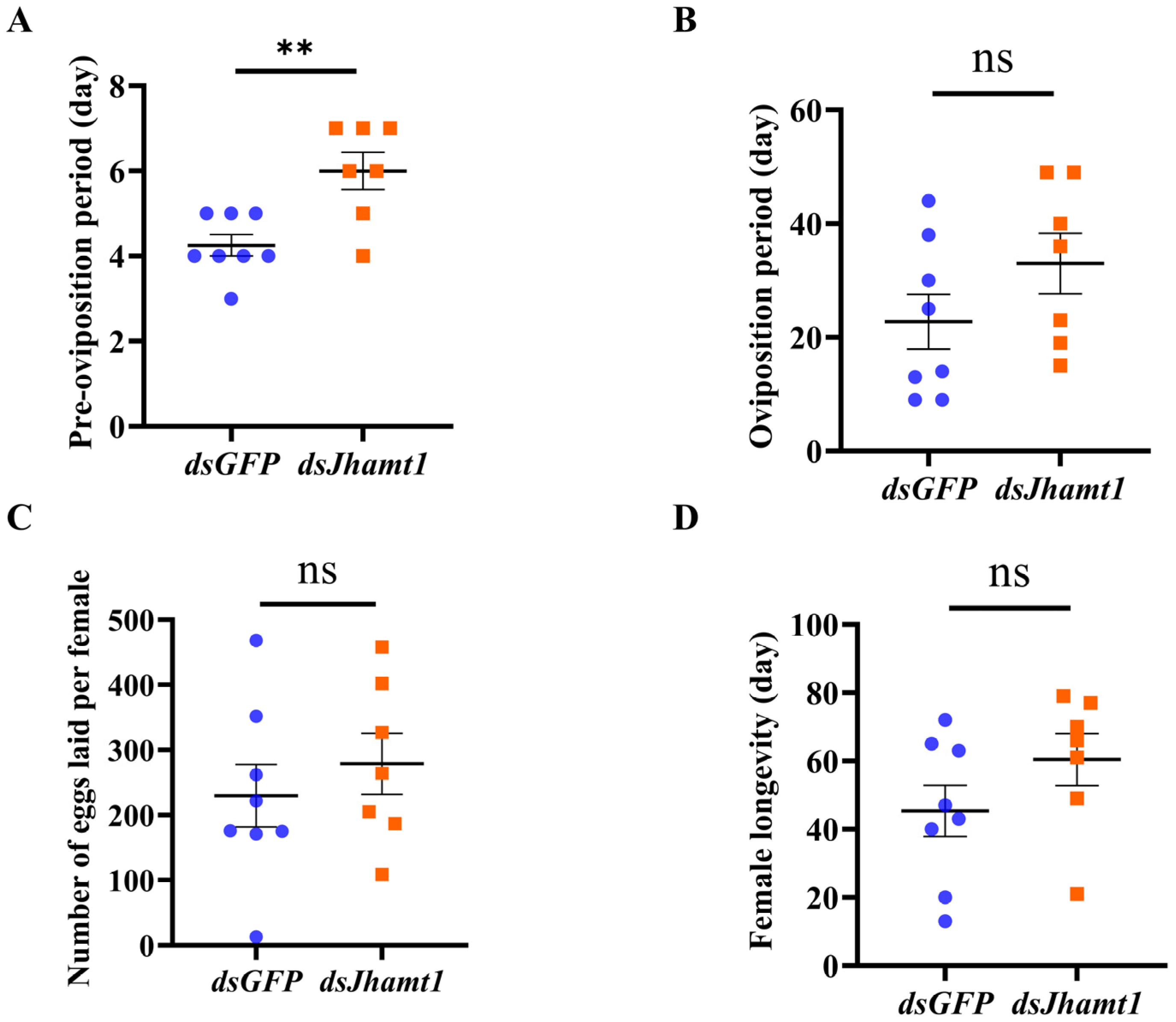
3.6. Functional Validation of Jhamt1 in Pre-Diapause Phase
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tauber, M.J.; Tauber, C.A.; Masaki, S. Seasonal Adaptations of Insects; Oxford University Press: New York, NY, USA, 1986. [Google Scholar]
- Denlinger, D.L.; Armbruster, P.A. Chapter eleven—Molecular physiology of mosquito diapause. In Advances in Insect Physiology; Raikhel, A.S., Ed.; Academic Press: Cambridge, MA, USA, 2016; Volume 51, pp. 329–361. [Google Scholar]
- Hahn, D.A.; Denlinger, D.L. Energetics of insect diapause. Annu. Rev. Entomol. 2010, 56, 103–121. [Google Scholar] [CrossRef]
- Wang, J.; Fan, H.; Xiong, K.-C.; Liu, Y.-H. Transcriptomic and metabolomic profiles of Chinese citrus fly, Bactrocera minax (Diptera: Tephritidae), along with pupal development provide insight into diapause program. PLoS ONE 2017, 12, e0181033. [Google Scholar] [CrossRef]
- Yang, M.; Wang, Z.; Wang, R.; Zhang, X.; Li, M.; Xin, J.; Qin, Y.; Zhang, C.; Meng, F. Transcriptomic and proteomic analyses of the mechanisms of overwintering diapause in soybean pod borer (Leguminivora glycinivorella). Pest Manag. Sci. 2020, 76, 4248–4257. [Google Scholar] [CrossRef]
- Harvey, W.R. Metabolic aspects of insect diapause. Annu. Rev. Entomol. 1962, 7, 57–80. [Google Scholar] [CrossRef]
- Denlinger, D.L. Why study diapause? Entomol. Res. 2008, 38, 1–9. [Google Scholar] [CrossRef]
- Denlinger, D.L.; Yocum, G.D.; Rinehart, J.P. 10—Hormonal control of diapause. In Insect Endocrinology; Gilbert, L.I., Ed.; Academic Press: San Diego, CA, USA, 2012; pp. 430–463. [Google Scholar]
- Hutfilz, C. Endocrine regulation of lifespan in insect diapause. Front. Physiol. 2022, 13, 825057. [Google Scholar] [CrossRef] [PubMed]
- Denlinger, D.L. Regulation of diapause. Annu. Rev. Entomol. 2002, 47, 93–122. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Shi, J.; Yang, L.; Cheng, Y.; Liu, X.; Sun, S. Diapause induction, color changes, and supercooling point of diapause larvae of Tetrastichus septentrionalis Yang (Hymenoptera: Eulophidae). Insects 2023, 14, 826. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Zhang, L.; Han, Y.; Ren, X.; Huang, J.; Chen, H. De novo transcriptome sequencing and analysis of Coccinella septempunctata L. in non-diapause, diapause and diapause-terminated states to identify diapause-associated genes. BMC Genom. 2015, 16, 1086. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, Y.; Zhu, L.; Zhu, F.; Lei, C.L.; Wang, X.P. Juvenile hormone facilitates the antagonism between adult reproduction and diapause through the methoprene-tolerant gene in the female Colaphellus bowringi. Insect Biochem. Mol. Biol. 2016, 74, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Wei, B.-X.; Liu, W.; Wang, J.-L.; Zhou, X.-M.; Wang, X.-P. Differences in the development of internal reproductive organs, feeding amount and nutrient storage between pre-diapause and pre-reproductive Harmonia axyridis adults. Insects 2019, 10, 243. [Google Scholar] [CrossRef]
- Huang, Q.; Zhang, G.; Nan, J.; Cheng, W.; Zhu-Salzman, K. Characterization of trehalose metabolic genes and corresponding enzymatic activities during diapause of Sitodiplosis mosellana. J. Insect Physiol. 2021, 135, 104324. [Google Scholar] [CrossRef]
- Riddiford, L.M. Cellular and molecular actions of juvenile hormone I. General considerations and premetamorphic actions. In Advances in Insect Physiology; Elsevier: Amsterdam, The Netherlands, 1994; Volume 24, pp. 213–274. [Google Scholar]
- Nijhout, H.F. Insect Hormones; Princeton University Press: Princeton, NJ, USA, 1998. [Google Scholar]
- Ishimaru, Y.; Tomonari, S.; Matsuoka, Y.; Watanabe, T.; Miyawaki, K.; Bando, T.; Tomioka, K.; Ohuchi, H.; Noji, S.; Mito, T. TGF-β signaling in insects regulates metamorphosis via juvenile hormone biosynthesis. Proc. Natl. Acad. Sci. USA 2016, 113, 5634–5639. [Google Scholar] [CrossRef]
- Mukai, A.; Mano, G.; Des Marteaux, L.; Shinada, T.; Goto, S.G. Juvenile hormone as a causal factor for maternal regulation of diapause in a wasp. Insect Biochem. Mol. Biol. 2022, 144, 103758. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.-Z.; Wu, Y.-F.; Yin, Z.-Y.; Guo, J.-J.; Li, H.-Y. Juvenile hormone is an important factor in regulating Aspongopus chinensis Dallas diapause. Front. Physiol. 2022, 13, 873580. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Sun, D.; Tian, Z.; Liu, W.; Zhu, F.; Wang, X.P. The limited regulatory roles of juvenile hormone degradation pathways in reproductive diapause preparation of the cabbage beetle, Colaphellus bowringi. J. Insect Physiol. 2019, 119, 103967. [Google Scholar] [CrossRef]
- Gao, Q.; Li, B.; Tian, Z.; De Loof, A.; Wang, J.L.; Wang, X.P.; Liu, W. Key role of juvenile hormone in controlling reproductive diapause in females of the asian lady beetle Harmonia axyridis. Pest. Manag. Sci. 2022, 78, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-Y.; Chen, J.-J.; Liu, M.-Y.; He, W.-W.; Reynolds, J.A.; Wang, Y.-N.; Wang, M.-Q.; Zhang, L.-S. Enhanced degradation of juvenile hormone promotes reproductive diapause in the predatory ladybeetle Coccinella septempunctata. Front. Physiol. 2022, 13, 877153. [Google Scholar] [CrossRef] [PubMed]
- Areiza, M.; Nouzova, M.; Rivera-Perez, C.; Noriega, F.G. 20-Hydroxyecdysone stimulation of juvenile hormone biosynthesis by the mosquito corpora allata. Insect Biochem. Mol. Biol. 2015, 64, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Meiselman, M.; Lee, S.S.; Tran, R.T.; Dai, H.; Ding, Y.; Rivera-Perez, C.; Wijesekera, T.P.; Dauwalder, B.; Noriega, F.G.; Adams, M.E. Endocrine network essential for reproductive success in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2017, 114, E3849–E3858. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Tian, Z.; Wu, Q.-W.; King-Jones, K.; Liu, W.; Zhu, F.; Wang, X.-P. Steroid hormone ecdysone deficiency stimulates preparation for photoperiodic reproductive diapause. PLoS Genet. 2021, 17, e1009352. [Google Scholar] [CrossRef]
- Sim, C.; Denlinger, D.L. Insulin signaling and FOXO regulate the overwintering diapause of the mosquito Culex pipiens. Proc. Natl. Acad. Sci. USA 2008, 105, 6777–6781. [Google Scholar] [CrossRef] [PubMed]
- Sim, C.; Denlinger, D.L. A shut-down in expression of an insulin-like peptide, ILP-1, halts ovarian maturation during the overwintering diapause of the mosquito Culex pipiens. Insect Mol. Biol. 2009, 18, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Sim, C.; Denlinger, D.L. Juvenile hormone III suppresses forkhead of transcription factor in the fat body and reduces fat accumulation in the diapausing mosquito, Culex pipiens. Insect Mol. Biol. 2013, 22, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sim, C.; Kang, D.S.; Kim, S.; Bai, X.; Denlinger, D.L. Identification of FOXO targets that generate diverse features of the diapause phenotype in the mosquito Culex pipiens. Proc. Natl. Acad. Sci. USA 2015, 112, 3811–3816. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Hedo, M.; Rivera-Perez, C.; Noriega, F.G. The insulin/TOR signal transduction pathway is involved in the nutritional regulation of juvenile hormone synthesis in Aedes aegypti. Insect Biochem. Mol. Biol. 2013, 43, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.-y.; Mu, J.-y.; Hu, C.; Wang, H.-g. Photoperiodic control of adult diapause in chrysoperla sinica (Tjeder) (neuroptera: Chrysopidae)—I. Critical photoperiod and sensitive stages of adult diapause induction. Insect Sci. 2004, 11, 191–198. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Romeis, J.; Chen, X.; Zhang, J.; Chen, H.; Peng, Y. Consumption of Bt rice pollen expressing Cry2Aa does not cause adverse effects on adult Chrysoperla sinica Tjeder (Neuroptera: Chrysopidae). Biol. Control 2012, 61, 246–251. [Google Scholar] [CrossRef]
- Huang, H.; Liu, Y.; Qi, Y.; Xu, Y.; Chen, Z. Predatory responses of Chrysoperla sinica (Tjeder) larvae to Spodoptera frugiperda (JE Smith) eggs and larvae. Chin. J. Appl. Entomol. 2020, 57, 1333–1340. [Google Scholar]
- Chen, Z.-Z.; Wang, X.; Kong, X.; Zhao, Y.-M.; Xu, M.-H.; Gao, Y.-Q.; Huang, H.-Y.; Liu, F.-H.; Wang, S.; Xu, Y.-Y.; et al. Quantitative transcriptomic and proteomic analyses reveal the potential maintenance mechanism of female adult reproductive diapause in Chrysoperla nipponensis. Pest Manag. Sci. 2023, 79, 1897–1911. [Google Scholar] [CrossRef]
- Chen, Z.Z.; Liu, L.Y.; Liu, S.Y.; Cheng, L.Y.; Wang, X.H.; Xu, Y.Y. Response of Chrysoperla nipponensis (Okamoto) (Neuroptera: Chrysopidae) under long and short photoperiods. J. Insect Sci. 2017, 17, 35. [Google Scholar] [CrossRef]
- Chen, Z.; Lu, H.; Wang, Y.; Cao, Y.; Yu, J.; Yin, X.; Xu, Y. Effects of two different photoperiods on cold hardiness of naturally overwintering adults and laboratory-bred larvae of Chrysoperla sinica (Neuroptera: Chrysopidae). Acta Entomol. Sin. 2014, 57, 52–60. [Google Scholar]
- Zhao, Z.; Huang, Y. The mechanisms of regulation and control of insect diapanse. J. Shanxi Univ. (Nat. Sci. Ed.) 1995, 18, 105–118. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Kong, X.; Liu, S.; Huang, H.; Chen, Z.; Xu, Y. Selection of reference genes for quantitative real-time pcr in Chrysoperla nipponensis (Neuroptera: Chrysopidae) under tissues in reproduction and diapause. J. Insect Sci. 2020, 20, 20. [Google Scholar] [CrossRef] [PubMed]
- Rodnina, M.V.; Wintermeyer, W. The ribosome as a molecular machine: The mechanism of tRNA-mRNA movement in translocation. Biochem. Soc. Trans. 2011, 39, 658–662. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.-Z.; Li, N.-Y.; Zeng, X.-D.; Song, J.-C.; Yu, X.-D.; Su, H.-N.; Chen, C.-X.; Yi, L.; Lu, Z.-J. Transcriptome analyses of Diaphorina citri midgut responses to Candidatus liberibacter asiaticus infection. Insects 2020, 11, 171. [Google Scholar] [CrossRef] [PubMed]
- Musolin, D.L.; Numata, H. Photoperiodic and temperature control of diapause induction and colour change in the southern green stink bug Nezara viridula. Physiol. Entomol. 2003, 28, 65–74. [Google Scholar] [CrossRef]
- Teyssier, J.; Saenko, S.V.; van der Marel, D.; Milinkovitch, M.C. Photonic crystals cause active colour change in chameleons. Nat. Commun. 2015, 6, 6368. [Google Scholar] [CrossRef]
- Xiong, G.; Tong, X.; Gai, T.; Li, C.; Qiao, L.; Monteiro, A.; Hu, H.; Han, M.; Ding, X.; Wu, S.; et al. Body shape and coloration of silkworm larvae are influenced by a novel cuticular protein. Genetics 2017, 207, 1053–1066. [Google Scholar] [CrossRef]
- Yang, M.; Wang, Y.; Liu, Q.; Liu, Z.; Jiang, F.; Wang, H.; Guo, X.; Zhang, J.; Kang, L. A β-carotene-binding protein carrying a red pigment regulates body-color transition between green and black in locusts. eLife 2019, 8, e41362. [Google Scholar] [CrossRef]
- Xu, Y.; Hu, C.; Mu, J.; Xu, H.; Wang, H. Relationship between adult diapause development and over-wintering coloration changes in Chrysoperla sinica (Neuroptera). Acta Ecol. Sin. 2002, 22, 1275–1280. [Google Scholar]
- Huang, H.-Y.; Zhao, Y.-M.; Wu, X.-L.; Chen, Z.-Z.; Xu, Y.-Y. Effects of exogenous juvenile hormone on the diapause termination and post-diapause development of Chrysoperla silica (Neuroptera Chrysopidae). Acta Entomol. Sin. 2021, 64, 392–399. [Google Scholar]
- Zhang, L.; Cheng, L.; Chapman, J.W.; Sappington, T.W.; Liu, J.; Cheng, Y.; Jiang, X. Juvenile hormone regulates the shift from migrants to residents in adult oriental armyworm, Mythimna separata. Sci. Rep. 2020, 10, 11626. [Google Scholar] [CrossRef]
- Lv, W.; Zeng, L.; Zhang, Z.; He, H.; Wang, F.; Xie, X. Effects of Juvenile Hormone Analog and Days after Emergence on the Reproduction of Oriental Armyworm, Mythimna separata (Lepidoptera: Noctuidae) Populations. Insects 2022, 13, 506. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhou, Y.; Ran, H.; Li, F. Effects of different hormones as dietary supplements on biological characteristics of Coccinella septempunctata L. J. Appl. Entomol. 2023, 147, 888–894. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Gao, Y.; Shi, R.; Huang, H.; Xu, Y.; Chen, Z. Transcriptomics Provide Insights into the Photoperiodic Regulation of Reproductive Diapause in the Green Lacewing, Chrysoperla nipponensis (Okamoto) (Neuroptera: Chrysopidae). Insects 2024, 15, 136. https://doi.org/10.3390/insects15020136
Liu S, Gao Y, Shi R, Huang H, Xu Y, Chen Z. Transcriptomics Provide Insights into the Photoperiodic Regulation of Reproductive Diapause in the Green Lacewing, Chrysoperla nipponensis (Okamoto) (Neuroptera: Chrysopidae). Insects. 2024; 15(2):136. https://doi.org/10.3390/insects15020136
Chicago/Turabian StyleLiu, Shaoye, Yuqing Gao, Rangjun Shi, Haiyi Huang, Yongyu Xu, and Zhenzhen Chen. 2024. "Transcriptomics Provide Insights into the Photoperiodic Regulation of Reproductive Diapause in the Green Lacewing, Chrysoperla nipponensis (Okamoto) (Neuroptera: Chrysopidae)" Insects 15, no. 2: 136. https://doi.org/10.3390/insects15020136
APA StyleLiu, S., Gao, Y., Shi, R., Huang, H., Xu, Y., & Chen, Z. (2024). Transcriptomics Provide Insights into the Photoperiodic Regulation of Reproductive Diapause in the Green Lacewing, Chrysoperla nipponensis (Okamoto) (Neuroptera: Chrysopidae). Insects, 15(2), 136. https://doi.org/10.3390/insects15020136







