The Potential Implications of Sex-Specific Differences in the Intestinal Bacteria of the Overwintering Wolf Spider Pardosa astrigera (Araneae: Lycosidae)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Spider Collection
2.2. Intestinal Extraction
2.3. DNA Extraction, PCR Amplification, and Sequencing
2.4. Bioinformatics and Sequence Analysis
2.5. Statistical Analyses
3. Results
3.1. Bacterial Sequence Data
3.2. Intestinal Bacterial Diversity
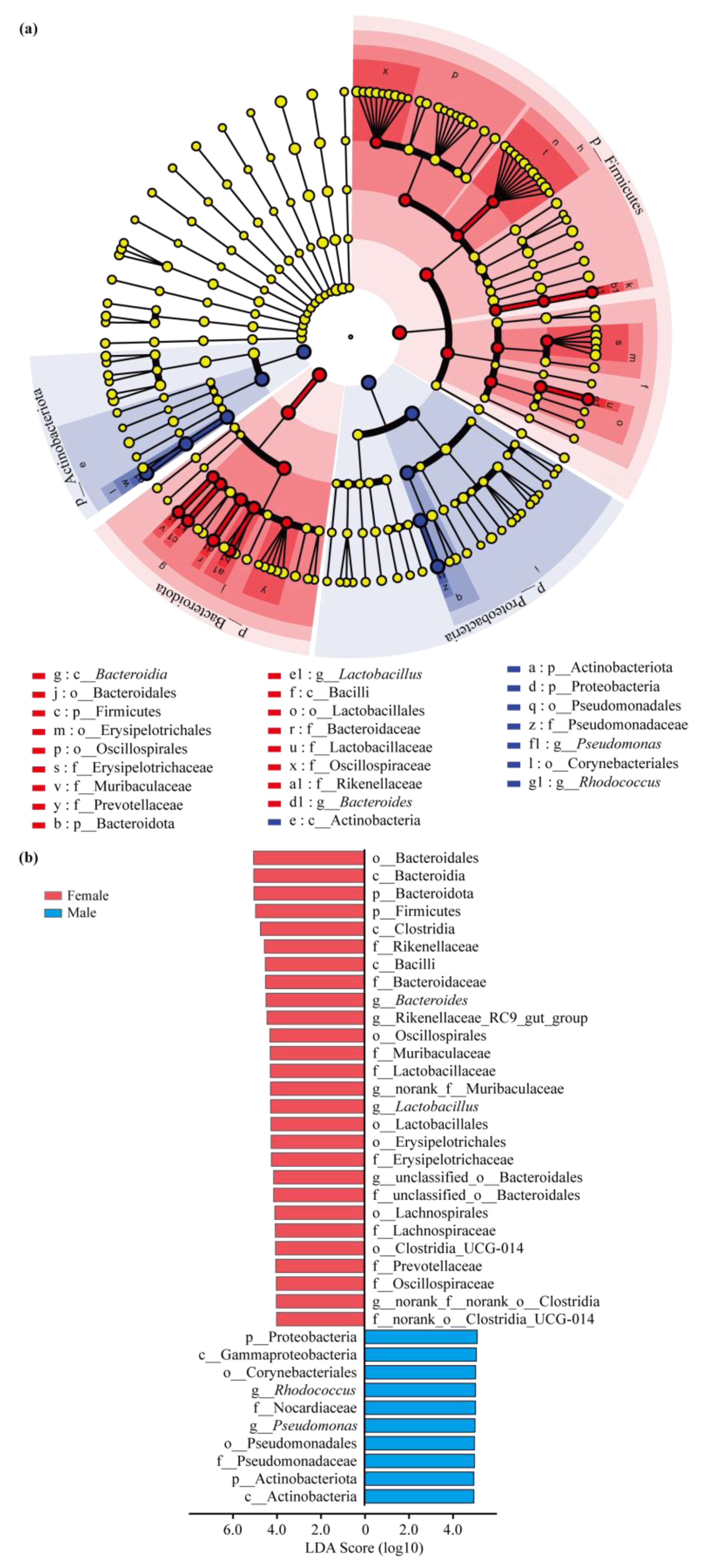
3.3. Intestinal Bacterial Community Structure
3.4. Functional Predictions with PICRUSt2
4. Discussion
4.1. Intestinal Bacterial Diversity
4.2. Intestinal Bacterial Community Structure
4.3. Potential Implications of the Intestinal Bacteria in the Cold Tolerance of Spiders
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Storey, K.B.; Storey, J.M. Insect cold hardiness: Metabolic, gene, and protein adaptation. Can. J. Zool. 2012, 90, 456–475. [Google Scholar] [CrossRef]
- Potts, L.J. Physiological Ecology of Overwintering and Cold-Adapted Arthropods. Ph.D. Thesis, University of Kentucky, Lexington, KY, USA, 2020. [Google Scholar]
- Gunnarsson, B. Body size and survival: Implications for an overwintering spider. Oikos 1988, 52, 274–282. [Google Scholar] [CrossRef]
- Whitney, T.D.; Philip, B.N.; Harwood, J.D. Tradeoff in two winter–active wolf spiders: Increased mortality for increased growth. Entomol. Exp. Appl. 2014, 153, 191–198. [Google Scholar] [CrossRef]
- Petráková, L.; Michalko, R.; Loverre, P.; Sentenská, L.; Korenko, S.; Pekár, S. Intraguild predation among spiders and their effect on the pear psylla during winter. Agric. Ecosyst. Environ. 2016, 233, 67–74. [Google Scholar] [CrossRef]
- Tanaka, K.; Ito, K. Accumulation of glycerol and myo-inositol in the overwintering nymphs of the wolf spider Pardosa astrigera (Araneae: Lycosidae). Acta Arachnol. 2015, 64, 1–4. [Google Scholar] [CrossRef]
- Korenko, S.; Pekár, S.; Honěk, A. Predation activity of two winter–active spiders (Araneae: Anyphaenidae, Philodromidae). J. Therm. Biol. 2010, 35, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.; Moran, N.A. The gut microbiota of insects—Diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef]
- Neelakanta, G.; Sultana, H.; Fish, D.; Anderson, J.F.; Fikrig, E. Anaplasma phagocytophilum induces Ixodes scapularis ticks to express an antifreeze glycoprotein gene that enhances their survival in the cold. J. Clin. Investig. 2010, 120, 3179–3190. [Google Scholar] [CrossRef]
- Tanaka, K.; Watanabe, M. Transmission of ice-nucleating active bacteria from a prey reduces cold hardiness of a predator (Araneae: Theridiidae). Naturwissenschaften 2003, 90, 449–451. [Google Scholar] [CrossRef]
- Tanaka, K. Supercooling ability in the house spider, Achaearanea tepidariorum: Effect of field-collected and laboratory-reared prey. Naturwissenschaften 2001, 88, 431–433. [Google Scholar] [CrossRef]
- Morgan-Richards, M.; Marshall, C.J.; Biggs, P.J.; Trewick, S.A. Insect freeze–tolerance downunder: The microbial connection. Insects 2023, 14, 89. [Google Scholar] [CrossRef] [PubMed]
- Moghadam, N.N.; Thorshauge, P.M.; Kristensen, T.N.; de Jonge, N.; Bahrndorff, S.; Kjeldal, H.; Nielsen, J.L. Strong responses of Drosophila melanogaster microbiota to developmental temperature. Fly 2018, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, H.; Tang, M. Community structure of gut bacteria of Dendroctonus armandi (Coleoptera: Curculionidae: Scolytinae) larvae during overwintering stage. Sci. Rep. 2017, 7, 14242. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Dong, Y.; Shi, F.; Xu, Y.; Ge, S.; Tao, J.; Ren, L.; Zong, S. Seasonal shifts in cold tolerance and the composition of the gut microbiome of Dendroctonus valens LeConte occur concurrently. Forests 2021, 12, 888. [Google Scholar] [CrossRef]
- Liu, P.; Zhu, Y.; Liang, Y.; Shi, T.; Li, L.; Cao, H.; Yu, L. Overwintering honeybees maintained dynamic and stable intestinal bacteria. Sci. Rep. 2021, 11, 22233. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, L.V.; Dhakal, P.; Lebenzon, J.E.; Heinrichs, D.E.; Bucking, C.; Sinclair, B.J. Seasonal shifts in the insect gut microbiome are concurrent with changes in cold tolerance and immunity. Funct. Ecol. 2018, 32, 2357–2368. [Google Scholar] [CrossRef]
- Raza, M.F.; Wang, Y.; Cai, Z.; Bai, S.; Yao, Z.; Awan, U.A.; Zhang, Z.; Zheng, W.; Zhang, H. Gut microbiota promotes host resistance to low–temperature stress by stimulating its arginine and proline metabolism pathway in adult Bactrocera dorsalis. PLoS Pathog. 2020, 16, e1008441. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, Z.; Liu, R.; Zhang, X.; Chen, J.; Peng, Y. Effects of the metals lead and zinc on the growth, development, and reproduction of Pardosa astrigera (Araneae: Lycosidae). Bull. Environm. Contam. Toxicol. 2011, 86, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Quan, X.; Wu, L.; Zhou, Q.; Yun, Y.; Peng, Y.; Chen, J. Identification of predation by spiders on the diamondback moth Plutella xylostella. Bull. Insectol. 2011, 64, 223–227. [Google Scholar]
- Yang, J.; Wu, Q.; Xiao, R.; Zhao, J.; Chen, J.; Jiao, X. Seasonal variations in body melanism and size of the wolf spider Pardosa astrigera (Araneae: Lycosidae). Ecol. Evol. 2018, 8, 4352–4359. [Google Scholar] [CrossRef]
- Liu, J. Mechanisms of Tolerance Induced by Temperature in Pardosa astrigera L. Koch. Ph.D. Thesis, Shanxi Agricultural University, Jinzhong, China, 2014. [Google Scholar]
- Gao, Y.; Wu, P.; Cui, S.; Ali, A.; Zheng, G. Divergence in gut bacterial community between females and males in the wolf spider Pardosa astrigera. Ecol. Evol. 2022, 12, e8823. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Jiang, Y.; Cao, Y.; Sun, B.; Xiang, X. Divergence in gut bacterial community structure between male and female stag beetles Odontolabis fallaciosa (Coleoptera, Lucanidae). Animals 2020, 10, 2352. [Google Scholar] [CrossRef] [PubMed]
- Foster, W.A. Mosquito sugar feeding and reproductive energetics. Annu. Rev. Entomol. 1995, 40, 443–474. [Google Scholar] [CrossRef] [PubMed]
- Zouache, K.; Raharimalala, F.N.; Raquin, V.; Tran-Van, V.; Raveloson, L.H.; Ravelonandro, P.; Mavingui, P. Bacterial diversity of field–caught mosquitoes, Aedes albopictus and Aedes aegypti, from different geographic regions of Madagascar. FEMS Microbiol. Ecol. 2011, 75, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Tyagi, I.; Tyagi, K.; Chandra, K. Diversity and structure of bacterial communities in the gut of spider: Thomisidae and Oxyopidae. Front. Ecol. Evol. 2020, 8, 588102. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra–fastall–in–one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Song, D. A review of the reproductive behavior of wolf spiders. Acta Arachnol. Sin. 1999, 8, 55–62. [Google Scholar]
- Hao, C.; de Jonge, N.; Zhu, D.; Feng, L.C.; Zhang, B.; Chen, T.W.; Wu, D.H.; Nielsen, J.L. Food origin influences microbiota and stable isotope enrichment profiles of cold–adapted Collembola (Desoria ruseki). Front. Microbiol. 2022, 13, 1030429. [Google Scholar] [CrossRef] [PubMed]
- Didion, E.M.; Sabree, Z.L.; Kenyon, L.; Nine, G.; Hagan, R.W.; Osman, S.; Benoit, J.B. Microbiome reduction prevents lipid accumulation during early diapause in the northern house mosquito, Culex pipiens pipiens. J. Insect Physiol. 2021, 134, 104295. [Google Scholar] [CrossRef] [PubMed]
- Naloka, K.; Kuntaveesuk, A.; Muangchinda, C.; Chavanich, S.; Viyakarn, V.; Chen, B.; Pinyakong, O. Pseudomonas and Pseudarthrobacter are the key players in synergistic phenanthrene biodegradation at low temperatures. Sci. Rep. 2024, 14, 11976. [Google Scholar] [CrossRef] [PubMed]
- Barbaro, S.E.; Trevors, J.T.; Inniss, W.E. Effects of low temperature, cold shock, and various carbon sources on esterase and lipase activities and exopolysaccharide production by a psychrotrophic Acinetobacter sp. Can. J. Microbiol. 2001, 47, 194–205. [Google Scholar] [CrossRef] [PubMed]
- Castrillo, L.A.; Lee, R.E., Jr.; Lee, M.R.; Wyman, J.A. Long-term retention of ice-nucleating active Pseudomonas fluorescens by overwintering colorado potato beetles. Cryo Lett. 2000, 21, 5–12. [Google Scholar]
- Zhou, J.; Wang, M.; Yi, X. Alteration of gut microbiota of a food–storing hibernator, siberian chipmunk Tamias sibiricus. Microb. Ecol. 2022, 84, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Png, C.W.; Linden, S.K.; Gilshenan, K.S.; Zoetendal, E.G.; McSweeney, C.S.; Sly, L.I.; McGuckin, M.A.; Florin, T.H. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 2010, 105, 2420–2428. [Google Scholar] [CrossRef]
- Rajilić-Stojanović, M.; Shanahan, F.; Guarner, F.; de Vos, W.M. Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm. Bowel. Dis. 2013, 19, 481–488. [Google Scholar] [CrossRef]
- See-Too, S.W.; Salazar, S.; Ee, R.; Convey, P.; Chan, K.G.; Peix, Á. Pseudomonas versuta sp. nov., isolated from Antarctic soil. Syst. Appl. Microbiol. 2017, 40, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Luong, T.M.; Ponamoreva, O.N.; Nechaeva, I.A.; Petrikov, K.V.; Delegan, Y.A.; Surin, A.K.; Linklater, D.; Filonov, A.E. Characterization of biosurfactants produced by the oil-degrading bacterium Rhodococcus erythropolis S67 at low temperature. World J. Microbiol. Biotechnol. 2018, 34, 20. [Google Scholar] [CrossRef] [PubMed]





| Sobs | Chao1 | Shannon | Simpson | Coverage | |
|---|---|---|---|---|---|
| Female | 162.67 ± 15.95 | 168.33 ± 13.30 | 3.13 ± 0.51 | 0.12 ± 0.07 | 0.9998 |
| Male | 142.33 ± 2.52 | 145.23 ± 3.53 | 1.60 ± 0.10 | 0.37 ± 0.02 | 0.9997 |
| p value * | 0.0946 | 0.0438 | 0.0069 | 0.0036 | 0.3050 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, N.; Yuan, Q.; Qi, Y.; Wu, P.; Cui, S.; Zheng, G. The Potential Implications of Sex-Specific Differences in the Intestinal Bacteria of the Overwintering Wolf Spider Pardosa astrigera (Araneae: Lycosidae). Insects 2024, 15, 490. https://doi.org/10.3390/insects15070490
Li N, Yuan Q, Qi Y, Wu P, Cui S, Zheng G. The Potential Implications of Sex-Specific Differences in the Intestinal Bacteria of the Overwintering Wolf Spider Pardosa astrigera (Araneae: Lycosidae). Insects. 2024; 15(7):490. https://doi.org/10.3390/insects15070490
Chicago/Turabian StyleLi, Ningkun, Quan Yuan, Yaru Qi, Pengfeng Wu, Shuyan Cui, and Guo Zheng. 2024. "The Potential Implications of Sex-Specific Differences in the Intestinal Bacteria of the Overwintering Wolf Spider Pardosa astrigera (Araneae: Lycosidae)" Insects 15, no. 7: 490. https://doi.org/10.3390/insects15070490
APA StyleLi, N., Yuan, Q., Qi, Y., Wu, P., Cui, S., & Zheng, G. (2024). The Potential Implications of Sex-Specific Differences in the Intestinal Bacteria of the Overwintering Wolf Spider Pardosa astrigera (Araneae: Lycosidae). Insects, 15(7), 490. https://doi.org/10.3390/insects15070490






