Fungal Colonization and Infections—Interactions with Other Human Diseases
Abstract
:1. Introduction
2. Association of Candida in Development and Progression of Cancer
3. Role of Candida in IBD
4. Candida and Other Fungal Infections in COVID-19
5. Established Immune Mechanisms in Candida Associated Co-Morbidities
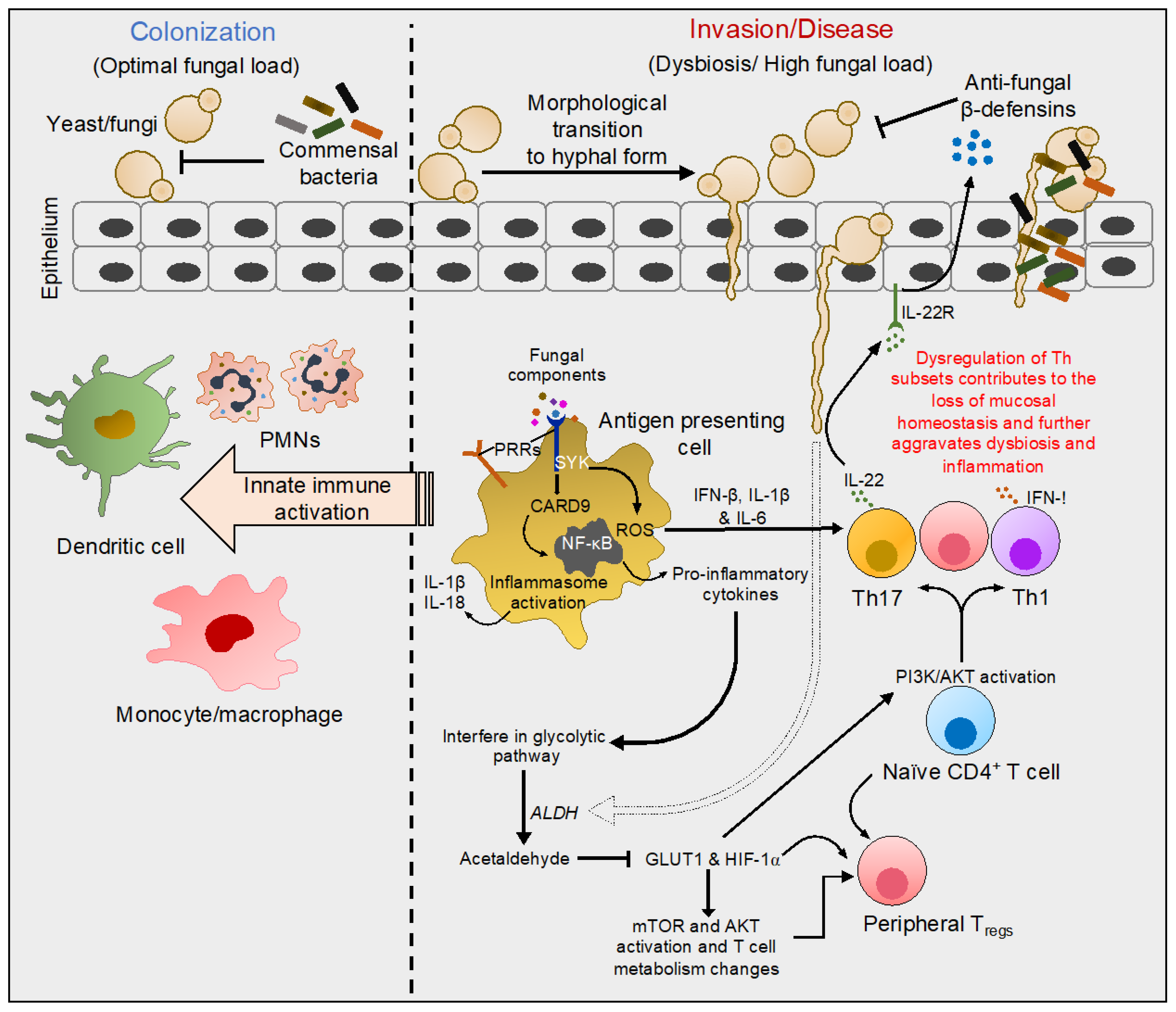
6. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and Multi-National Prevalence of Fungal Diseases-Estimate Precision. J. Fungi. 2017, 3, 57. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, E.; Smith, P.B.; Jacqz-Aigrain, E.; Kaguelidou, F.; Cohen-Wolkowiez, M.; Manzoni, P.; Benjamin, D.K. Neonatal fungal infections: When to treat? Early Hum. Dev. 2012, 88 (Suppl. S2), S6–S10. [Google Scholar] [CrossRef] [Green Version]
- Eisi, H.; Ibraheem, S.; Hisham, T.; Al-Harbi, A.; Saidy, K.; Ali, I.; Nour, I.; Nasef, N. Risk factors and outcomes of deep tissue Candida invasion in neonates with invasive candidiasis. Mycoses 2021, 65, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Perlin, D.S.; Rautemaa-Richardson, R.; Alastruey-Izquierdo, A. The global problem of antifungal resistance: Prevalence, mechanisms, and management. Lancet Infect. Dis. 2017, 17, e383–e392. [Google Scholar] [CrossRef]
- Benedict, K.; Richardson, M.; Vallabhaneni, S.; Jackson, B.R.; Chiller, T. Emerging issues, challenges, and changing epidemiology of fungal disease outbreaks. Lancet Infect. Dis. 2017, 17, e403–e411. [Google Scholar] [CrossRef]
- Gavaldà, J.; Meije, Y.; Fortún, J.; Roilides, E.; Saliba, F.; Lortholary, O.; Muñoz, P.; Grossi, P.; Cuenca-Estrella, M.; ESCMID Study Group for Infections in Compromised Hosts. Invasive fungal infections in solid organ transplant recipients. Clin. Microbiol. Infect. 2014, 20 (Suppl. 7), 27–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tönshoff, B. Immunosuppressants in Organ Transplantation. Handb. Exp. Pharmacol. 2020, 261, 441–469. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.W.; Collins, S.A.B.; Resneck, J.S.; Bolognia, J.L.; Hodge, J.A.; Rohrer, T.A.; Van Beek, M.J.; Margolis, D.J.; Sober, A.J.; Weinstock, M.A.; et al. The burden of skin disease in the United States. J. Am. Acad. Dermatol. 2017, 76, 958–972.e2. [Google Scholar] [CrossRef] [Green Version]
- Ortman, J.M.; Velkoff, V.A.; Hogan, H. An Aging Nation: The Older Population in the United States. Available online: https://www.census.gov/library/publications/2014/demo/p25-1140.html (accessed on 1 November 2021).
- Dekkers, B.G.J.; Veringa, A.; Marriott, D.J.E.; Boonstra, J.M.; van der Elst, K.C.M.; Doukas, F.F.; McLachlan, A.J.; Alffenaar, J.C. Invasive Candidiasis in the Elderly: Considerations for Drug Therapy. Drugs Aging 2018, 35, 781–789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, T.; Kishi, M.; Suda, M.; Sakata, K.; Shimoda, H.; Miura, H.; Ogawa, A.; Kobayashi, S. Prevalence of Candida albicans and non-albicans on the tongue dorsa of elderly people living in a post-disaster area: A cross-sectional survey. BMC Oral Health 2017, 17, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barchiesi, F.; Orsetti, E.; Mazzanti, S.; Trave, F.; Salvi, A.; Nitti, C.; Manso, E. Candidemia in the elderly: What does it change? PLoS ONE 2017, 12, e0176576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miceli, M.H.; Díaz, J.A.; Lee, S.A. Emerging opportunistic yeast infections. Lancet Infect. Dis. 2011, 11, 142–151. [Google Scholar] [CrossRef]
- Kabir, M.A.; Ahmad, Z. Candida infections and their prevention. ISRN Prev. Med. 2013, 2013, 763628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kullberg, B.J.; Arendrup, M.C. Invasive Candidiasis. N. Engl. J. Med. 2015, 373, 1445–1456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cleveland, A.A.; Harrison, L.H.; Farley, M.M.; Hollick, R.; Stein, B.; Chiller, T.M.; Lockhart, S.R.; Park, B.J. Declining incidence of candidemia and the shifting epidemiology of Candida resistance in two US metropolitan areas, 2008-2013: Results from population-based surveillance. PLoS ONE 2015, 10, e0120452. [Google Scholar] [CrossRef] [Green Version]
- Pappas, P.G.; Lionakis, M.S.; Arendrup, M.C.; Ostrosky-Zeichner, L.; Kullberg, B.J. Invasive candidiasis. Nat. Rev. Dis. Primers 2018, 4, 18026. [Google Scholar] [CrossRef] [PubMed]
- McCarty, T.P.; White, C.M.; Pappas, P.G. Candidemia and Invasive Candidiasis. Infect. Dis. Clin. North. Am. 2021, 35, 389–413. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Diekema, D.J.; Turnidge, J.D.; Castanheira, M.; Jones, R.N. Twenty Years of the SENTRY Antifungal Surveillance Program: Results for Candida Species From 1997–2016. Open Forum Infect. Dis. 2019, 6, S79–S94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patin, E.C.; Thompson, A.; Orr, S.J. Pattern recognition receptors in fungal immunity. Semin. Cell Dev. Biol. 2019, 89, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Netea, M.G.; Joosten, L.A.; van der Meer, J.W.; Kullberg, B.J.; van de Veerdonk, F.L. Immune defence against Candida fungal infections. Nat. Rev. Immunol. 2015, 15, 630–642. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vautier, S.; MacCallum, D.M.; Brown, G.D. C-type lectin receptors and cytokines in fungal immunity. Cytokine 2012, 58, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Geijtenbeek, T.B.; Gringhuis, S.I. Signalling through C-type lectin receptors: Shaping immune responses. Nat. Rev. Immunol. 2009, 9, 465–479. [Google Scholar] [CrossRef] [PubMed]
- Geijtenbeek, T.B.; Gringhuis, S.I. C-type lectin receptors in the control of T helper cell differentiation. Nat. Rev. Immunol. 2016, 16, 433–448. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Pan, D.; Zhou, Z.; You, Y.; Jiang, C.; Zhao, X.; Lin, X. Dectin-3 Deficiency Promotes Colitis Development due to Impaired Antifungal Innate Immune Responses in the Gut. PLoS Pathog. 2016, 12, e1005662. [Google Scholar] [CrossRef] [Green Version]
- Rogers, N.C.; Slack, E.C.; Edwards, A.D.; Nolte, M.A.; Schulz, O.; Schweighoffer, E.; Williams, D.L.; Gordon, S.; Tybulewicz, V.L.; Brown, G.D.; et al. Syk-dependent cytokine induction by Dectin-1 reveals a novel pattern recognition pathway for C type lectins. Immunity 2005, 22, 507–517. [Google Scholar] [CrossRef] [Green Version]
- Gross, O.; Gewies, A.; Finger, K.; Schäfer, M.; Sparwasser, T.; Peschel, C.; Förster, I.; Ruland, J. Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature 2006, 442, 651–656. [Google Scholar] [CrossRef]
- Gringhuis, S.I.; den Dunnen, J.; Litjens, M.; van der Vlist, M.; Wevers, B.; Bruijns, S.C.; Geijtenbeek, T.B. Dectin-1 directs T helper cell differentiation by controlling noncanonical NF-kappaB activation through Raf-1 and Syk. Nat. Immunol. 2009, 10, 203–213. [Google Scholar] [CrossRef]
- LeibundGut-Landmann, S.; Gross, O.; Robinson, M.J.; Osorio, F.; Slack, E.C.; Tsoni, S.V.; Schweighoffer, E.; Tybulewicz, V.; Brown, G.D.; Ruland, J.; et al. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat. Immunol. 2007, 8, 630–638. [Google Scholar] [CrossRef]
- Yu, M.; Song, X.T.; Liu, B.; Luan, T.T.; Liao, S.L.; Zhao, Z.T. The Emerging Role of Mast Cells in Response to Fungal Infection. Front. Immunol. 2021, 12, 688659. [Google Scholar] [CrossRef]
- Falk, P.G.; Hooper, L.V.; Midtvedt, T.; Gordon, J.I. Creating and maintaining the gastrointestinal ecosystem: What we know and need to know from gnotobiology. Microbiol. Mol. Biol. Rev. 1998, 62, 1157–1170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonnenburg, J.L.; Angenent, L.T.; Gordon, J.I. Getting a grip on things: How do communities of bacterial symbionts become established in our intestine? Nat. Immunol. 2004, 5, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Ghannoum, M.A.; Jurevic, R.J.; Mukherjee, P.K.; Cui, F.; Sikaroodi, M.; Naqvi, A.; Gillevet, P.M. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010, 6, e1000713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, P.K.; Sendid, B.; Hoarau, G.; Colombel, J.F.; Poulain, D.; Ghannoum, M.A. Mycobiota in gastrointestinal diseases. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.K.; Wang, H.; Retuerto, M.; Zhang, H.; Burkey, B.; Ghannoum, M.A.; Eng, C. Bacteriome and mycobiome associations in oral tongue cancer. Oncotarget 2017, 8, 97273–97289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, G.D. Innate antifungal immunity: The key role of phagocytes. Annu. Rev. Immunol. 2011, 29, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Berman, J.; Sudbery, P.E. Candida Albicans: A molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 2002, 3, 918–930. [Google Scholar] [CrossRef]
- Qin, Y.; Zhang, L.; Xu, Z.; Zhang, J.; Jiang, Y.Y.; Cao, Y.; Yan, T. Innate immune cell response upon Candida albicans infection. Virulence 2016, 7, 512–526. [Google Scholar] [CrossRef] [Green Version]
- Richardson, J.P.; Moyes, D.L. Adaptive immune responses to Candida albicans infection. Virulence 2015, 6, 327–337. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Cheng, L.; Lei, Y.L.; Ren, B.; Zhou, X. The Interactions between Candida albicans and mucosal immunity. Front. Microbiol. 2021, 12, 652725. [Google Scholar] [CrossRef]
- Villar, C.C.; Dongari-Bagtzoglou, A. Fungal diseases: Oral dysbiosis in susceptible hosts. Periodontol 2000 2021, 87, 166–180. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Weiderpass, E.; Soerjomataram, I. The ever-increasing importance of cancer as a leading cause of premature death worldwide. Cancer 2021, 127, 3029–3030. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Garcia, A.; Rementeria, A.; Aguirre-Urizar, J.M.; Moragues, M.D.; Antoran, A.; Pellon, A.; Abad-Diaz-de-Cerio, A.; Hernando, F.L. Candida albicans and cancer: Can this yeast induce cancer development or progression? Crit. Rev. Microbiol. 2016, 42, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Dolgin, E. Fighting cancer with microbes. Nature 2020, 577, S16–S18. [Google Scholar] [CrossRef] [PubMed]
- Whisner, C.M.; Athena Aktipis, C. The Role of the Microbiome in Cancer Initiation and Progression: How Microbes and Cancer Cells Utilize Excess Energy and Promote One Another’s Growth. Curr. Nutr. Rep. 2019, 8, 42–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhaskaran, N.; Jayaraman, S.; Quigley, C.; Mamileti, P.; Ghannoum, M.; Weinberg, A.; Thuener, J.; Pan, Q.; Pandiyan, P. The Role of Dectin-1 Signaling in Altering Tumor Immune Microenvironment in the Context of Aging. Front. Oncol. 2021, 11, 669066. [Google Scholar] [CrossRef] [PubMed]
- O’Higgins, C.; Ward, F.J.; Abu Eid, R. Deciphering the Role of Regulatory CD4 T Cells in Oral and Oropharyngeal Cancer: A Systematic Review. Front. Oncol. 2018, 8, 442. [Google Scholar] [CrossRef]
- Lee, C.H.; Chang, J.S.; Syu, S.H.; Wong, T.S.; Chan, J.Y.; Tang, Y.C.; Yang, Z.P.; Yang, W.C.; Chen, C.T.; Lu, S.C.; et al. IL-1β promotes malignant transformation and tumor aggressiveness in oral cancer. J. Cell Physiol. 2015, 230, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Perera, M.; Al-Hebshi, N.N.; Perera, I.; Ipe, D.; Ulett, G.C.; Speicher, D.J.; Chen, T.; Johnson, N.W. A dysbiotic mycobiome dominated by. J. Oral Microbiol. 2017, 9, 1385369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, L.M.; Liang, J.A.; Lin, C.L.; Sun, L.M.; Kao, C.H. Cancer risk in patients with candidiasis: A nationwide population-based cohort study. Oncotarget 2017, 8, 63562–63573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mellinghoff, S.C.; Thelen, M.; Bruns, C.; Garcia-Marquez, M.; Hartmann, P.; Lammertz, T.; Lehmann, J.; Nowag, A.; Stemler, J.; Wennhold, K.; et al. T-cells of invasive candidiasis patients show patterns of T-cell-exhaustion suggesting checkpoint blockade as treatment option. J. Infect. 2021. [Google Scholar] [CrossRef]
- Pellon, A.; Sadeghi Nasab, S.D.; Moyes, D.L. New Insights in Candida albicans Innate Immunity at the Mucosa: Toxins, Epithelium, Metabolism, and Beyond. Front. Cell Infect. Microbiol. 2020, 10, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Cosola, M.; Cazzolla, A.P.; Charitos, I.A.; Ballini, A.; Inchingolo, F.; Santacroce, L. Candida albicans and Oral Carcinogenesis. A Brief Review. J. Fungi. 2021, 7, 476. [Google Scholar] [CrossRef] [PubMed]
- Alnuaimi, A.D.; Wiesenfeld, D.; O’Brien-Simpson, N.M.; Reynolds, E.C.; McCullough, M.J. Oral Candida colonization in oral cancer patients and its relationship with traditional risk factors of oral cancer: A matched case-control study. Oral Oncol. 2015, 51, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Kaźmierczak-Siedlecka, K.; Dvořák, A.; Folwarski, M.; Daca, A.; Przewłócka, K.; Makarewicz, W. Fungal Gut Microbiota Dysbiosis and Its Role in Colorectal, Oral, and Pancreatic Carcinogenesis. Cancers 2020, 12, 1326. [Google Scholar] [CrossRef]
- Mäkinen, A.; Nawaz, A.; Mäkitie, A.; Meurman, J.H. Role of Non-Albicans Candida and Candida Albicans in Oral Squamous Cell Cancer Patients. J. Oral Maxillofac. Surg. 2018, 76, 2564–2571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayachandran, A.L.; Katragadda, R.; Thyagarajan, R.; Vajravelu, L.; Manikesi, S.; Kaliappan, S.; Jayachandran, B. Oral Candidiasis among Cancer Patients Attending a Tertiary Care Hospital in Chennai, South India: An Evaluation of Clinicomycological Association and Antifungal Susceptibility Pattern. Can. J. Infect. Dis. Med. Microbiol. 2016, 2016, 8758461. [Google Scholar] [CrossRef] [Green Version]
- Mäkinen, A.I.; Mäkitie, A.; Meurman, J.H. Candida prevalence in saliva before and after oral cancer treatment. Surgeon 2021, 19, e446–e451. [Google Scholar] [CrossRef] [PubMed]
- Hongal, B.P.; Kulkarni, V.V.; Deshmukh, R.S.; Joshi, P.S.; Karande, P.P.; Shroff, A.S. Prevalence of fungal hyphae in potentially malignant lesions and conditions-does its occurrence play a role in epithelial dysplasia? J. Oral Maxillofac. Pathol. 2015, 19, 10–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, S.K.; Astekar, M.; Sapra, G.; Chitlangia, R.K.; Raj, N. Evaluation of candidal species among individuals with oral potentially malignant disorders and oral squamous cell carcinoma. J. Oral Maxillofac. Pathol. 2019, 23, 302. [Google Scholar] [CrossRef] [PubMed]
- Isacco, C.G.; Ballini, A.; De Vito, D.; Nguyen, K.C.D.; Cantore, S.; Bottalico, L.; Quagliuolo, L.; Boccellino, M.; Di Domenico, M.; Santacroce, L.; et al. Rebalancing the Oral Microbiota as an Efficient Tool in Endocrine, Metabolic and Immune Disorders. Endocr. Metab. Immune Disord. Drug. Targets 2021, 21, 777–784. [Google Scholar] [CrossRef] [PubMed]
- Santacroce, L.; Carlaio, R.G.; Bottalico, L. Does it make sense that diabetes is reciprocally associated with periodontal disease? Endocr. Metab. Immune Disord. Drug. Targets 2010, 10, 57–70. [Google Scholar] [CrossRef]
- Hawkins, B.L.; Heniford, B.W.; Ackermann, D.M.; Leonberger, M.; Martinez, S.A.; Hendler, F.J. 4NQO carcinogenesis: A mouse model of oral cavity squamous cell carcinoma. Head Neck 1994, 16, 424–432. [Google Scholar] [CrossRef] [PubMed]
- O’Grady, J.F.; Reade, P.C. Candida albicans as a promoter of oral mucosal neoplasia. Carcinogenesis 1992, 13, 783–786. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, K.; Fifita, S.F.; Kuyama, K. The Cytological Findings of Oral Inflammatory Lesions, Lichen Planus and Leukoplakia Coexisted with and without Candida: With Special Reference to Clinical, Histopathological, Immunohistochemical and Flow Cytometrical Analyses. Int. J. Oral-Med. Sci. 2007, 6, 81–90. [Google Scholar] [CrossRef]
- Rivera, C. Essentials of oral cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 11884–11894. [Google Scholar]
- Rivera, C.; Venegas, B. Histological and molecular aspects of oral squamous cell carcinoma (Review). Oncol. Lett. 2014, 8, 7–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engku Nasrullah Satiman, E.A.F.; Ahmad, H.; Ramzi, A.B.; Abdul Wahab, R.; Kaderi, M.A.; Wan Harun, W.H.A.; Dashper, S.; McCullough, M.; Arzmi, M.H. The role of Candida albicans candidalysin ECE1 gene in oral carcinogenesis. J. Oral Pathol. Med. 2020, 49, 835–841. [Google Scholar] [CrossRef] [PubMed]
- Whiley, R.A.; Cruchley, A.T.; Gore, C.; Hagi-Pavli, E. Candida albicans strain-dependent modulation of pro-inflammatory cytokine release by in vitro oral and vaginal mucosal models. Cytokine 2012, 57, 89–97. [Google Scholar] [CrossRef]
- Gupta, S.R.; Gupta, N.; Sharma, A.; Xess, I.; Singh, G.; Mani, K. The association of Candida and antifungal therapy with pro-inflammatory cytokines in oral leukoplakia. Clin. Oral Investig. 2021, 25, 6287–6296. [Google Scholar] [CrossRef] [PubMed]
- Seitz, H.K.; Stickel, F. Molecular mechanisms of alcohol-mediated carcinogenesis. Nat. Rev. Cancer 2007, 7, 599–612. [Google Scholar] [CrossRef] [PubMed]
- Niu, B.; Liao, K.; Zhou, Y.; Wen, T.; Quan, G.; Pan, X.; Wu, C. Application of glutathione depletion in cancer therapy: Enhanced ROS-based therapy, ferroptosis, and chemotherapy. Biomaterials 2021, 277, 121110. [Google Scholar] [CrossRef] [PubMed]
- Perillo, B.; Di Donato, M.; Pezone, A.; Di Zazzo, E.; Giovannelli, P.; Galasso, G.; Castoria, G.; Migliaccio, A. ROS in cancer therapy: The bright side of the moon. Exp. Mol. Med. 2020, 52, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Zhao, Y. Acetaldehyde induces phosphorylation of dynamin-related protein 1 and mitochondrial dysfunction via elevating intracellular ROS and Ca2+ levels. Redox Biol. 2020, 28, 101381. [Google Scholar] [CrossRef]
- Stornetta, A.; Guidolin, V.; Balbo, S. Alcohol-Derived Acetaldehyde Exposure in the Oral Cavity. Cancers 2018, 10, 20. [Google Scholar] [CrossRef] [Green Version]
- Cohen, S.; Danzaki, K.; MacIver, N.J. Nutritional effects on T-cell immunometabolism. Eur. J. Immunol. 2017, 47, 225–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacIver, N.J.; Michalek, R.D.; Rathmell, J.C. Metabolic regulation of T lymphocytes. Annu. Rev. Immunol. 2013, 31, 259–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Zhou, Z.; Ren, T.; Kim, S.J.; He, Y.; Seo, W.; Guillot, A.; Ding, Y.; Wu, R.; Shao, S.; et al. Alcohol inhibits T-cell glucose metabolism and hepatitis in ALDH2-deficient mice and humans: Roles of acetaldehyde and glucocorticoids. Gut 2019, 68, 1311–1322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dang, E.V.; Barbi, J.; Yang, H.Y.; Jinasena, D.; Yu, H.; Zheng, Y.; Bordman, Z.; Fu, J.; Kim, Y.; Yen, H.R.; et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell 2011, 146, 772–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clambey, E.T.; McNamee, E.N.; Westrich, J.A.; Glover, L.E.; Campbell, E.L.; Jedlicka, P.; de Zoeten, E.F.; Cambier, J.C.; Stenmark, K.R.; Colgan, S.P.; et al. Hypoxia-inducible factor-1 alpha-dependent induction of FoxP3 drives regulatory T-cell abundance and function during inflammatory hypoxia of the mucosa. Proc. Natl. Acad. Sci. USA 2012, 109, E2784–E2793. [Google Scholar] [CrossRef] [Green Version]
- Bhaskaran, N.; Faddoul, F.; Paes da Silva, A.; Jayaraman, S.; Schneider, E.; Mamileti, P.; Weinberg, A.; Pandiyan, P. IL-1β-MyD88-mTOR Axis Promotes Immune-Protective IL-17A. Front. Immunol. 2020, 11, 595936. [Google Scholar] [CrossRef] [PubMed]
- de Souza, H.S.; Fiocchi, C. Immunopathogenesis of IBD: Current state of the art. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.C.; Shi, H.Y.; Hamidi, N.; Underwood, F.E.; Tang, W.; Benchimol, E.I.; Panaccione, R.; Ghosh, S.; Wu, J.C.Y.; Chan, F.K.L.; et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: A systematic review of population-based studies. Lancet 2017, 390, 2769–2778. [Google Scholar] [CrossRef]
- Li, X.V.; Leonardi, I.; Iliev, I.D. Gut Mycobiota in Immunity and Inflammatory Disease. Immunity 2019, 50, 1365–1379. [Google Scholar] [CrossRef]
- Paterson, M.J.; Oh, S.; Underhill, D.M. Host-microbe interactions: Commensal fungi in the gut. Curr. Opin. Microbiol. 2017, 40, 131–137. [Google Scholar] [CrossRef]
- Bhaskaran, N.; Quigley, C.; Paw, C.; Butala, S.; Schneider, E.; Pandiyan, P. Role of Short Chain Fatty Acids in Controlling Tregs and Immunopathology During Mucosal Infection. Front. Microbiol. 2018, 9, 1995. [Google Scholar] [CrossRef]
- Pandiyan, P.; Bhaskaran, N.; Zou, M.; Schneider, E.; Jayaraman, S.; Huehn, J. Microbiome Dependent Regulation of Tregs and Th17 Cells in Mucosa. Front. Immunol. 2019, 10, 426. [Google Scholar] [CrossRef] [Green Version]
- Mason, K.L.; Erb Downward, J.R.; Mason, K.D.; Falkowski, N.R.; Eaton, K.A.; Kao, J.Y.; Young, V.B.; Huffnagle, G.B. Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infect. Immun. 2012, 80, 3371–3380. [Google Scholar] [CrossRef] [Green Version]
- Ben-Ami, R.; Olshtain-Pops, K.; Krieger, M.; Oren, I.; Bishara, J.; Dan, M.; Wiener-Well, Y.; Weinberger, M.; Zimhony, O.; Chowers, M.; et al. Antibiotic exposure as a risk factor for fluconazole-resistant Candida bloodstream infection. Antimicrob. Agents Chemother. 2012, 56, 2518–2523. [Google Scholar] [CrossRef] [Green Version]
- Sokol, H.; Leducq, V.; Aschard, H.; Pham, H.P.; Jegou, S.; Landman, C.; Cohen, D.; Liguori, G.; Bourrier, A.; Nion-Larmurier, I.; et al. Fungal microbiota dysbiosis in IBD. Gut 2017, 66, 1039–1048. [Google Scholar] [CrossRef] [Green Version]
- Wheeler, M.L.; Limon, J.J.; Bar, A.S.; Leal, C.A.; Gargus, M.; Tang, J.; Brown, J.; Funari, V.A.; Wang, H.L.; Crother, T.R.; et al. Immunological Consequences of Intestinal Fungal Dysbiosis. Cell Host Microbe 2016, 19, 865–873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iliev, I.D.; Funari, V.A.; Taylor, K.D.; Nguyen, Q.; Reyes, C.N.; Strom, S.P.; Brown, J.; Becker, C.A.; Fleshner, P.R.; Dubinsky, M.; et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 2012, 336, 1314–1317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chehoud, C.; Albenberg, L.G.; Judge, C.; Hoffmann, C.; Grunberg, S.; Bittinger, K.; Baldassano, R.N.; Lewis, J.D.; Bushman, F.D.; Wu, G.D. Fungal Signature in the Gut Microbiota of Pediatric Patients With Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2015, 21, 1948–1956. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, C.; Tang, C.; He, Q.; Li, N.; Li, J. Dysbiosis of gut fungal microbiota is associated with mucosal inflammation in Crohn’s disease. J. Clin. Gastroenterol. 2014, 48, 513–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liguori, G.; Lamas, B.; Richard, M.L.; Brandi, G.; da Costa, G.; Hoffmann, T.W.; Di Simone, M.P.; Calabrese, C.; Poggioli, G.; Langella, P.; et al. Fungal Dysbiosis in Mucosa-associated Microbiota of Crohn’s Disease Patients. J. Crohns Colitis 2016, 10, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhya, I.; Hansen, R.; Meharg, C.; Thomson, J.M.; Russell, R.K.; Berry, S.H.; El-Omar, E.M.; Hold, G.L. The fungal microbiota of de-novo paediatric inflammatory bowel disease. Microbes Infect. 2015, 17, 304–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, T.; Ng, S.C. The Gut Microbiota in the Pathogenesis and Therapeutics of Inflammatory Bowel Disease. Front. Microbiol. 2018, 9, 2247. [Google Scholar] [CrossRef]
- Ott, S.J.; Kühbacher, T.; Musfeldt, M.; Rosenstiel, P.; Hellmig, S.; Rehman, A.; Drews, O.; Weichert, W.; Timmis, K.N.; Schreiber, S. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scand. J. Gastroenterol. 2008, 43, 831–841. [Google Scholar] [CrossRef]
- Colombel, J.F.; Sendid, B.; Jouault, T.; Poulain, D. Secukinumab failure in Crohn’s disease: The yeast connection? Gut 2013, 62, 800–801. [Google Scholar] [CrossRef]
- Li, J.; Chen, D.; Yu, B.; He, J.; Zheng, P.; Mao, X.; Yu, J.; Luo, J.; Tian, G.; Huang, Z.; et al. Fungi in Gastrointestinal Tracts of Human and Mice: From Community to Functions. Microb. Ecol. 2018, 75, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Zhang, F.; Yang, X.; Wu, N.; Jiang, W.; Li, X.; Liu, Y. Changes in the composition of intestinal fungi and their role in mice with dextran sulfate sodium-induced colitis. Sci. Rep. 2015, 5, 10416. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Yoshida, M.; Ishikawa, H.; Kameyama, K.; Wakabayashi, G.; Otani, Y.; Shimazu, M.; Tanabe, M.; Kawachi, S.; Kumai, K.; et al. Candida albicans aggravates duodenal ulcer perforation induced by administration of cysteamine in rats. J. Gastroenterol. Hepatol. 2007, 22, 749–756. [Google Scholar] [CrossRef]
- Chiffoleau, E. C-Type Lectin-Like Receptors As Emerging Orchestrators of Sterile Inflammation Represent Potential Therapeutic Targets. Front. Immunol. 2018, 9, 227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021, 19, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Wiersinga, W.J.; Rhodes, A.; Cheng, A.C.; Peacock, S.J.; Prescott, H.C. Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19): A Review. JAMA 2020, 324, 782–793. [Google Scholar] [CrossRef]
- World Health Organization. Available online: https://covid19.who.int (accessed on 25 January 2022).
- Sharma, A.; Ahmad Farouk, I.; Lal, S.K. COVID-19: A Review on the Novel Coronavirus Disease Evolution, Transmission, Detection, Control and Prevention. Viruses 2021, 13, 202. [Google Scholar] [CrossRef]
- Soltani, S.; Zakeri, A.; Zandi, M.; Kesheh, M.M.; Tabibzadeh, A.; Dastranj, M.; Faramarzi, S.; Didehdar, M.; Hafezi, H.; Hosseini, P.; et al. The Role of Bacterial and Fungal Human Respiratory Microbiota in COVID-19 Patients. Biomed. Res. Int. 2021, 2021, 6670798. [Google Scholar] [CrossRef] [PubMed]
- Chowdhary, A.; Sharma, A. The lurking scourge of multidrug resistant Candida auris in times of COVID-19 pandemic. J. Glob. Antimicrob. Resist. 2020, 22, 175–176. [Google Scholar] [CrossRef] [PubMed]
- Talento, A.F.; Hoenigl, M. Fungal Infections Complicating COVID-19: With the Rain Comes the Spores. J. Fungi. 2020, 6, 279. [Google Scholar] [CrossRef] [PubMed]
- Mulet Bayona, J.V.; Tormo Palop, N.; Salvador García, C.; Fuster Escrivá, B.; Chanzá Aviñó, M.; Ortega García, P.; Gimeno Cardona, C. Impact of the SARS-CoV-2 Pandemic in Candidaemia, Invasive Aspergillosis and Antifungal Consumption in a Tertiary Hospital. J. Fungi. 2021, 7, 440. [Google Scholar] [CrossRef]
- Roudbary, M.; Kumar, S.; Kumar, A.; Černáková, L.; Nikoomanesh, F.; Rodrigues, C.F. Overview on the Prevalence of Fungal Infections, Immune Response, and Microbiome Role in COVID-19 Patients. J. Fungi. 2021, 7, 720. [Google Scholar] [CrossRef] [PubMed]
- Moser, D.; Biere, K.; Han, B.; Hoerl, M.; Schelling, G.; Choukér, A.; Woehrle, T. COVID-19 Impairs Immune Response to Candida albicans. Front. Immunol. 2021, 12, 640644. [Google Scholar] [CrossRef]
- Chen, X.; Liao, B.; Cheng, L.; Peng, X.; Xu, X.; Li, Y.; Hu, T.; Li, J.; Zhou, X.; Ren, B. The microbial coinfection in COVID-19. Appl. Microbiol. Biotechnol. 2020, 104, 7777–7785. [Google Scholar] [CrossRef] [PubMed]
- Hanson, B.M.; Dinh, A.Q.; Tran, T.T.; Arenas, S.; Pronty, D.; Gershengorn, H.B.; Ferreira, T.; Arias, C.A.; Shukla, B.S. Invasive Infections during a COVID-19 Case Surge. Antimicrob. Agents Chemother. 2021, 65, e0114621. [Google Scholar] [CrossRef] [PubMed]
- Casalini, G.; Giacomelli, A.; Ridolfo, A.; Gervasoni, C.; Antinori, S. Invasive Fungal Infections Complicating COVID-19: A Narrative Review. J. Fungi. 2021, 7, 921. [Google Scholar] [CrossRef]
- Wong, L.R.; Perlman, S. Immune dysregulation and immunopathology induced by SARS-CoV-2 and related coronaviruses-are we our own worst enemy? Nat. Rev. Immunol. 2021, 22, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Arastehfar, A.; Carvalho, A.; Nguyen, M.H.; Hedayati, M.T.; Netea, M.G.; Perlin, D.S.; Hoenigl, M. COVID-19-Associated Candidiasis (CAC): An Underestimated Complication in the Absence of Immunological Predispositions? J. Fungi. 2020, 6, 211. [Google Scholar] [CrossRef] [PubMed]
- Riad, A.; Gomaa, E.; Hockova, B.; Klugar, M. Oral candidiasis of COVID-19 patients: Case report and review of evidence. J. Cosmet. Dermatol. 2021, 20, 1580–1584. [Google Scholar] [CrossRef]
- Nucci, M.; Barreiros, G.; Guimarães, L.F.; Deriquehem, V.A.S.; Castiñeiras, A.C.; Nouér, S.A. Increased incidence of candidemia in a tertiary care hospital with the COVID-19 pandemic. Mycoses 2021, 64, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Katz, J. Prevalence of candidiasis and oral candidiasis in COVID-19 patients: A cross-sectional pilot study from the patients’ registry in a large health center. Quintessence Int. 2021, 52, 714–718. [Google Scholar] [CrossRef]
- Frías-De-León, M.G.; Pinto-Almazán, R.; Hernández-Castro, R.; García-Salazar, E.; Meza-Meneses, P.; Rodríguez-Cerdeira, C.; Arenas, R.; Conde-Cuevas, E.; Acosta-Altamirano, G.; Martínez-Herrera, E. Epidemiology of Systemic Mycoses in the COVID-19 Pandemic. J. Fungi. 2021, 7, 556. [Google Scholar] [CrossRef]
- Alanio, A.; Dellière, S.; Fodil, S.; Bretagne, S.; Mégarbane, B. Prevalence of putative invasive pulmonary aspergillosis in critically ill patients with COVID-19. Lancet Respir. Med. 2020, 8, e48–e49. [Google Scholar] [CrossRef]
- Gangneux, J.P.; Reizine, F.; Guegan, H.; Pinceaux, K.; Le Balch, P.; Prat, E.; Pelletier, R.; Belaz, S.; Le Souhaitier, M.; Le Tulzo, Y.; et al. Is the COVID-19 Pandemic a Good Time to Include Aspergillus Molecular Detection to Categorize Aspergillosis in ICU Patients? A Monocentric Experience. J. Fungi. 2020, 6, 105. [Google Scholar] [CrossRef] [PubMed]
- Wheat, L.J.; Azar, M.M.; Bahr, N.C.; Spec, A.; Relich, R.F.; Hage, C. Histoplasmosis. Infect. Dis. Clin. North. Am. 2016, 30, 207–227. [Google Scholar] [CrossRef]
- Azar, M.M.; Hage, C.A. Clinical Perspectives in the Diagnosis and Management of Histoplasmosis. Clin. Chest Med. 2017, 38, 403–415. [Google Scholar] [CrossRef]
- Messina, F.A.; Marin, E.; Caceres, D.H.; Romero, M.; Depardo, R.; Priarone, M.M.; Rey, L.; Vázquez, M.; Verweij, P.E.; Chiller, T.M.; et al. Coronavirus Disease 2019 (COVID-19) in a Patient with Disseminated Histoplasmosis and HIV-A Case Report from Argentina and Literature Review. J. Fungi. 2020, 6, 275. [Google Scholar] [CrossRef]
- Basso, R.P.; Poester, V.R.; Benelli, J.L.; Stevens, D.A.; Zogbi, H.E.; Vasconcellos, I.C.D.S.; Pasqualotto, A.C.; Xavier, M.O. COVID-19-Associated Histoplasmosis in an AIDS Patient. Mycopathologia 2021, 186, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Stasiak, C.E.S.; Nigri, D.H.; Cardoso, F.R.; Mattos, R.S.A.R.; Gonçalves Martins, P.A.; Carvalho, A.R.S.; Altino de Almeida, S.; Rodrigues, R.S.; Rosado-de-Castro, P.H. Case Report: Incidental Finding of COVID-19 Infection after Positron Emission Tomography/CT Imaging in a Patient with a Diagnosis of Histoplasmosis and Recurring Fever. Am. J. Trop. Med. Hyg. 2021, 104, 1651. [Google Scholar] [CrossRef]
- De Macedo, P.M.; Freitas, A.D.; Bártholo, T.P.; Bernardes-Engemann, A.R.; Almeida, M.A.; Almeida-Silva, F.; Zancopé-Oliveira, R.M.; Almeida-Paes, R. Acute Pulmonary Histoplasmosis Following COVID-19: Novel Laboratorial Methods Aiding Diagnosis. J. Fungi. 2021, 7, 346. [Google Scholar] [CrossRef]
- Bertolini, M.; Mutti, M.F.; Barletta, J.A.; Falak, A.; Cuatz, D.; Sisto, A.; Ragusa, M.A.; Fernandez Claros, N.O.; Rolón, M.J. COVID-19 associated with AIDS-related disseminated histoplasmosis: A case report. Int. J. STD AIDS 2020, 31, 1222–1224. [Google Scholar] [CrossRef]
- Khatib, M.Y.; Ahmed, A.A.; Shaat, S.B.; Mohamed, A.S.; Nashwan, A.J. Cryptococcemia in a patient with COVID-19: A case report. Clin. Case Rep. 2021, 9, 853–855. [Google Scholar] [CrossRef]
- Aranjani, J.M.; Manuel, A.; Abdul Razack, H.I.; Mathew, S.T. COVID-19-associated mucormycosis: Evidence-based critical review of an emerging infection burden during the pandemic’s second wave in India. PLoS Negl. Trop. Dis. 2021, 15, e0009921. [Google Scholar] [CrossRef]
- Chao, C.M.; Lai, C.C.; Yu, W.L. COVID-19 associated mucormycosis—An emerging threat. J. Microbiol. Immunol. Infect. 2022. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Yu, Y.; Xu, J.; Shu, H.; Xia, J.; Liu, H.; Wu, Y.; Zhang, L.; Yu, Z.; Fang, M.; et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study. Lancet Respir. Med. 2020, 8, 475–481. [Google Scholar] [CrossRef] [Green Version]
- Mang, S.; Kaddu-Mulindwa, D.; Metz, C.; Becker, A.; Seiler, F.; Smola, S.; Maßmann, A.; Becker, S.L.; Papan, C.; Bals, R.; et al. Pneumocystis jirovecii Pneumonia and Severe Acute Respiratory Syndrome Coronavirus 2 Coinfection in a Patient With Newly Diagnosed HIV-1 Infection. Clin. Infect. Dis. 2021, 72, 1487–1489. [Google Scholar] [CrossRef]
- Kim, D.; Kim, S.B.; Jeon, S.; Kim, S.; Lee, K.H.; Lee, H.S.; Han, S.H. No Change of Pneumocystis jirovecii Pneumonia after the COVID-19 Pandemic: Multicenter Time-Series Analyses. J. Fungi. 2021, 7, 990. [Google Scholar] [CrossRef]
- Viceconte, G.; Buonomo, A.R.; Lanzardo, A.; Pinchera, B.; Zappulo, E.; Scotto, R.; Schiano Moriello, N.; Vargas, M.; Iacovazzo, C.; Servillo, G.; et al. Pneumocystis jirovecii pneumonia in an immunocompetent patient recovered from COVID-19. Infect. Dis. 2021, 53, 382–385. [Google Scholar] [CrossRef]
- Nasrullah, A.; Javed, A.; Malik, K. Coronavirus Disease-Associated Pulmonary Aspergillosis: A Devastating Complication of COVID-19. Cureus 2021, 13, e13004. [Google Scholar] [CrossRef]
- Bacher, P.; Hohnstein, T.; Beerbaum, E.; Röcker, M.; Blango, M.G.; Kaufmann, S.; Röhmel, J.; Eschenhagen, P.; Grehn, C.; Seidel, K.; et al. Human Anti-fungal Th17 Immunity and Pathology Rely on Cross-Reactivity against Candida albicans. Cell 2019, 176, 1340–1355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leonardi, I.; Li, X.; Semon, A.; Li, D.; Doron, I.; Putzel, G.; Bar, A.; Prieto, D.; Rescigno, M.; McGovern, D.P.B.; et al. CX3CR1+ mononuclear phagocytes control immunity to intestinal fungi. Science 2018, 359, 232–236. [Google Scholar] [CrossRef] [Green Version]
- Witchley, J.N.; Penumetcha, P.; Abon, N.V.; Woolford, C.A.; Mitchell, A.P.; Noble, S.M. Candida albicans Morphogenesis Programs Control the Balance between Gut Commensalism and Invasive Infection. Cell Host Microbe 2019, 25, 432–443.e436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staab, J.F.; Bradway, S.D.; Fidel, P.L.; Sundstrom, P. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 1999, 283, 1535–1538. [Google Scholar] [CrossRef]
- Naglik, J.R.; König, A.; Hube, B.; Gaffen, S.L. Candida albicans-epithelial interactions and induction of mucosal innate immunity. Curr. Opin. Microbiol. 2017, 40, 104–112. [Google Scholar] [CrossRef]
- Liu, Y.; Filler, S.G. Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot. Cell 2011, 10, 168–173. [Google Scholar] [CrossRef] [Green Version]
- Moyes, D.L.; Wilson, D.; Richardson, J.P.; Mogavero, S.; Tang, S.X.; Wernecke, J.; Höfs, S.; Gratacap, R.L.; Robbins, J.; Runglall, M.; et al. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 2016, 532, 64–68. [Google Scholar] [CrossRef] [Green Version]
- Verma, A.H.; Richardson, J.P.; Zhou, C.; Coleman, B.M.; Moyes, D.L.; Ho, J.; Huppler, A.R.; Ramani, K.; McGeachy, M.J.; Mufazalov, I.A.; et al. Oral epithelial cells orchestrate innate type 17 responses to Candida albicans through the virulence factor candidalysin. Sci. Immunol. 2017, 2. [Google Scholar] [CrossRef] [Green Version]
- Allert, S.; Förster, T.M.; Svensson, C.M.; Richardson, J.P.; Pawlik, T.; Hebecker, B.; Rudolphi, S.; Juraschitz, M.; Schaller, M.; Blagojevic, M.; et al. Candida albicans-Induced Epithelial Damage Mediates Translocation through Intestinal Barriers. mBio 2018, 9, e00915-18. [Google Scholar] [CrossRef] [Green Version]
- Naglik, J.R.; Richardson, J.P.; Moyes, D.L. Candida albicans pathogenicity and epithelial immunity. PLoS Pathog. 2014, 10, e1004257. [Google Scholar] [CrossRef]
- Tang, S.X.; Moyes, D.L.; Richardson, J.P.; Blagojevic, M.; Naglik, J.R. Epithelial discrimination of commensal and pathogenic Candida albicans. Oral Dis. 2016, 22 (Suppl. 1), 114–119. [Google Scholar] [CrossRef]
- Kalia, N.; Singh, J.; Kaur, M. The role of dectin-1 in health and disease. Immunobiology 2021, 226, 152071. [Google Scholar] [CrossRef]
- Taylor, P.R.; Tsoni, S.V.; Willment, J.A.; Dennehy, K.M.; Rosas, M.; Findon, H.; Haynes, K.; Steele, C.; Botto, M.; Gordon, S.; et al. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat. Immunol. 2007, 8, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Hise, A.G.; Tomalka, J.; Ganesan, S.; Patel, K.; Hall, B.A.; Brown, G.D.; Fitzgerald, K.A. An essential role for the NLRP3 inflammasome in host defense against the human fungal pathogen Candida albicans. Cell Host Microbe 2009, 5, 487–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chi, H. Regulation and function of mTOR signalling in T cell fate decisions. Nat. Rev. Immunol. 2012, 12, 325–338. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xiao, Y.; Su, L.; Cui, N.; Liu, D. mTOR Modulates CD8+ T Cell Differentiation in Mice with Invasive Pulmonary Aspergillosis. Open Life Sci. 2018, 13, 129–136. [Google Scholar] [CrossRef]
- Wang, H.; Bai, G.; Cui, N.; Han, W.; Long, Y. T-cell-specific mTOR deletion in mice ameliorated CD4 + T-cell survival in lethal sepsis induced by severe invasive candidiasis. Virulence 2019, 10, 892–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Cui, N.; Wang, H.; Han, W.; Bai, G.; Cheng, W. Impact of mTOR signaling pathway on CD8+ T cell immunity through Eomesodermin in response to invasive candidiasis. J. Microbiol. Immunol. Infect. 2021, 54, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Pollizzi, K.N.; Patel, C.H.; Sun, I.H.; Oh, M.H.; Waickman, A.T.; Wen, J.; Delgoffe, G.M.; Powell, J.D. mTORC1 and mTORC2 selectively regulate CD8⁺ T cell differentiation. J. Clin. Investig. 2015, 125, 2090–2108. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Tschumi, B.O.; Lopez-Mejia, I.C.; Oberle, S.G.; Meyer, M.; Samson, G.; Rüegg, M.A.; Hall, M.N.; Fajas, L.; Zehn, D.; et al. Mammalian Target of Rapamycin Complex 2 Controls CD8 T Cell Memory Differentiation in a Foxo1-Dependent Manner. Cell Rep. 2016, 14, 1206–1217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salmond, R.J. mTOR Regulation of Glycolytic Metabolism in T Cells. Front. Cell Dev. Biol. 2018, 6, 122. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahalingam, S.S.; Jayaraman, S.; Pandiyan, P. Fungal Colonization and Infections—Interactions with Other Human Diseases. Pathogens 2022, 11, 212. https://doi.org/10.3390/pathogens11020212
Mahalingam SS, Jayaraman S, Pandiyan P. Fungal Colonization and Infections—Interactions with Other Human Diseases. Pathogens. 2022; 11(2):212. https://doi.org/10.3390/pathogens11020212
Chicago/Turabian StyleMahalingam, Shanmuga S., Sangeetha Jayaraman, and Pushpa Pandiyan. 2022. "Fungal Colonization and Infections—Interactions with Other Human Diseases" Pathogens 11, no. 2: 212. https://doi.org/10.3390/pathogens11020212
APA StyleMahalingam, S. S., Jayaraman, S., & Pandiyan, P. (2022). Fungal Colonization and Infections—Interactions with Other Human Diseases. Pathogens, 11(2), 212. https://doi.org/10.3390/pathogens11020212






