Temporal Patterns of Phenotypic Antimicrobial Resistance and Coinfecting Pathogens in Glaesserella parasuis Strains Isolated from Diseased Swine in Germany from 2006 to 2021
Abstract
:1. Introduction
2. Results
2.1. Meta Data of Sample Set and Microbiological Findings for G. parasuis
2.2. Distribution of Antimicrobial Resistance
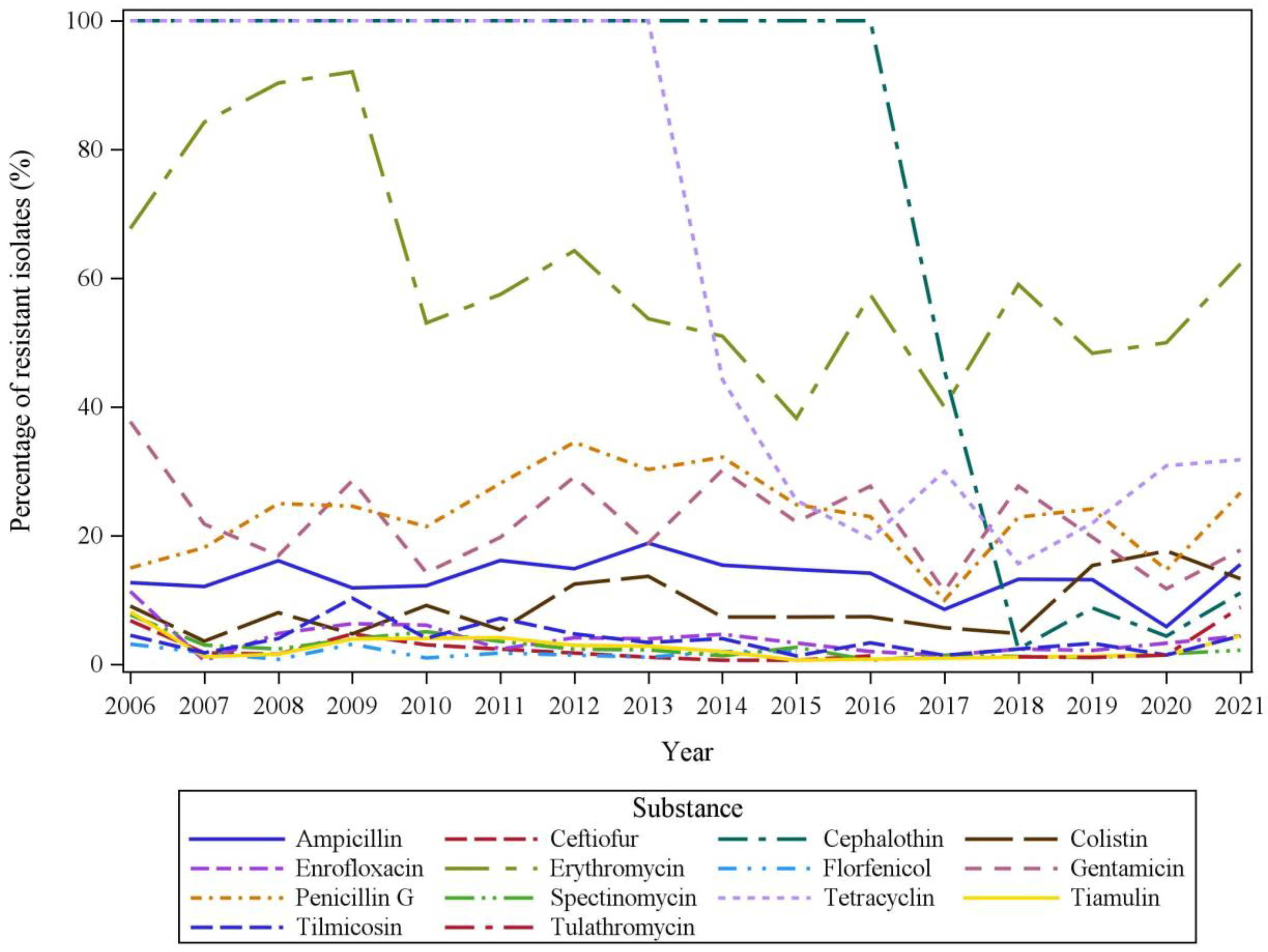
2.3. Temporal Trends in AMR
2.4. Comparison of AMR in Gps Isolates from Different Age Groups and Sampling Sites
2.5. Parallel Detection of Other Microorganisms
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Isolation of Bacterial Strains
4.2. Molecular Biological Detection of Pathogens by PCR, Sero- and Pathotyping of Gps
4.3. Antimicrobial Susceptibility Testing
4.4. Data Management and Statistical (Evaluation)
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Glässer, K. Untersuchungen über die Schweineseuche mit besonderer Berücksichtigung ihrer Aetiologie und Pathologie Dtsch. Tierärztliche Wochenschr. 1910, 18, 729–733. [Google Scholar]
- Howell, K.J.; Peters, S.E.; Wang, J.; Hernandez-Garcia, J.; Weinert, L.A.; Luan, S.L.; Chaudhuri, R.R.; Angen, Ø.; Aragon, V.; Williamson, S.M.; et al. Development of a Multiplex PCR Assay for Rapid Molecular Serotyping of Haemophilus parasuis. J. Clin. Microbiol. 2015, 53, 3812–3821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kielstein, P.; Rapp-Gabrielson, V.J. Designation of 15 serovars of Haemophilus parasuis on the basis of immunodiffusion using heat-stable antigen extracts. J. Clin. Microbiol. 1992, 30, 862–865. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Zhang, C.; Aragon, V.; Zhou, X.; Zou, M.; Wu, C.; Shen, Z. Investigation of Haemophilus parasuis from healthy pigs in China. Vet. Microbiol. 2019, 231, 40–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, D.; Dalla-Costa, F.; Ferroni, L.; Moraes, C.; Schocken-Iturrino, R.; Oliveira, L. The challenges with Glässer’s disease in technified pig production. Austral J. Vet. Sci. 2017, 49, 63–69. [Google Scholar] [CrossRef] [Green Version]
- Schuwerk, L.; Hoeltig, D.; Waldmann, K.H.; Strutzberg-Minder, K.; Valentin-Weigand, P.; Rohde, J. Serotyping and pathotyping of Glaesserella parasuis isolated 2012-2019 in Germany comparing different PCR-based methods. Vet. Res. 2020, 51, 137. [Google Scholar] [CrossRef] [PubMed]
- Galofré-Milà, N.; Correa-Fiz, F.; Lacouture, S.; Gottschalk, M.; Strutzberg-Minder, K.; Bensaid, A.; Pina-Pedrero, S.; Aragon, V. A robust PCR for the differentiation of potential virulent strains of Haemophilus parasuis. BMC Vet. Res. 2017, 13, 124. [Google Scholar] [CrossRef] [Green Version]
- Olvera, A.; Cerdà-Cuéllar, M.; Aragon, V. Study of the population structure of Haemophilus parasuis by multilocus sequence typing. Microbiology 2006, 152, 3683–3690. [Google Scholar] [CrossRef] [Green Version]
- Rapp-Gabrielson, V.; Oliveira, S.; Pijoan, C. Haemophilus parasuis. In Disease of Swine; Straw, B.E., Zimmermann, J.J., D’Allaire, S., Taylor, D.J., Eds.; Blackwell Publishing: Ames, IA, USA, 2006; pp. 681–690. [Google Scholar]
- Correa-Fiz, F.; Fraile, L.; Aragon, V. Piglet nasal microbiota at weaning may influence the development of Glässer’s disease during the rearing period. BMC Genom. 2016, 17, 404. [Google Scholar] [CrossRef] [Green Version]
- Samuelson, D.R.; Welsh, D.A.; Shellito, J.E. Regulation of lung immunity and host defense by the intestinal microbiota. Front. Microbiol. 2015, 6, 1085. [Google Scholar] [CrossRef] [Green Version]
- Charlson, E.S.; Bittinger, K.; Haas, A.R.; Fitzgerald, A.S.; Frank, I.; Yadav, A.; Bushman, F.D.; Collman, R.G. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am. J. Respir. Crit. Care Med. 2011, 184, 957–963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogers, G.B. The lung microbiome. Emerg. Top. Life Sci. 2017, 1, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Kernaghan, S.; Bujold, A.R.; MacInnes, J.I. The microbiome of the soft palate of swine. Anim. Health Res. Rev. 2012, 13, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Lowe, B.A.; Marsh, T.L.; Isaacs-Cosgrove, N.; Kirkwood, R.N.; Kiupel, M.; Mulks, M.H. Defining the “core microbiome” of the microbial communities in the tonsils of healthy pigs. BMC Microbiol. 2012, 12, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickson, R.P.; Erb-Downward, J.R.; Freeman, C.M.; McCloskey, L.; Falkowski, N.R.; Huffnagle, G.B.; Curtis, J.L.; Clemente, J.C.; Molyneaux, P.; Bogaert, D. Bacterial Topography of the Healthy Human Lower Respiratory Tract. mBio 2017, 8, e02287-16. [Google Scholar] [CrossRef]
- Bello-Orti, B.; Deslandes, V.; Tremblay, Y.D.; Labrie, J.; Howell, K.J.; Tucker, A.W.; Maskell, D.J.; Aragon, V.; Jacques, M. Biofilm formation by virulent and non-virulent strains of Haemophilus parasuis. Vet. Res. 2014, 45, 104. [Google Scholar] [CrossRef] [Green Version]
- Jiang, R.; Xiang, M.; Chen, W.; Zhang, P.; Wu, X.; Zhu, G.; Tu, T.; Jiang, D.; Yao, X.; Luo, Y.; et al. Biofilm characteristics and transcriptomic analysis of Haemophilus parasuis. Vet. Microbiol. 2021, 258, 109073. [Google Scholar] [CrossRef]
- Macedo, N.; Cheeran, M.C.; Rovira, A.; Holtcamp, A.; Torremorell, M. Effect of enrofloxacin on Haemophilus parasuis infection, disease and immune response. Vet. Microbiol. 2017, 199, 91–99. [Google Scholar] [CrossRef]
- Hemme, M.; Ruddat, I.; Hartmann, M.; Werner, N.; van Rennings, L.; Käsbohrer, A.; Kreienbrock, L. Antibiotic use on German pig farms—A longitudinal analysis for 2011, 2013 and 2014. PLoS ONE 2018, 13, e0199592. [Google Scholar] [CrossRef]
- Schaekel, F.; May, T.; Seiler, J.; Hartmann, M.; Kreienbrock, L. Antibiotic drug usage in pigs in Germany-Are the class profiles changing? PLoS ONE 2017, 12, e0182661. [Google Scholar] [CrossRef] [Green Version]
- Wallmann, J.; Bode, C.; Heberer, T. Abgabemengenerfassung von Antibiotika in Deutschland 2018. Dtsch. Tierärzteblatt 2019, 67, 1082–1090. [Google Scholar]
- van Rennings, L.; von Münchhausen, C.; Ottilie, H.; Hartmann, M.; Merle, R.; Honscha, W.; Käsbohrer, A.; Kreienbrock, L. Cross-sectional study on antibiotic usage in pigs in Germany. PLoS ONE 2015, 10, e0119114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vahle, J.L.; Haynes, J.S.; Andrews, J.J. Experimental reproduction of Haemophilus parasuis infection in swine: Clinical, bacteriological, and morphologic findings. J. Vet. Diagn. Investig. 1995, 7, 476–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vahle, J.L.; Haynes, J.S.; Andrews, J.J. Interaction of Haemophilus parasuis with nasal and tracheal mucosa following intranasal inoculation of cesarean derived colostrum deprived (CDCD) swine. Can. J. Vet. Res. 1997, 61, 200–206. [Google Scholar]
- Wang, H.; Wei, W.; Cao, Q.; Xu, M.; Chen, Q.; Lv, Y.; Tan, C.; Dai, M.; Xu, X.; Chen, H.; et al. Sialylated Lipooligosaccharide Contributes to Glaesserella parasuis Penetration of Porcine Respiratory Epithelial Barrier. ACS Infect. Dis. 2021, 7, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Costa-Hurtado, M.; Ballester, M.; Galofré-Milà, N.; Darji, A.; Aragon, V. VtaA8 and VtaA9 from Haemophilus parasuis delay phagocytosis by alveolar macrophages. Vet. Res. 2012, 43, 57. [Google Scholar] [CrossRef] [Green Version]
- Olvera, A.; Ballester, M.; Nofrarías, M.; Sibila, M.; Aragon, V. Differences in phagocytosis susceptibility in Haemophilus parasuis strains. Vet. Res. 2009, 40, 24. [Google Scholar] [CrossRef] [Green Version]
- Olvera, A.; Pina, S.; Pérez-Simó, M.; Oliveira, S.; Bensaid, A. Virulence-associated trimeric autotransporters of Haemophilus parasuis are antigenic proteins expressed in vivo. Vet. Res. 2010, 41, 26. [Google Scholar] [CrossRef] [Green Version]
- Pina, S.; Olvera, A.; Barceló, A.; Bensaid, A. Trimeric autotransporters of Haemophilus parasuis: Generation of an extensive passenger domain repertoire specific for pathogenic strains. J. Bacteriol. 2009, 191, 576–587. [Google Scholar] [CrossRef] [Green Version]
- Wan, X.; Wang, G.; Gao, W.; Li, C.; Yan, H. Comparative genome analysis of multidrug-resistant and susceptible Glaesserella parasuis strains isolated from diseased pigs in China. Vet. Microbiol. 2021, 254, 109002. [Google Scholar] [CrossRef]
- Zhang, K.; Chu, P.; Song, S.; Yang, D.; Bian, Z.; Li, Y.; Gou, H.; Jiang, Z.; Cai, R.; Li, C. Proteome Analysis of Outer Membrane Vesicles From a Highly Virulent Strain of Haemophilus parasuis. Front. Vet. Sci. 2021, 8, 756764. [Google Scholar] [CrossRef] [PubMed]
- Macedo, N.; Gottschalk, M.; Strutzberg-Minder, K.; Van, C.N.; Zhang, L.; Zou, G.; Zhou, R.; Marostica, T.; Clavijo, M.J.; Tucker, A.; et al. Molecular characterization of Glaesserella parasuis strains isolated from North America, Europe and Asia by serotyping PCR and LS-PCR. Vet. Res. 2021, 52, 68. [Google Scholar] [CrossRef] [PubMed]
- Rahbauer, M. Vergleich Unterschiedlicher Berechnungsmodelle zum Monitoring von Antibiotikaverbrauchsmengen, sowie Überprüfung des Einflusses Betriebsspezifischer Parameter auf den Therapieindex, die Animal Daily Dose und die Netherlands Animal Daily Dose in süddeutschen Schweinemastbetrieben; Tierärztliche Fakultät der Ludwig-Maximilian Universität München: Landshut, Germany, 2015. [Google Scholar]
- Burow, E.; Kasbohrer, A. Risk Factors for Antimicrobial Resistance in Escherichia coli in Pigs Receiving Oral Antimicrobial Treatment: A Systematic Review. Microb. Drug Resist. 2017, 23, 194–205. [Google Scholar] [CrossRef]
- Dorey, L.; Pelligand, L.; Cheng, Z.; Lees, P. Pharmacokinetic/pharmacodynamic integration and modelling of florfenicol for the pig pneumonia pathogens Actinobacillus pleuropneumoniae and Pasteurella multocida. PLoS ONE 2017, 12, e0177568. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente, A.J.; Tucker, A.W.; Navas, J.; Blanco, M.; Morris, S.J.; Gutiérrez-Martín, C.B. Antimicrobial susceptibility patterns of Haemophilus parasuis from pigs in the United Kingdom and Spain. Vet. Microbiol. 2007, 120, 184–191. [Google Scholar] [CrossRef] [Green Version]
- El Garch, F.; de Jong, A.; Simjee, S.; Moyaert, H.; Klein, U.; Ludwig, C.; Marion, H.; Haag-Diergarten, S.; Richard-Mazet, A.; Thomas, V.; et al. Monitoring of antimicrobial susceptibility of respiratory tract pathogens isolated from diseased cattle and pigs across Europe, 2009–2012: VetPath results. Vet. Microbiol. 2016, 194, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Wallmann, J.; Bode, C.; Köper, L.; Heberer, T. Abgabemengenerfassung von Antibiotika in Deutschland 2019 Auswertung der nach DIMDI-AMV übermittelten Daten 2019 und Vergleich zu den Vorjahren. Dtsch. Tierärzteblatt 2020, 68, 1102–1109. [Google Scholar]
- Li, J.; Wang, S.; Li, C.; Wang, C.; Liu, Y.; Wang, G.; He, X.; Hu, L.; Liu, Y.; Cui, M.; et al. Secondary Haemophilus parasuis infection enhances highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV) infection-mediated inflammatory responses. Vet. Microbiol. 2017, 204, 35–42. [Google Scholar] [CrossRef]
- Costa-Hurtado, M.; Barba-Vidal, E.; Maldonado, J.; Aragon, V. Update on Glässer’s disease: How to control the disease under restrictive use of antimicrobials. Vet. Microbiol. 2020, 242, 108595. [Google Scholar] [CrossRef]
- Hau, S.J.; Eberle, K.C.; Brockmeier, S.L. Importance of strain selection in the generation of heterologous immunity to Glaesserella (Haemophilus) parasuis. Vet. Immunol. Immunopathol. 2021, 234, 110205. [Google Scholar] [CrossRef]
- Liu, H.; Xue, Q.; Zeng, Q.; Zhao, Z. Haemophilus parasuis vaccines. Vet. Immunol. Immunopathol. 2016, 180, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Fuertes, M.; Correa-Fiz, F.; López-Serrano, S.; Sibila, M.; Aragon, V. Sow vaccination against virulent Glaesserella parasuis shapes the nasal microbiota of their offspring. Sci. Rep. 2022, 12, 3357. [Google Scholar] [CrossRef] [PubMed]
- López-Serrano, S.; Neila-Ibáñez, C.; Costa-Hurtado, M.; Mahmmod, Y.; Martínez-Martínez, J.; Galindo-Cardiel, I.J.; Darji, A.; Rodríguez, F.; Sibila, M.; Aragon, V. Sow vaccination with a protein fragment against virulent Glaesserella (Haemophilus) parasuis modulates immunity traits in their offspring. Vaccines 2021, 9, 534. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, S.; Pijoan, C. Haemophilus parasuis: New trends on diagnosis, epidemiology and control. Vet. Microbiol. 2004, 99, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Saade, G.; Deblanc, C.; Bougon, J.; Marois-Créhan, C.; Fablet, C.; Auray, G.; Belloc, C.; Leblanc-Maridor, M.; Gagnon, C.A.; Zhu, J.; et al. Coinfections and their molecular consequences in the porcine respiratory tract. Vet. Res. 2020, 51, 80. [Google Scholar] [CrossRef]
- Mathieu-Denoncourt, A.; Letendre, C.; Auger, J.P.; Segura, M.; Aragon, V.; Lacouture, S.; Gottschalk, M. Limited interactions between Streptococcus suis and Haemophilus parasuis in in vitro co-infection studies. Pathogens 2018, 7, 7. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Wu, J.; Zhang, Y.; Guo, L.; Cong, X.; Du, Y.; Li, J.; Sun, W.; Shi, J.; Peng, J.; et al. Concurrent highly pathogenic porcine reproductive and respiratory syndrome virus infection accelerates Haemophilus parasuis infection in conventional pigs. Vet. Microbiol. 2012, 158, 316–321. [Google Scholar] [CrossRef]
- Liu, S.; Li, W.; Wang, Y.; Gu, C.; Liu, X.; Charreyre, C.; Fan, S.; He, Q. Coinfection with Haemophilus parasuis serovar 4 increases the virulence of porcine circovirus type 2 in piglets. Virol. J. 2017, 14, 227. [Google Scholar] [CrossRef] [Green Version]
- Dubosson, C.R.; Conzelmann, C.; Miserez, R.; Boerlin, P.; Frey, J.; Zimmermann, W.; Häni, H.; Kuhnert, P. Development of two real-time PCR assays for the detection of Mycoplasma hyopneumoniae in clinical samples. Vet. Microbiol. 2004, 102, 55–65. [Google Scholar] [CrossRef]
- Luehrs, A.; Siegenthaler, S.; Grützner, N.; Grosse Beilage, E.; Kuhnert, P.; Nathues, H. Occurrence of Mycoplasma hyorhinis infections in fattening pigs and association with clinical signs and pathological lesions of Enzootic Pneumonia. Vet. Microbiol. 2017, 203, 1–5. [Google Scholar] [CrossRef]
- Brunborg, I.M.; Moldal, T.; Jonassen, C.M. Quantitation of porcine circovirus type 2 isolated from serum/plasma and tissue samples of healthy pigs and pigs with postweaning multisystemic wasting syndrome using a TaqMan-based real-time PCR. J. Virol. Methods 2004, 122, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Tabynov, K.; Sansyzbay, A.; Tulemissova, Z.; Tabynov, K.; Dhakal, S.; Samoltyrova, A.; Renukaradhya, G.J.; Mambetaliyev, M. Inactivated porcine reproductive and respiratory syndrome virus vaccine adjuvanted with Montanide™ Gel 01 ST elicits virus-specific cross-protective inter-genotypic response in piglets. Vet. Microbiol. 2016, 192, 81–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodell, C.K.; Zhang, J.; Strait, E.; Harmon, K.; Patnayak, D.; Otterson, T.; Culhane, M.; Christopher-Hennings, J.; Clement, T.; Leslie-Steen, P.; et al. Ring test evaluation of the detection of influenza A virus in swine oral fluids by real-time reverse-transcription polymerase chain reaction and virus isolation. Can. J. Vet. Res. 2016, 80, 12–20. [Google Scholar] [PubMed]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; 2nd Informational Supplement; CLSI Document VET01-S2; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2013. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 4th ed.; CLSI Document VET01-A4; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2013. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing; CLSI Document M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 4th ed.; CLSI Supplement VET08; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Fessler, A.T.; Böttner, A.; Fehr, M.; Kaspar, H.; Kehrenberg, C.; Kietzmann, M.; Klarmann, D.; Klein, G.; Peters, T.A.; Richter, A.; et al. Mikrotiterplattenlayouts für Kleintiere, Großtiere und Mastitis: Aktualisierung der Layouts des DVG-Arbeitskreises “Antibiotikaresistenz”. Dtsch. Tierärzteblatt 2017, 65, 472–481. [Google Scholar]
- Dayao, D.A.; Kienzle, M.; Gibson, J.S.; Blackall, P.J.; Turni, C. Use of a proposed antimicrobial susceptibility testing method for Haemophilus parasuis. Vet. Microbiol. 2014, 172, 586–589. [Google Scholar] [CrossRef] [Green Version]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 5th ed.; CLSI Document VET01S ED5:2020; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
| Gps Serotype | Number of Isolates | Proportion of Isolates (%) |
|---|---|---|
| 1 | 8 | 7.9 |
| 2 | 1 | 1.0 |
| 1/2 | 2 | 2.0 |
| 4 | 30 | 29.7 |
| 5/12 | 13 | 12.9 |
| 7 | 13 | 12.9 |
| 9 | 7 | 6.9 |
| 10 | 1 | 1.0 |
| 11 | 1 | 1.0 |
| 13 | 22 | 21.8 |
| 14 | 1 | 1.2 |
| 15 | 2 | 2.0 |
| Sampling Site | Suckling Piglets | Nursery Pigs | Fatteners | Sows | No Age Group | All Samples | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | n | % | n | % | |
| Respiratory tract | 118 | 73.3 | 1252 | 83.7 | 60 | 76.9 | 2 | 40.0 | 265 | 86.6 | 1697 | 82.9 |
| Pericardium | 18 | 11.2 | 104 | 7.0 | 5 | 6.4 | 2 | 40.0 | 10 | 3.3 | 139 | 6.8 |
| Pleural cavity | 8 | 5.0 | 59 | 3.9 | 4 | 5.1 | − | − | 13 | 4.2 | 84 | 4.1 |
| Brain | 7 | 4.3 | 44 | 2.9 | 7 | 9.0 | − | − | 12 | 3.9 | 70 | 3.4 |
| Joint | 6 | 3.7 | 18 | 1.2 | 2 | 2.6 | 1 | 20.0 | 1 | 0.3 | 28 | 1.4 |
| Abdominal cavity | 3 | 1.9 | 19 | 1.3 | − | − | − | − | 1 | 0.3 | 23 | 1.1 |
| Nose | 1 | 0.6 | − | − | − | − | − | − | 4 | 1.3 | ||
| Total | 161 | 100 | 1496 | 100 | 78 | 100 | 5 | 100 | 306 | 100 | 2046 | 100 |
| Gps Isolates Positive for LS vta | Serotypes | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 4 | 5/12 | 7 | 9 | 11 | 13 | 14 | 15 | n.s. * | Total | |
| Respiratory tract | 1 | 16 | 7 | 6 | 1 | 1 | 6 | 1 | 1 | 5 | 45 |
| Other sites | − | − | 2 | 2 | 1 | − | 2 | − | − | 2 | 9 |
| Total | 1 (1.9%) | 16 (29.6%) | 9 (16.7%) | 8 (14.8%) | 2 (3.7%) | 1 (1.9%) | 8 (14.8%) | 1 (1.9%) | 1 (1.9%) | 7 (13.0%) | 54 (100%) |
| Substance (Clinical Breakpoint (mg/L)) | Number of Isolates with MIC Values (mg/L) of | MIC50 | MIC90 | s | i | r | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | ≤0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | ≥64 | (mg/L) | % | % | % | ||
| Ampicillin (≤0.5) | 2046 | − | − | 1372 | 260 | 128 | 57 | 48 | 29 | 28 | 39 | 33 | 52 | ≤0.13 | 2.00 | 86.02 | 2.79 | 11.19 |
| Ceftiofur (≤2) | 2046 | − | − | 313 | 5 | 3 | 1670 | 13 | 6 | − | 36 | − | − | 1.00 | 1.00 | 97.95 | 0.29 | 1.80 |
| Cephalothin 1 (≤8) | 2046 | − | − | − | − | − | 291 | 16 | 1637 | 46 | 42 | 6 | 8 | 4.00 | 4.00 | 15.00 | 80.00 | 5.00 |
| Colistin (<0.5) | 2046 | − | − | − | − | 1868 | 91 | 16 | 4 | 67 | − | − | − | 0.50 | 0.50 | 91.30 | 5.20 | 3.50 |
| Enrofloxacin (≤0.25) | 2046 | 281 | 1503 | 108 | 68 | 63 | 12 | 8 | 3 | − | − | − | − | 0.06 | 0.13 | 95.80 | 3.10 | 1.10 |
| Erythromycin (≤0.25) | 2046 | − | − | 558 | 221 | 382 | 412 | 249 | 94 | 78 | 52 | − | − | 0.50 | 4.00 | 38.10 | 18.70 | 43.30 |
| Florfenicol (≤2) | 2046 | − | − | − | − | − | 1956 | 63 | 23 | 3 | 1 | − | − | 1.00 | 1.00 | 98.70 | 1.10 | 0.20 |
| Gentamicin (≤2) | 2046 | − | − | − | 66 | 28 | 79 | 1384 | 337 | 32 | 24 | 96 | − | 2.00 | 4.00 | 76.10 | 16.50 | 7.40 |
| Penicillin G 2 (≤0.25) | 2046 | − | 969 | 214 | 370 | 214 | 62 | 39 | 23 | 25 | 124 | 6 | − | 0.13 | 2.00 | 75.90 | 10.50 | 13.60 |
| Spectinomycin 3 (≤32) | 2046 | − | − | − | − | − | − | − | 277 | 1562 | 101 | 47 | 59 | 8.00 | 16.00 | 97.10 | 0.30 | 2.50 |
| Tiamulin (≤16) | 2046 | − | − | − | − | − | − | − | 1518 | 422 | 51 | 20 | 35 | 4.00 | 8.00 | 97.30 | − | 2.70 |
| Tilmicosin (≤16) | 2046 | − | − | − | − | − | 230 | 38 | 1557 | 73 | 65 | 30 | 53 | 4.00 | 8.00 | 95.94 | − | 4.10 |
| Tetracycline (≤0.5) | 2046 | − | − | 125 | 64 | 391 | 1121 | 100 | 66 | 48 | 65 | 65 | − | 1.00 | 4.00 | 28.40 | 54.80 | 16.80 |
| Tulathromycin 4 (≤16) | 325 | − | − | − | − | − | − | 272 | 25 | 10 | 11 | 5 | 2 | 2.00 | 4.00 | 97.80 | 1.50 | 0.60 |
| Risk Categories | Resistant | Susceptible | Univariable Model | |||||
|---|---|---|---|---|---|---|---|---|
| n | % | n | % | OR | 95% CI | p | ||
| Lower | Upper | |||||||
| Ampicillin | ||||||||
| 2006–2013 (ref) | 180 | 14.48 | 1063 | 85.52 | 1 | x | x | x |
| 2014–2021 | 106 | 13.20 | 697 | 86.80 | 0.898 | 0.694 | 1.163 | 0.4148 |
| Ceftiofur | ||||||||
| 2006–2013 (ref) | 38 | 53.06 | 1250 | 96.94 | 1 | x | x | x |
| 2014–2021 | 4 | 0.50 | 799 | 99.50 | 0.159 | 0.056 | 0.447 | 0.0004 |
| Cephalothin * | ||||||||
| 2006–2013 (ref) | 1243 | 100.00 | 0 | 0.00 | 1 | x | x | x |
| 2014–2021 | 496 | 61.77 | 307 | 38.23 | n.d. | n.d. | n.d. | * |
| Colistin | ||||||||
| 2006–2013 (ref) | 105 | 8.45 | 1138 | 91.55 | 1 | x | x | x |
| 2014–2021 | 73 | 9.09 | 730 | 90.91 | 1.084 | 0.793 | 1.482 | 0.6141 |
| Enrofloxacin | ||||||||
| 2006–2013 (ref) | 64 | 5.15 | 1179 | 94.85 | 1 | x | x | x |
| 2014–2021 | 22 | 2.74 | 781 | 97.26 | 0.519 | 0.317 | 0.849 | 0.009 |
| Erythromycin | ||||||||
| 2006–2013 (ref) | 866 | 69.67 | 377 | 30.33 | 1 | x | x | x |
| 2014–2021 | 401 | 49.94 | 402 | 50.06 | 0.434 | 0.361 | 0.522 | <0.0001 |
| Florfenicol | ||||||||
| 2006–2013 (ref) | 21 | 1.69 | 1722 | 98.31 | 1 | x | x | x |
| 2014–2021 | 6 | 0.75 | 797 | 99.25 | 0.438 | 0.176 | 1.090 | 0.0760 |
| Gentamicin | ||||||||
| 2006–2013 (ref) | 305 | 24.54 | 938 | 75.46 | 1 | x | x | x |
| 2014–2021 | 184 | 22.91 | 619 | 77.09 | 0.914 | 0.742 | 1.127 | 0.4006 |
| Penicillin | ||||||||
| 2006–2013 (ref) | 304 | 24.46 | 939 | 75.54 | 1 | x | x | x |
| 2014–2021 | 189 | 23.54 | 614 | 76.46 | 0.951 | 0.772 | 1.171 | 0.6356 |
| Spectinomycin | ||||||||
| 2006–2013 (ref) | 49 | 3.94 | 1194 | 96.06 | 1 | x | x | x |
| 2014–2021 | 10 | 1.25 | 793 | 98.75 | 0.307 | 0.155 | 0.610 | 0.0007 |
| Tetracyclin * | ||||||||
| 2006–2013 (ref) | 1243 | 100.00 | 0 | 0.00 | 1 | x | x | x |
| 2014–2021 | 222 | 27.68 | 580 | 72.32 | n.d. | n.d. | n.d. | * |
| Tiamulin | ||||||||
| 2006–2013 (ref) | 48 | 3.86 | 1195 | 96.14 | 1 | x | x | x |
| 2014–2021 | 7 | 0.87 | 796 | 99.13 | 0.219 | 0.099 | 0.486 | 0.0002 |
| Tilmicosin | ||||||||
| 2006–2013 (ref) | 61 | 4.91 | 1182 | 95.09 | 1 | x | x | x |
| 2014–2021 | 22 | 2.74 | 781 | 97.26 | 0.546 | 0.332 | 0.896 | 0.0167 |
| Colistin | Resistant Isolates | Susceptible Isolates | Univariable log. reg. | Multivariable log. reg. | |||||
| n | % | n | % | OR | p | OR | p | ||
| Time period | 2006–2013 (Ref.) | 105 | 8.45 | 1138 | 91.55 | 1 | − | 1 | − |
| 2014–2021 | 73 | 9.09 | 730 | 90.91 | 1.08 | 0.614 | 1.00 | 0.989 | |
| Age group | Nursery pigs (Ref.) | 124 | 8.29 | 1372 | 91.71 | 1 | − | 1 | − |
| Suckling piglet | 13 | 8.07 | 148 | 91.93 | 0.97 | 0.925 | 0.91 | 0.755 | |
| Fattening Pigs | 6 | 7.69 | 72 | 92.31 | 0.92 | 0.852 | 0.87 | 0.759 | |
| not specified | 35 | 11.44 | 271 | 88.56 | 1.43 | 0.078 | 1.46 | 0.072 | |
| Sampling site | Respiratory tract (Ref.) | 131 | 7.72 | 1566 | 92.28 | 1 | − | 1 | − |
| Pericardium | 22 | 15.83 | 117 | 84.17 | 2.25 | 0.001 | 2.38 | 0.001 | |
| Pleural cavity | 11 | 13.10 | 73 | 86.90 | 1.80 | 0.080 | 1.81 | 0.079 | |
| Brain | 9 | 12.86 | 61 | 87.14 | 1.76 | 0.124 | 1.77 | 0.124 | |
| Joints | 3 | 10.71 | 25 | 89.29 | 1.43 | 0.559 | 1.60 | 0.450 | |
| Abdominal cavity | 1 | 4.35 | 22 | 95.65 | 0.54 | 0.552 | 0.57 | 0.585 | |
| Erythromycin | Resistant Isolates | Susceptible Isolates | Univariable log. reg. | Multivariable log. reg. | |||||
| n | % | n | % | OR | p | OR | p | ||
| Time period | 2006–2013 (Ref.) | 866 | 69.67 | 377 | 30.33 | 1 | − | 1 | − |
| 2014–2021 | 401 | 49.94 | 402 | 50.06 | 0.43 | <0.001 | 0.45 | <0.001 | |
| Age group | Nursery pigs (Ref.) | 917 | 61.30 | 579 | 38.70 | 1 | − | 1 | − |
| Suckling piglet | 103 | 63.98 | 58 | 36.02 | 1.12 | 0.507 | 1.12 | 0.510 | |
| Fattening Pigs | 51 | 65.38 | 27 | 34.62 | 1.19 | 0.470 | 1.26 | 0.355 | |
| not specified | 193 | 63.07 | 113 | 36.93 | 1.08 | 0.561 | 0.946 | 0.682 | |
| Sampling site | Respiratory tract (Ref.) | 1073 | 63.23 | 624 | 36.77 | 1 | − | 1 | − |
| Pericardium | 75 | 53.96 | 64 | 46.04 | 0.68 | 0.031 | 0.79 | 0.201 | |
| Pleural cavity | 48 | 57.14 | 36 | 42.86 | 0.78 | 0.261 | 0.87 | 0.545 | |
| Brain | 39 | 55.71 | 31 | 44.29 | 0.73 | 0.204 | 0.80 | 0.385 | |
| Joints | 12 | 42.86 | 16 | 57.14 | 0.44 | 0.031 | 0.42 | 0.033 | |
| Abdominal cavity | 17 | 73.91 | 6 | 26.09 | 1.65 | 0.296 | 1.47 | 0.431 | |
| Florfenicol | Resistant Isolates | Susceptible Isolates | Univariable log. reg. | Multivariable log. reg. | |||||
| n | % | n | % | OR | p | OR | p | ||
| Time period | 2006–2013 (Ref.) | 21 | 1.69 | 1222 | 98.31 | 1 | − | 1 | − |
| 2014–2021 | 6 | 0.75 | 797 | 99.25 | 0.44 | 0.076 | 0.42 | 0.062 | |
| Age group | Nursery pigs (Ref.) | 19 | 1.27 | 1477 | 98.73 | 1 | − | 1 | − |
| Suckling piglet | 1 | 0.62 | 160 | 99.38 | 0.49 | 0.483 | 0.48 | 0.471 | |
| Fattening Pigs | 4 | 5.13 | 74 | 94.87 | 4.20 | 0.011 | 4.29 | 0.009 | |
| Not specified | 3 | 0.98 | 303 | 99.02 | 0.77 | 0.675 | 0.696 | 0.564 | |
| Sampling site | Respiratory tract (Ref.) | 25 | 1.47 | 1672 | 98.53 | 1 | − | 1 | − |
| Pericardium | − | − | 139 | 100.00 | − | − | −- | − | |
| Pleural cavity | 2 | 2.38 | 82 | 97.62 | 1.63 | 0.510 | − | − | |
| Brain | − | − | 70 | 100.00 | − | − | − | − | |
| Joints | − | − | 28 | 100.00 | − | − | − | − | |
| Abdominal cavity | − | − | 23 | 100.00 | − | − | − | − | |
| Tetracyclin | Resistant Isolates | Susceptible Isolates | Univariable log. reg. | Multivariable log. reg. | |||||
| n | % | n | % | OR | p | OR | p | ||
| Time period | 2006–2013 (Ref.) | 1243 | 100.00 | − | − | − | − | − | − |
| 2014–2021 | 222 | 27.68 | 580 | 72.32 | 1 | − | 1 | − | |
| Age group | Nursery pigs (Ref.) | 1044 | 69.83 | 451 | 30.17 | 1 | − | 1 | − |
| Suckling piglet | 120 | 74.53 | 41 | 25.47 | 1.26 | 0.216 | 1.32 | 0.146 | |
| Fattening Pigs | 56 | 71.79 | 22 | 28.21 | 1.10 | 0.713 | 1.16 | 0.560 | |
| Not specified | 240 | 78.43 | 66 | 21.57 | 1.57 | 0.003 | 1.66 | 0.001 | |
| Sampling site | Respiratory tract (Ref.) | 1248 | 73.58 | 448 | 26.42 | 1 | − | 1 | − |
| Pericardium | 81 | 58.27 | 58 | 41.73 | 0.50 | <0.001 | 0.50 | 0.0001 | |
| Pleural cavity | 55 | 65.48 | 29 | 34.52 | 0.68 | 0.103 | 0.68 | 0.097 | |
| Brain | 41 | 58.57 | 29 | 41.43 | 0.51 | 0.006 | 0.49 | 0.005 | |
| Joints | 20 | 71.43 | 8 | 28.57 | 0.90 | 0.798 | 0.86 | 0.725 | |
| Abdominal cavity | 19 | 82.61 | 4 | 17.39 | 1.71 | 0.334 | 1.78 | 0.297 | |
| Antimicrobial Substance | 2006–2013 | 2014–2021 | p * | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Resistant Isolates | Sensitive Isolates | Resistant Isolates | Sensitive Isolates | ||||||
| Number | (%) | Number | (%) | Number | (%) | Number | (%) | ||
| Ampicillin | 105 | 13.74 | 659 | 86.26 | 64 | 13.11 | 424 | 86.89 | 0.751 |
| Ceftiofur | 24 | 3.14 | 740 | 96–86 | 4 | 0.82 | 484 | 99.18 | 0.012 |
| Cephalothin | 764 | 100.00 | − | − | 288 | 59.02 | 200 | 40.98 | n.d. |
| Colistin | 50 | 6.54 | 714 | 93.46 | 40 | 8.20 | 448 | 91.80 | 0.271 |
| Enrofloxacin | 33 | 4.32 | 731 | 95.68 | 15 | 3.07 | 473 | 69.93 | 0.265 |
| Erythromycin | 540 | 70.68 | 224 | 29.32 | 244 | 50.00 | 244 | 50.00 | <0.0001 |
| Florfenicol | 13 | 1.70 | 751 | 98.30 | 5 | 1.02 | 483 | 98.98 | 0.331 |
| Gentamicin | 186 | 24.35 | 578 | 75.65 | 119 | 24.39 | 369 | 75.61 | 0.987 |
| Penicillin | 173 | 22.64 | 591 | 77.36 | 112 | 22.95 | 376 | 77.05 | 0.899 |
| Spectinomycin | 28 | 3.66 | 736 | 96.34 | 6 | 1.23 | 482 | 98.77 | 0.014 |
| Tetracyclin | 764 | 100.00 | − | − | 133 | 27.31 | 354 | 72.69 | n.d. |
| Tiamulin | 33 | 4.32 | 731 | 95.68 | 4 | 0,82 | 484 | 99.18 | 0.001 |
| Tilmicosin | 38 | 4.97 | 726 | 95.03 | 15 | 3.07 | 473 | 96.93 | 0.107 |
| Additional Microorganisms in Samples Positive for Gps | Combinations with Gps | Number of Samples Tested | ||||
|---|---|---|---|---|---|---|
| Negative | Positive | |||||
| n | % | n | % | n | ||
| Type | Pathogen | |||||
| Virus a | Influenza | 730 | 77.00 | 218 | 23.00 | 948 |
| PCV2 | 633 | 82.00 | 139 | 18.00 | 772 | |
| PRRSV-EU | 818 | 74.9 | 274 | 25.10 | 1092 | |
| PRSV-US | 977 | 90.70 | 100 | 9.30 | 1077 | |
| Bacteria | Actinobacillus pleuropneumoniae * | 2000 | 97.80 | 46 | 2.20 | 2046 |
| Actinobacillus suis * | 2045 | 100.00 | 1 | 0.00 | 2046 | |
| Beta-haemolytic streptococcus * | 1992 | 97.40 | 54 | 2.60 | 2046 | |
| Bordetella bronchiseptica * | 1824 | 89.10 | 222 | 10.90 | 2046 | |
| Mycoplasma hyopneumoniae a | 739 | 86.80 | 112 | 13.20 | 851 | |
| Mycoplasma hyorhinis a | 308 | 54.90 | 253 | 45.10 | 561 | |
| Staphylococcus aureus * | 2016 | 98.50 | 30 | 1.50 | 2046 | |
| Streptococcus suis * | 1526 | 74.60 | 520 | 25.40 | 2046 | |
| Pasteurella multocida * | 1873 | 91.20 | 173 | 8.50 | 2046 | |
| Antimicrobial Substance | Sensitive | Intermediate | Resistant |
|---|---|---|---|
| Ampicillin | ≤0.5 | 1 | ≥ 2 |
| Ceftiofur | ≤2 | 4 | ≥8 |
| Cephalotin | ≤2 | 4 | ≥8 |
| Colistin | ≤0.5 | 1–2 | ≥4 |
| Enrofloxacin | ≤0.25 | 0.5 | ≥1 |
| Erythromycin | ≤0.25 | 0.5 | ≥1 |
| Florfenicol | ≤2 | 4 | ≥8 |
| Gentamicin | ≤2 | 4 | ≥8 |
| Penicillin | ≤0.25 | 0.5 | ≥1 |
| Spectinomycin | ≤32 | 64 | >64 |
| Tetracyclin | ≤0.5 | 1 | ≥2 |
| Tiamulin | ≤16 | − | ≥32 |
| Tilmicosin | ≤16 | − | ≥32 |
| Tulathromycin | ≤16 | 32 | ≥64 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wiencek, I.; Hartmann, M.; Merkel, J.; Trittmacher, S.; Kreienbrock, L.; Hennig-Pauka, I. Temporal Patterns of Phenotypic Antimicrobial Resistance and Coinfecting Pathogens in Glaesserella parasuis Strains Isolated from Diseased Swine in Germany from 2006 to 2021. Pathogens 2022, 11, 721. https://doi.org/10.3390/pathogens11070721
Wiencek I, Hartmann M, Merkel J, Trittmacher S, Kreienbrock L, Hennig-Pauka I. Temporal Patterns of Phenotypic Antimicrobial Resistance and Coinfecting Pathogens in Glaesserella parasuis Strains Isolated from Diseased Swine in Germany from 2006 to 2021. Pathogens. 2022; 11(7):721. https://doi.org/10.3390/pathogens11070721
Chicago/Turabian StyleWiencek, Isabeau, Maria Hartmann, Jörg Merkel, Sara Trittmacher, Lothar Kreienbrock, and Isabel Hennig-Pauka. 2022. "Temporal Patterns of Phenotypic Antimicrobial Resistance and Coinfecting Pathogens in Glaesserella parasuis Strains Isolated from Diseased Swine in Germany from 2006 to 2021" Pathogens 11, no. 7: 721. https://doi.org/10.3390/pathogens11070721






