In Vitro and In Silico Analyses of New Cinnamid and Rosmarinic Acid-Derived Compounds Biosynthesized in Escherichia coli as Leishmania amazonensis Arginase Inhibitors
Abstract
1. Introduction
2. Results
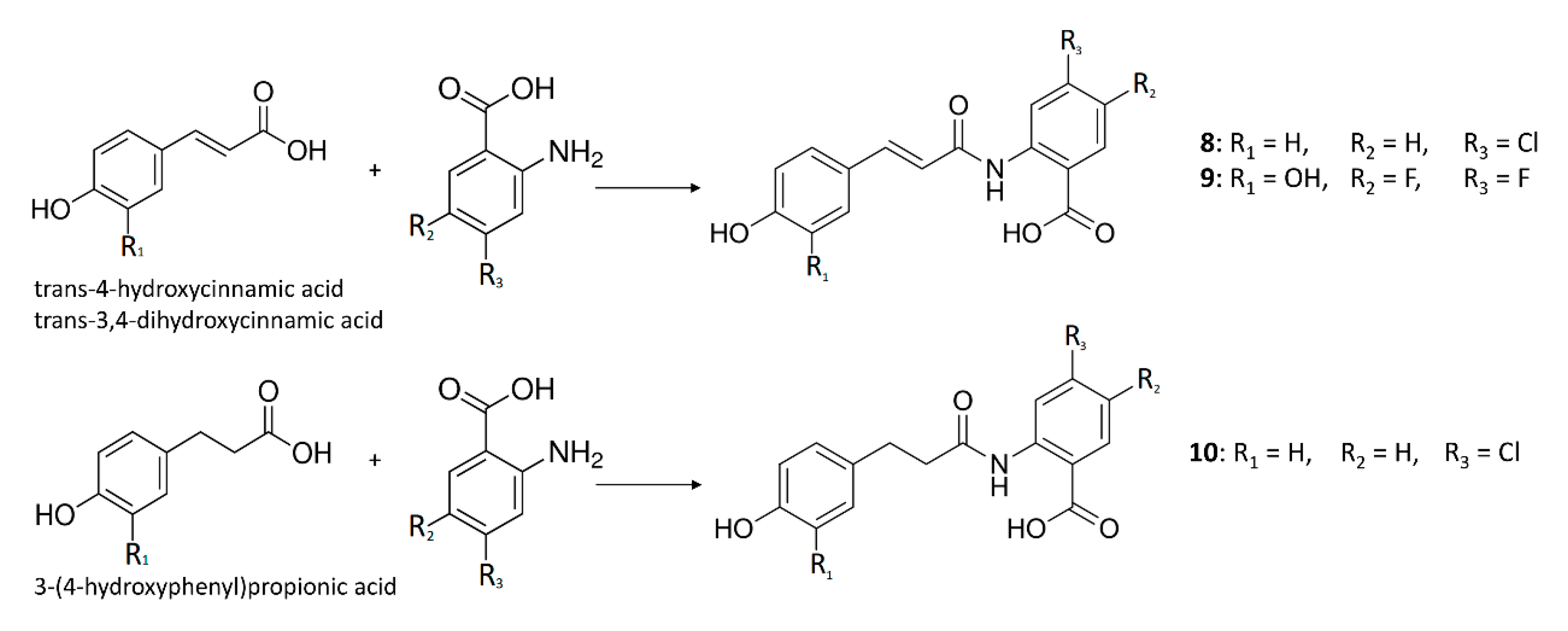
2.1. Production of Compounds 8–10 in E. coli via Feeding Experiments
2.2. Arginase Inhibition
2.3. Kinetics of Arginase Inhibition
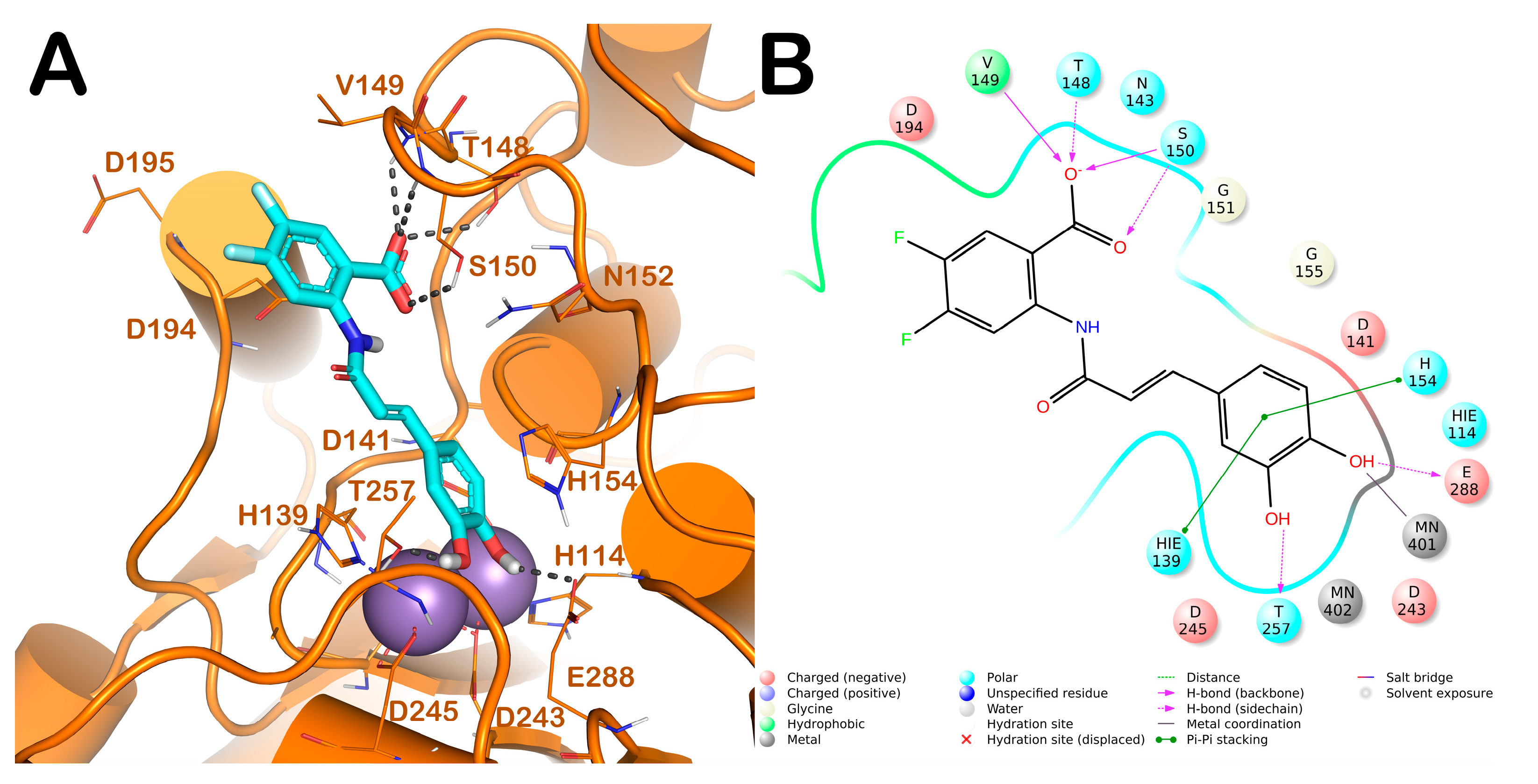
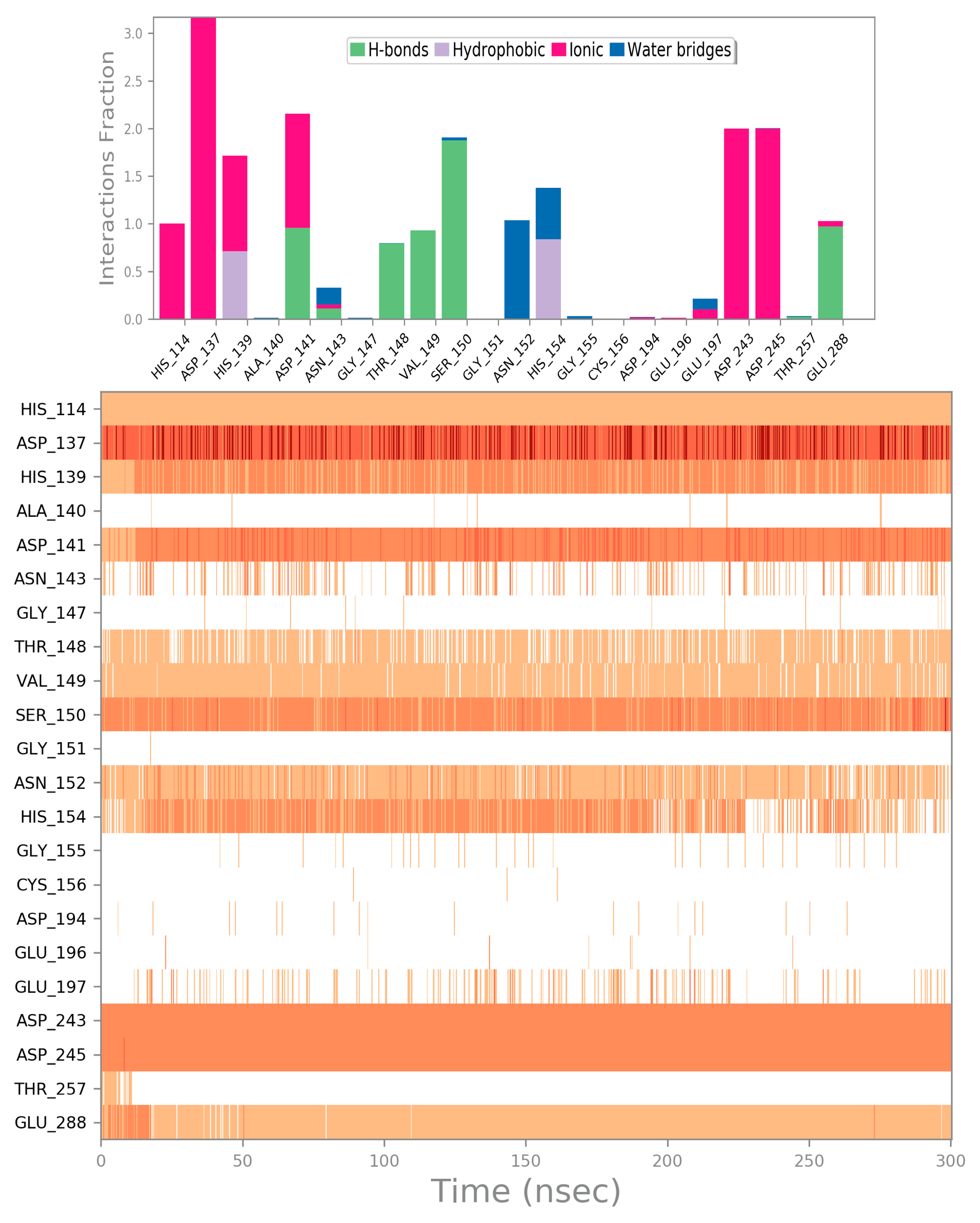
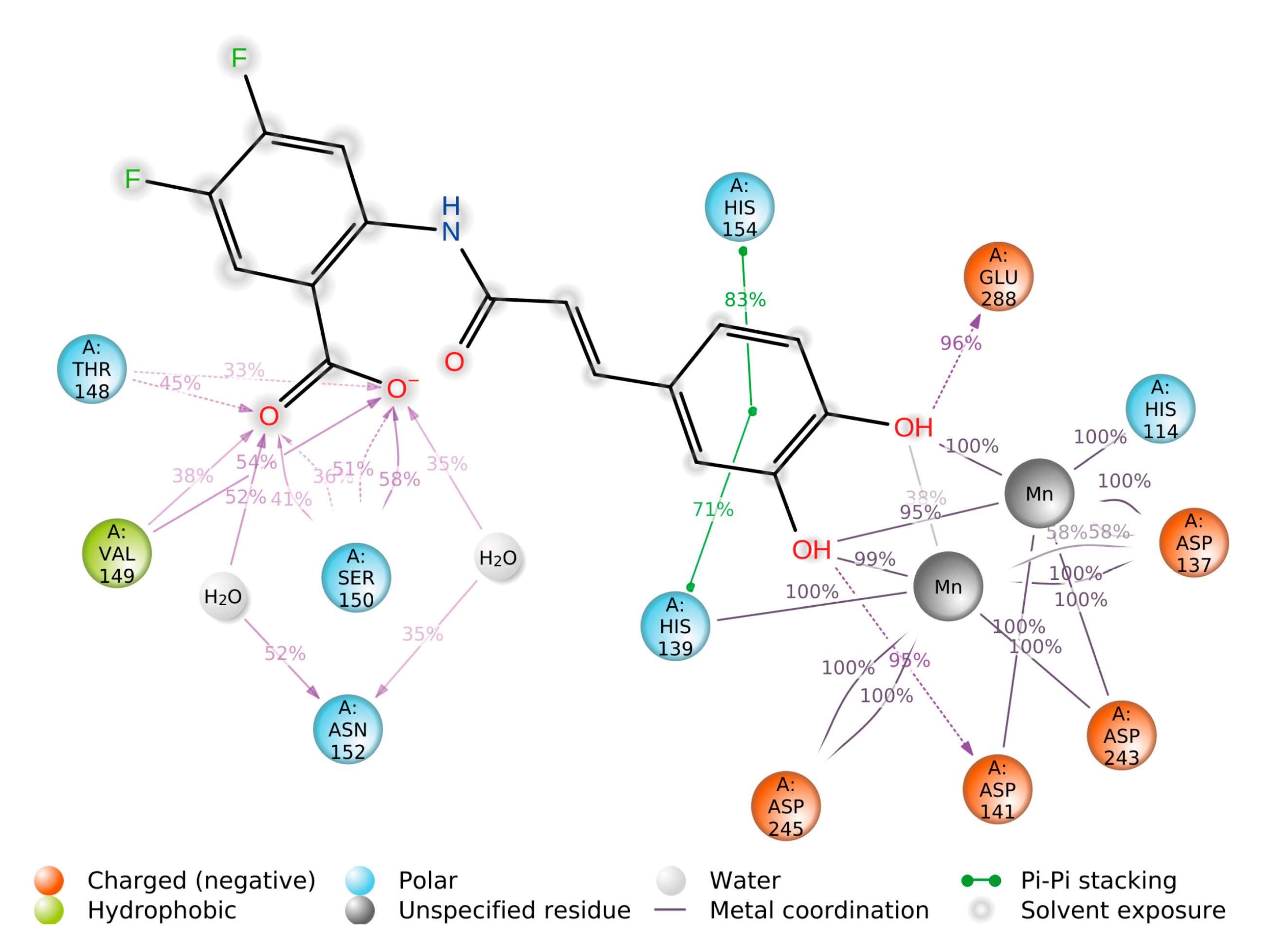
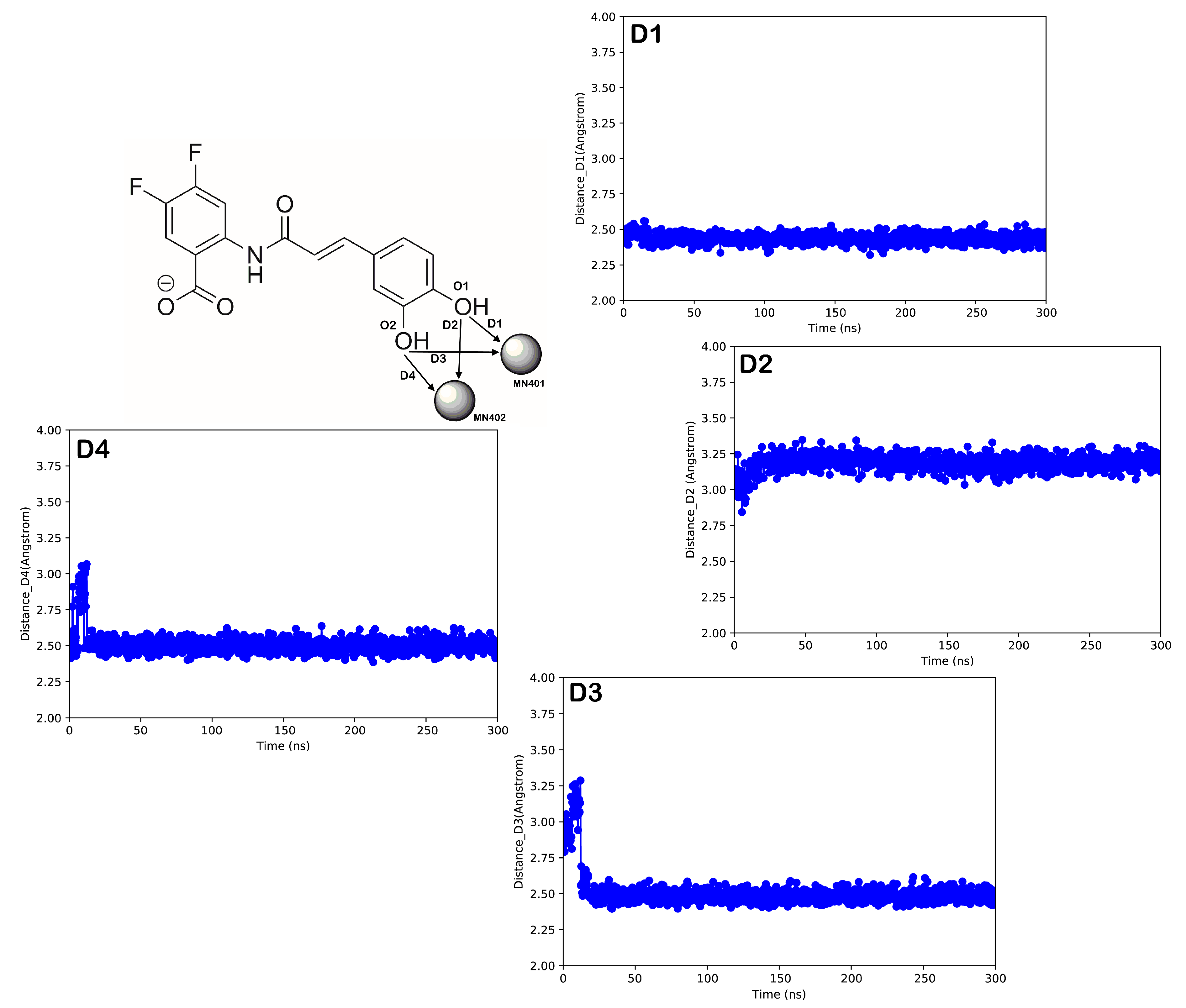
2.4. Computational Studies
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Bacterial Strain, Cultivation, and Chemicals
4.3. Extraction, Isolation, and Identification of the Biotransformation Products
4.4. Arginase Inhibition and Kinetics
4.5. Computational Details
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO|Leishmaniasis. WHO 2018. Available online: https://www.who.int/leishmaniasis/en/ (accessed on 25 January 2019).
- Sundar, S.; Chakravarty, J. An update on pharmacotherapy for leishmaniasis. Expert. Opin. Pharm. 2015, 16, 237–252. [Google Scholar] [CrossRef] [PubMed]
- Colotti, G.; Ilari, A. Polyamine metabolism in Leishmania: From arginine to trypanothione. Amino Acids 2011, 40, 269–285. [Google Scholar] [CrossRef]
- Da Silva, M.F.; Zampieri, R.A.; Muxel, S.M.; Beverley, S.M.; Floeter-Winter, L.M. Leishmania amazonensis arginase compartmentalization in the glycosome is important for parasite infectivity. PLoS ONE 2012, 7, e34022. [Google Scholar] [CrossRef] [PubMed]
- Mandal, A.; Das, S.; Roy, S.; Ghosh, A.K.; Sardar, A.H.; Verma, S.; Saini, S.; Singh, R.; Abhishek, K.; Kumar, A.; et al. Deprivation of L-Arginine Induces Oxidative Stress Mediated Apoptosis in Leishmania donovani Promastigotes: Contribution of the Polyamine Pathway. PLoS Negl. Trop. Dis. 2016, 10, e0004373. [Google Scholar] [CrossRef]
- Bocedi, A.; Dawood, K.F.; Fabrini, R.; Federici, G.; Gradoni, L.; Pedersen, J.Z.; Ricci, G. Trypanothione efficiently intercepts nitric oxide as a harmless iron complex in trypanosomatid parasites. FASEB J. 2010, 24, 1035–1042. [Google Scholar] [CrossRef]
- Fairlamb, A.H.; Cerami, A. Metabolism and functions of trypanothione in the Kinetoplastida. Annu. Rev. Microbiol. 1992, 46, 695–729. [Google Scholar] [CrossRef]
- Iniesta, V.; Gomez-Nieto, L.C.; Corraliza, I. The inhibition of arginase by N(omega)-hydroxy-l-arginine controls the growth of Leishmania inside macrophages. J. Exp. Med. 2001, 193, 777–784. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, E.R.; do Carmo Maquiaveli, C.; Magalhaes, P.P. The leishmanicidal flavonols quercetin and quercitrin target Leishmania (Leishmania) amazonensis arginase. Exp. Parasitol. 2012, 130, 183–188. [Google Scholar] [CrossRef]
- Manjolin, L.C.; dos Reis, M.B.; Maquiaveli Cdo, C.; Santos-Filho, O.A.; da Silva, E.R. Dietary flavonoids fisetin, luteolin and their derived compounds inhibit arginase, a central enzyme in Leishmania (Leishmania) amazonensis infection. Food Chem. 2013, 141, 2253–2262. [Google Scholar] [CrossRef]
- Dos Reis, M.B.; Manjolin, L.C.; Maquiaveli Cdo, C.; Santos-Filho, O.A.; da Silva, E.R. Inhibition of Leishmania (Leishmania) amazonensis and rat arginases by green tea EGCG, (+)-catechin and (-)-epicatechin: A comparative structural analysis of enzyme-inhibitor interactions. PLoS ONE 2013, 8, e78387. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, E.R.; Boechat, N.; Pinheiro, L.C.; Bastos, M.M.; Costa, C.C.; Bartholomeu, J.C.; da Costa, T.H. Novel selective inhibitor of Leishmania (Leishmania) amazonensis arginase. Chem. Biol. Drug Des. 2015, 86, 969–978. [Google Scholar] [CrossRef] [PubMed]
- Moreira Rda, C.; Rebelo, J.M.; Gama, M.E.; Costa, J.M. Knowledge level about of American tegumentary leishmaniasis (ATL) and use of alternative therapies in an endemic area in the Amazon Region in the State of Maranhao, Brazil. Cad. Saúde Pública 2002, 18, 187–195. [Google Scholar] [CrossRef][Green Version]
- Kvist, L.P.; Christensen, S.B.; Rasmussen, H.B.; Mejia, K.; Gonzalez, A. Identification and evaluation of Peruvian plants used to treat malaria and leishmaniasis. J. Ethnopharmacol. 2006, 106, 390–402. [Google Scholar] [CrossRef] [PubMed]
- Maquiaveli, C.C.; Lucon-Junior, J.F.; Brogi, S.; Campiani, G.; Gemma, S.; Vieira, P.C.; Silva, E.R. Verbascoside Inhibits Promastigote Growth and Arginase Activity of Leishmania amazonensis. J. Nat. Prod. 2016, 79, 1459–1463. [Google Scholar] [CrossRef]
- Maquiaveli, C.D.C.; Rochetti, A.L.; Fukumasu, H.; Vieira, P.C.; da Silva, E.R. Antileishmanial activity of verbascoside: Selective arginase inhibition of intracellular amastigotes of Leishmania (Leishmania) amazonensis with resistance induced by LPS plus IFN-gamma. Biochem. Pharm. 2017, 127, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Froelich, S.; Gupta, M.P.; Siems, K.; Jenett-Siems, K. Phenylethanoid glycosides from Stachytarpheta cayennensis (Rich.) Vahl, Verbenaceae, a traditional antimalarial medicinal plant. Rev. Bras. Farm. 2008, 18, 517–520. [Google Scholar] [CrossRef]
- Montrieux, E.; Perera, W.H.; Garcia, M.; Maes, L.; Cos, P.; Monzote, L. In vitro and in vivo activity of major constituents from Pluchea carolinensis against Leishmania amazonensis. Parasitol. Res. 2014, 113, 2925–2932. [Google Scholar] [CrossRef]
- Da Silva, E.R.; Brogi, S.; Grillo, A.; Campiani, G.; Gemma, S.; Vieira, P.C.; Maquiaveli, C.D.C. Cinnamic acids derived compounds with antileishmanial activity target Leishmania amazonensis arginase. Chem. Biol. Drug Des. 2019, 93, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Bi, H.; Zhuang, Y.; Liu, S.; Liu, T.; Ma, Y. Engineered synthesis of rosmarinic acid in Escherichia coli resulting production of a new intermediate, caffeoyl-phenyllactate. Biotechnol. Lett. 2016, 38, 81–88. [Google Scholar] [CrossRef]
- Zhuang, Y.; Jiang, J.; Bi, H.; Yin, H.; Liu, S.; Liu, T. Synthesis of rosmarinic acid analogues in Escherichia coli. Biotechnol. Lett. 2016, 38, 619–627. [Google Scholar] [CrossRef]
- Dixon, M. The determination of enzyme inhibitor constants. Biochem. J. 1953, 55, 170–171. [Google Scholar] [CrossRef]
- Cornish-Bowden, A. A simple graphical method for determining the inhibition constants of mixed, uncompetitive and non-competitive inhibitors. Biochem. J. 1974, 137, 143–144. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, E.R.; Brogi, S.; Lucon-Junior, J.F.; Campiani, G.; Gemma, S.; Maquiaveli, C.D.C. Dietary polyphenols rutin, taxifolin and quercetin related compounds target Leishmania amazonensis arginase. Food Funct. 2019, 10, 3172–3180. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, E.R.; Come, J.; Brogi, S.; Calderone, V.; Chemi, G.; Campiani, G.; Oliveira, T.; Pham, T.N.; Pudlo, M.; Girard, C.; et al. Cinnamides Target Leishmania amazonensis Arginase Selectively. Molecules 2020, 25, 5271. [Google Scholar] [CrossRef]
- Pham, T.N.; Bordage, S.; Pudlo, M.; Demougeot, C.; Thai, K.M.; Girard-Thernier, C. Cinnamide Derivatives as Mammalian Arginase Inhibitors: Synthesis, Biological Evaluation and Molecular Docking. Int. J. Mol. Sci. 2016, 17, 1656. [Google Scholar] [CrossRef] [PubMed]
- Castilho-Martins, E.A.; Laranjeira da Silva, M.F.; dos Santos, M.G.; Muxel, S.M.; Floeter-Winter, L.M. Axenic Leishmania amazonensis promastigotes sense both the external and internal arginine pool distinctly regulating the two transporter-coding genes. PLoS ONE 2011, 6, e27818. [Google Scholar] [CrossRef] [PubMed]
- Battah, B.; Chemi, G.; Butini, S.; Campiani, G.; Brogi, S.; Delogu, G.; Gemma, S. A Repurposing Approach for Uncovering the Anti-Tubercular Activity of FDA-Approved Drugs with Potential Multi-Targeting Profiles. Molecules 2019, 24, 4373. [Google Scholar] [CrossRef] [PubMed]
- D’Antonio, E.L.; Ullman, B.; Roberts, S.C.; Dixit, U.G.; Wilson, M.E.; Hai, Y.; Christianson, D.W. Crystal structure of arginase from Leishmania mexicana and implications for the inhibition of polyamine biosynthesis in parasitic infections. Arch. Biochem. Biophys. 2013, 535, 163–176. [Google Scholar] [CrossRef]
- Paolino, M.; Brindisi, M.; Vallone, A.; Butini, S.; Campiani, G.; Nannicini, C.; Giuliani, G.; Anzini, M.; Lamponi, S.; Giorgi, G.; et al. Development of Potent Inhibitors of the Mycobacterium tuberculosis Virulence Factor Zmp1 and Evaluation of Their Effect on Mycobacterial Survival inside Macrophages. ChemMedChem 2018, 13, 422–430. [Google Scholar] [CrossRef]
- Sirous, H.; Chemi, G.; Gemma, S.; Butini, S.; Debyser, Z.; Christ, F.; Saghaie, L.; Brogi, S.; Fassihi, A.; Campiani, G.; et al. Identification of Novel 3-Hydroxy-pyran-4-One Derivatives as Potent HIV-1 Integrase Inhibitors Using in silico Structure-Based Combinatorial Library Design Approach. Front. Chem. 2019, 7, 574. [Google Scholar] [CrossRef] [PubMed]
- Nickolls, J.; Buck, I.; Garland, M.; Skadron, K. Scalable Parallel Programming with CUDA. Queue 2008, 6, 40–53. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and Testing of the OPLS All-Atom Force Field on Conformational Energetics and Properties of Organic Liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Humphreys, D.D.; Friesner, R.A.; Berne, B.J. A Multiple-Time-Step Molecular Dynamics Algorithm for Macromolecules. J. Phys. Chem. 2002, 98, 6885–6892. [Google Scholar] [CrossRef]
- Hoover, W.G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 1985, 31, 1695–1697. [Google Scholar] [CrossRef] [PubMed]
- Martyna, G.J.; Tobias, D.J.; Klein, M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994, 101, 4177–4189. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]


| Compound | Structures MW (g/mol) | IC50 (μM) * | Ki (μM) Mechanism of Inhibition |
|---|---|---|---|
| 1 | 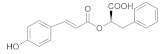 312.10 | Inactive | - |
| 2 |  330.11 | 2.8 ± 0.9 | 5.7 ± 0.7 Noncompetitive |
| 3 | 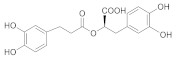 362.10 | 3.0 ± 0.2 | 3.9 ± 0.4 Noncompetitive |
| 4 |  346.11 | 1.9 ± 0.7 | 2.0 ± 0.6 Noncompetitive |
| 5 |  330.11 | >200 | - |
| 6 | 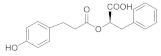 314.12 | Inactive | - |
| 7 |  296.05 | 36 ± 6 | 74 ± 8 Noncompetitive |
| 8 |  317.05 | Inactive | - |
| 9 |  335.06 | 5.5 ± 0.5 | 4.6 ± 1.7 Competitive |
| 10 | 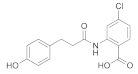 319.06 | Inactive | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Come, J.A.A.d.S.S.; Zhuang, Y.; Li, T.; Brogi, S.; Gemma, S.; Liu, T.; da Silva, E.R. In Vitro and In Silico Analyses of New Cinnamid and Rosmarinic Acid-Derived Compounds Biosynthesized in Escherichia coli as Leishmania amazonensis Arginase Inhibitors. Pathogens 2022, 11, 1020. https://doi.org/10.3390/pathogens11091020
Come JAAdSS, Zhuang Y, Li T, Brogi S, Gemma S, Liu T, da Silva ER. In Vitro and In Silico Analyses of New Cinnamid and Rosmarinic Acid-Derived Compounds Biosynthesized in Escherichia coli as Leishmania amazonensis Arginase Inhibitors. Pathogens. 2022; 11(9):1020. https://doi.org/10.3390/pathogens11091020
Chicago/Turabian StyleCome, Julio Abel Alfredo dos Santos Simone, Yibin Zhuang, Tianzhen Li, Simone Brogi, Sandra Gemma, Tao Liu, and Edson Roberto da Silva. 2022. "In Vitro and In Silico Analyses of New Cinnamid and Rosmarinic Acid-Derived Compounds Biosynthesized in Escherichia coli as Leishmania amazonensis Arginase Inhibitors" Pathogens 11, no. 9: 1020. https://doi.org/10.3390/pathogens11091020
APA StyleCome, J. A. A. d. S. S., Zhuang, Y., Li, T., Brogi, S., Gemma, S., Liu, T., & da Silva, E. R. (2022). In Vitro and In Silico Analyses of New Cinnamid and Rosmarinic Acid-Derived Compounds Biosynthesized in Escherichia coli as Leishmania amazonensis Arginase Inhibitors. Pathogens, 11(9), 1020. https://doi.org/10.3390/pathogens11091020










