Immune Responses in Oral Papillomavirus Clearance in the MmuPV1 Mouse Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Viral Infection, Oral Swab, and Oral Tissue Collection
2.2. Viral Load Assessments
2.3. Cell Disassociation for Flow Cytometry
2.4. Flow Cytometry
2.5. Peptide Stimulation and Intracellular Cytokine Assay
2.6. Anti-MmuPV1 Antibody Detection by Enzyme-Linked Immunosorbent Assay (ELISA)
2.7. Statistical Analyses
3. Results
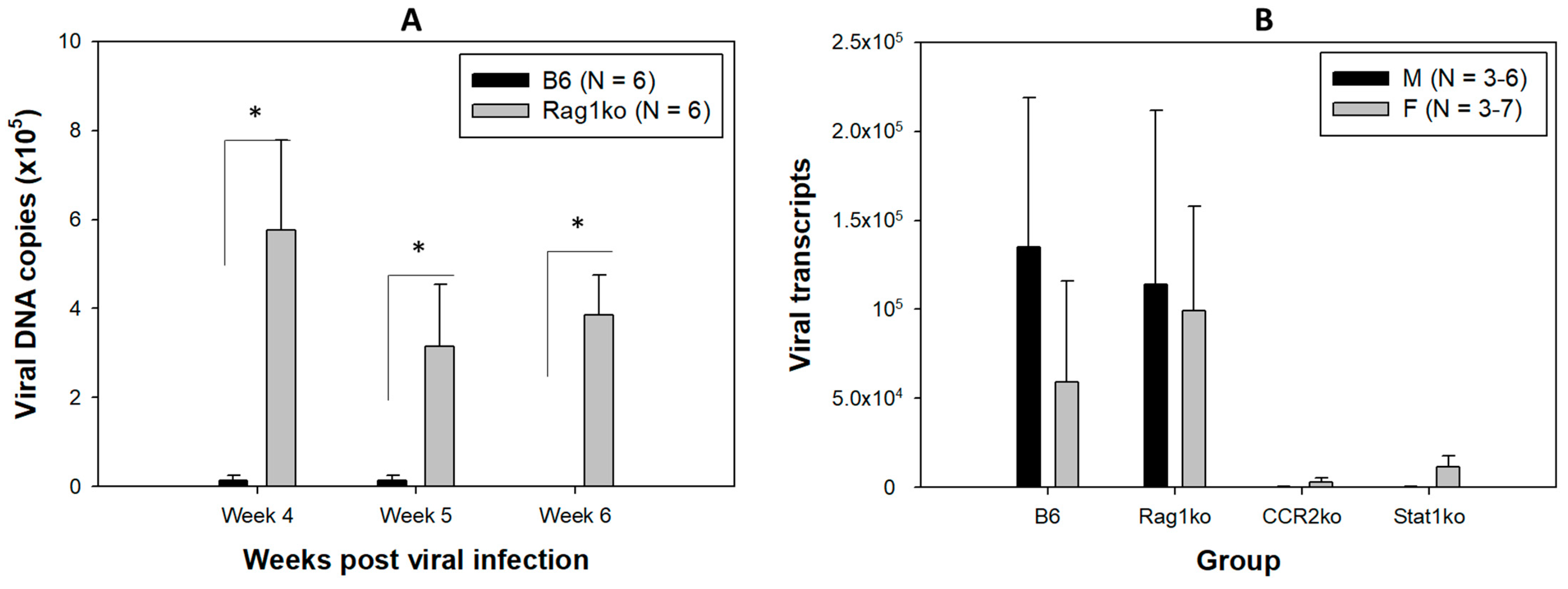
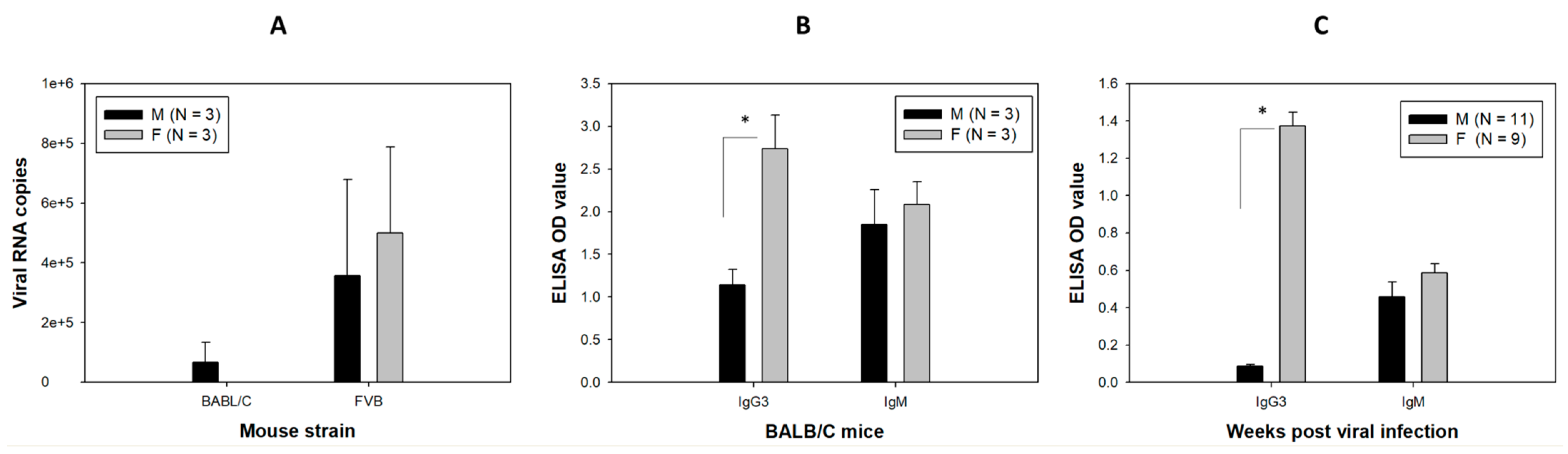
3.1. Viral DNA and RNA Transcripts Were Detected in the Orally Infected Mice
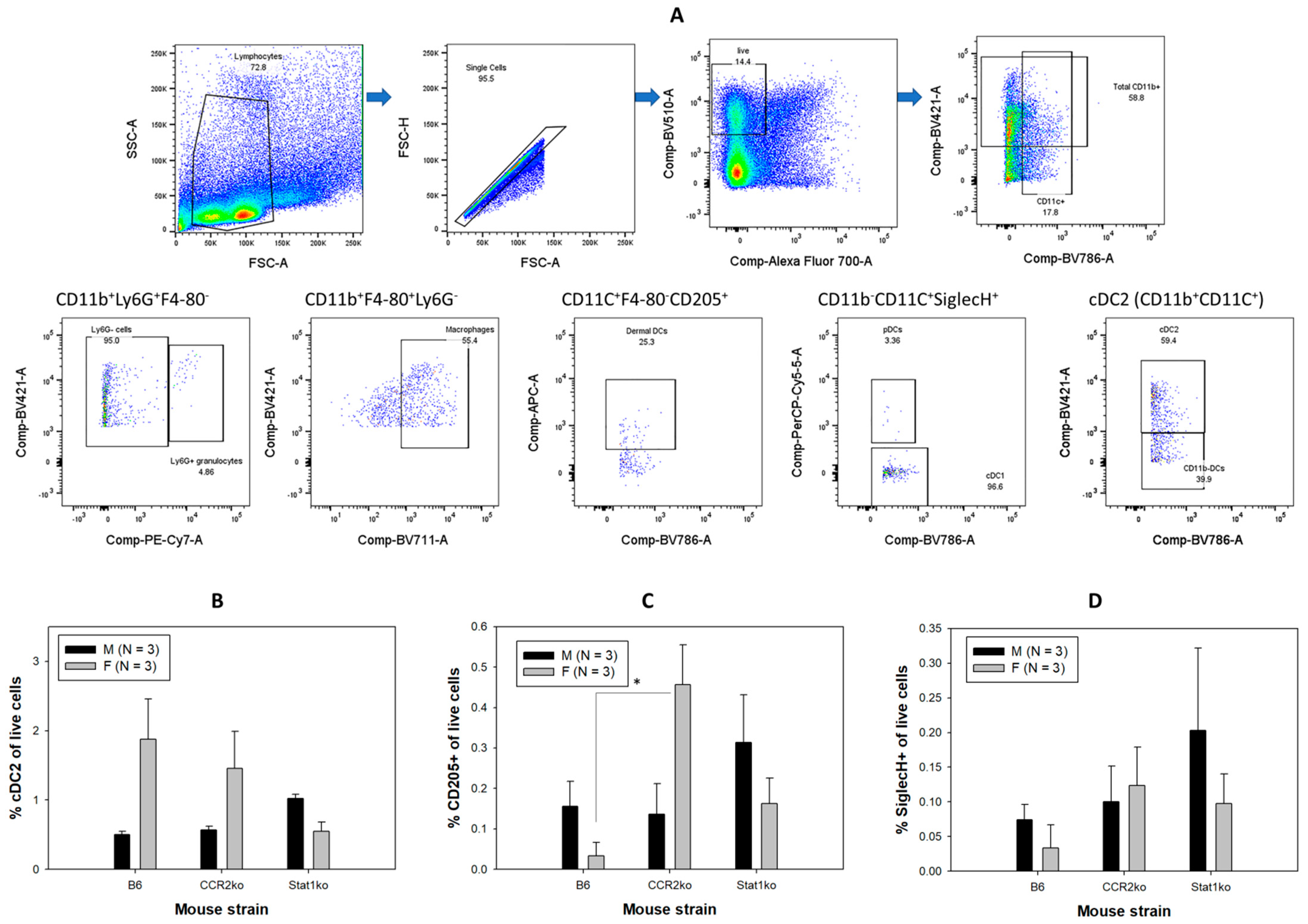
3.2. Higher Numbers of Dermal Dendritic Cells Were Found in Female B6 Mice
3.3. Significantly Higher Numbers of Macrophages and Granulocytes Were Recruited to the Infected Tongues of Males
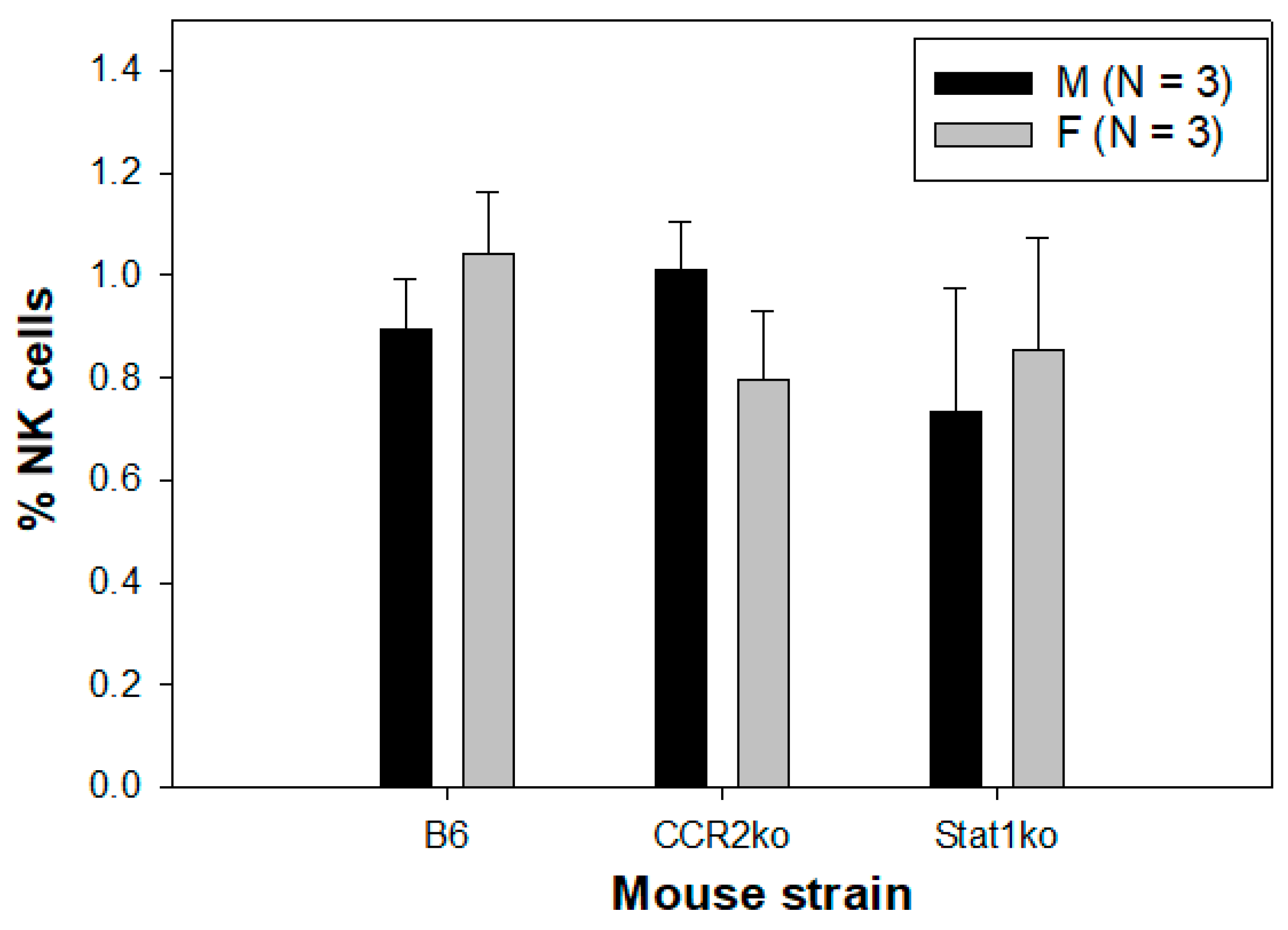
3.4. Similar Numbers of NK Cells Were Found in All Tested Mice
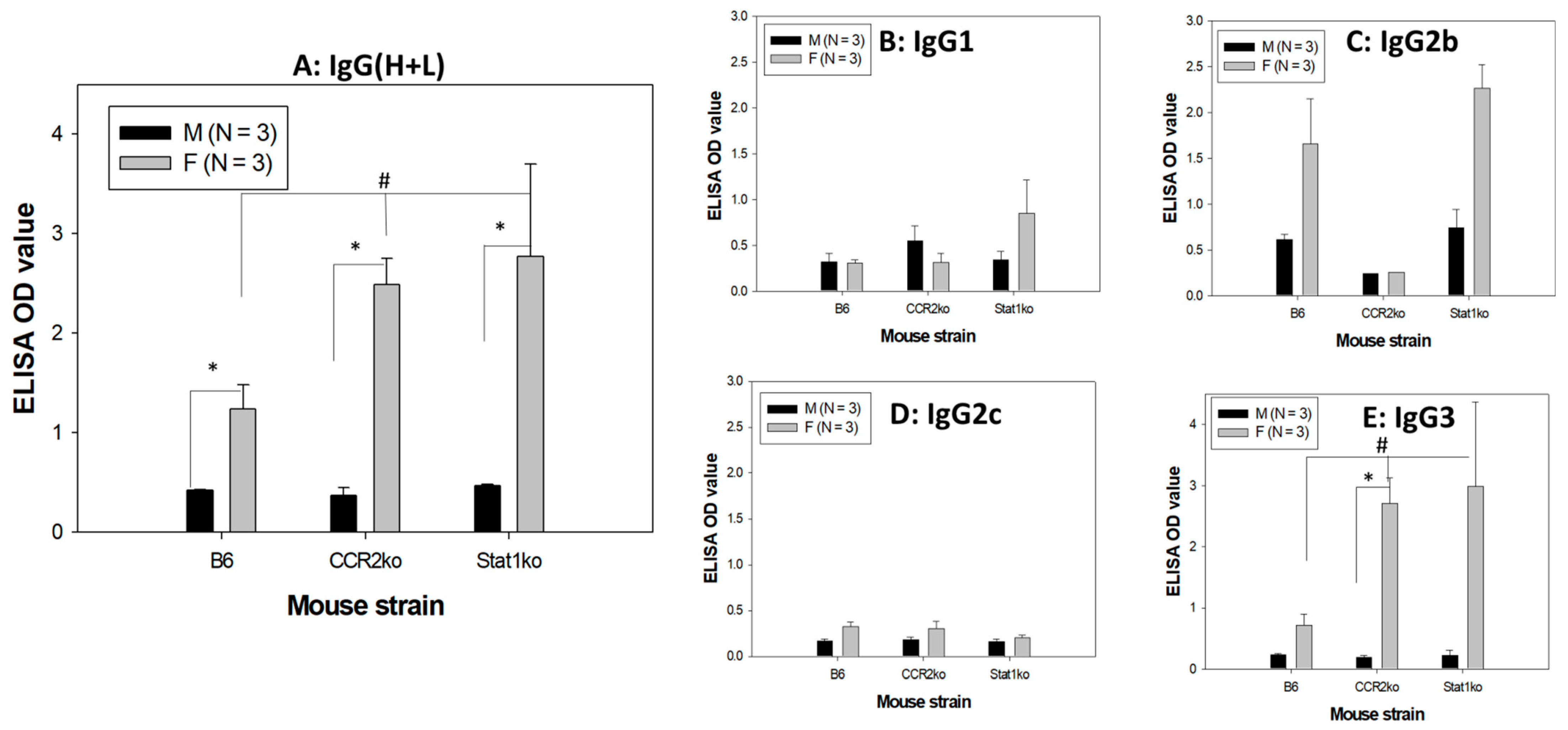
3.5. Higher Levels of Anti-MmuPV1 E4 Antibodies Are Detected in Female Mice, Regardless of Strain
3.6. Anti-MmuPV1 Antibodies Are Skewed toward IgG3 in Females of Additional Inbred Mouse Strains
3.7. Comparable T-Cell-Mediated Immune Responses to MmuPV1 E6 and E7 in Both Sexes of Infected B6 Mice
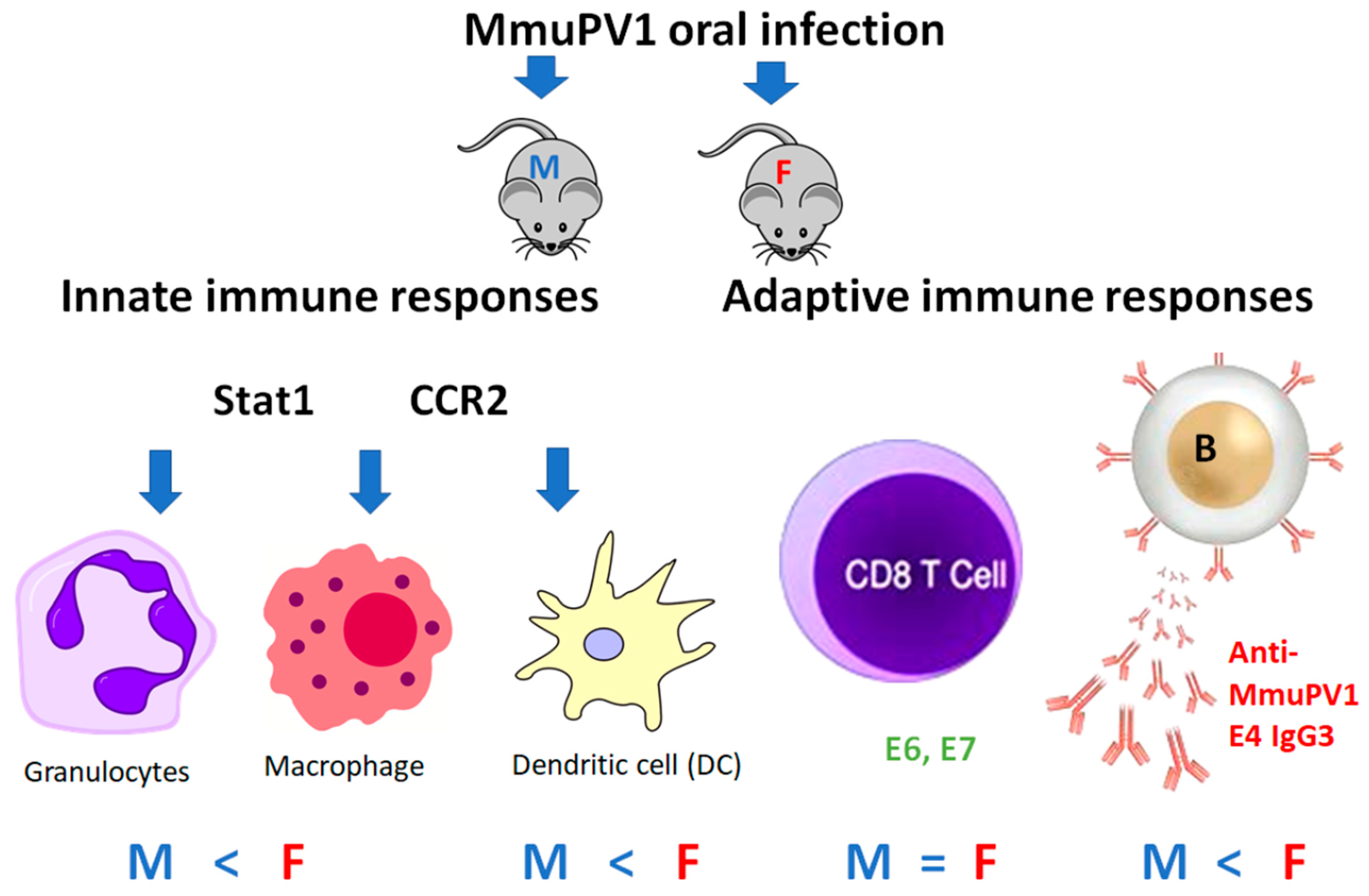
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lechner, M.; Liu, J.; Masterson, L.; Fenton, T.R. HPV-associated oropharyngeal cancer: Epidemiology, molecular biology and clinical management. Nat. Rev. Clin. Oncol. 2022, 19, 306–327. [Google Scholar] [CrossRef]
- Qureishi, A.; Mawby, T.; Fraser, L.; Shah, K.A.; Moller, H.; Winter, S. Current and future techniques for human papilloma virus (HPV) testing in oropharyngeal squamous cell carcinoma. Eur. Arch. Otorhinolaryngol. 2017, 274, 2675–2683. [Google Scholar] [CrossRef]
- Okami, K. Clinical features and treatment strategy for HPV-related oropharyngeal cancer. Int. J. Clin. Oncol. 2016, 21, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Trosman, S.J.; Koyfman, S.A.; Ward, M.C.; Al-Khudari, S.; Nwizu, T.; Greskovich, J.F.; Lamarre, E.D.; Scharpf, J.; Khan, M.J.; Lorenz, R.R.; et al. Effect of human papillomavirus on patterns of distant metastatic failure in oropharyngeal squamous cell carcinoma treated with chemoradiotherapy. JAMA Otolaryngol. Head Neck Surg. 2015, 141, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Dalianis, T. Human papillomavirus and oropharyngeal cancer, the epidemics, and significance of additional clinical biomarkers for prediction of response to therapy (Review). Int. J. Oncol. 2014, 44, 1799–1805. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J. Host control of human papillomavirus infection and disease. Best Pract. Res. Clin. Obstet. Gynaecol. 2018, 47, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Ingle, A.; Ghim, S.; Joh, J.; Chepkoech, I.; Bennett Jenson, A.; Sundberg, J.P. Novel laboratory mouse papillomavirus (MusPV) infection. Vet. Pathol. 2011, 48, 500–505. [Google Scholar] [CrossRef] [PubMed]
- Cladel, N.M.; Budgeon, L.R.; Cooper, T.K.; Balogh, K.K.; Hu, J.; Christensen, N.D. Secondary infections, expanded tissue tropism, and evidence for malignant potential in immunocompromised mice infected with Mus musculus papillomavirus 1 DNA and virus. J. Virol. 2013, 87, 9391–9395. [Google Scholar] [CrossRef] [PubMed]
- Cladel, N.M.; Budgeon, L.R.; Balogh, K.K.; Cooper, T.K.; Hu, J.; Christensen, N.D. Mouse papillomavirus MmuPV1 infects oral mucosa and preferentially targets the base of the tongue. Virology 2016, 488, 73–80. [Google Scholar] [CrossRef]
- Hu, J.; Budgeon, L.R.; Cladel, N.M.; Balogh, K.; Myers, R.; Cooper, T.K.; Christensen, N.D. Tracking vaginal, anal and oral infection in a mouse papillomavirus infection model. J. Gen. Virol. 2015, 96, 3554–3565. [Google Scholar] [CrossRef]
- Hu, J.; Cladel, N.M.; Budgeon, L.R.; Balogh, K.K.; Christensen, N.D. The Mouse Papillomavirus Infection Model. Viruses 2017, 9, 246. [Google Scholar] [CrossRef]
- Christensen, N.D.; Chen, K.M.; Hu, J.; Stairs, D.B.; Sun, Y.W.; Aliaga, C.; Balogh, K.K.; Atkins, H.; Shearer, D.; Li, J.; et al. The environmental pollutant and tobacco smoke constituent dibenzo[def,p]chrysene is a co-factor for malignant progression of mouse oral papillomavirus infections. Chem. Biol. Interact. 2020, 109321. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Buehler, D.; Ward-Shaw, E.; Lambert, P.F. An Infection-Based Murine Model for Papillomavirus-Associated Head and Neck Cancer. mBio 2020, 11, e00908-20. [Google Scholar] [CrossRef] [PubMed]
- Bilger, A.; King, R.E.; Schroeder, J.P.; Piette, J.T.; Hinshaw, L.A.; Kurth, A.D.; AlRamahi, R.W.; Barthel, M.V.; Ward-Shaw, E.T.; Buehler, D.; et al. A Mouse Model of Oropharyngeal Papillomavirus-Induced Neoplasia Using Novel Tools for Infection and Nasal Anesthesia. Viruses 2020, 12, 450. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.W.; Jiang, R.; Peng, S.; Chang, Y.N.; Hung, C.F.; Roden, R.B. Immunologic Control of Mus musculus Papillomavirus Type 1. PLoS Pathog. 2015, 11, e1005243. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.T.; Wang, J.W.; Peng, S.; Huang, T.C.; Wang, C.; Cannella, F.; Chang, Y.N.; Viscidi, R.P.; Best, S.R.A.; Hung, C.F.; et al. Spontaneous and Vaccine-Induced Clearance of Mus Musculus Papillomavirus 1 Infection. J. Virol. 2017, 91, e00699-17. [Google Scholar] [CrossRef] [PubMed]
- Handisurya, A.; Day, P.M.; Thompson, C.D.; Bonelli, M.; Lowy, D.R.; Schiller, J.T. Strain-Specific Properties and T Cells Regulate the Susceptibility to Papilloma Induction by Mus musculus Papillomavirus 1. PLoS. Pathog. 2014, 10, e1004314. [Google Scholar] [CrossRef]
- Cladel, N.M.; Budgeon, L.R.; Balogh, K.K.; Cooper, T.K.; Brendle, S.A.; Christensen, N.D.; Schell, T.D.; Hu, J. Mouse papillomavirus infection persists in mucosal tissues of an immunocompetent mouse strain and progresses to cancer. Sci. Rep. 2017, 7, 16932. [Google Scholar] [CrossRef]
- Spurgeon, M.E.; Lambert, P.F. Sexual transmission of murine papillomavirus (MmuPV1) in Mus musculus. eLife 2019, 8, e50056. [Google Scholar] [CrossRef]
- Scagnolari, C.; Cannella, F.; Pierangeli, A.; Mellinger Pilgrim, R.; Antonelli, G.; Rowley, D.; Wong, M.; Best, S.; Xing, D.; Roden, R.B.S.; et al. Insights into the Role of Innate Immunity in Cervicovaginal Papillomavirus Infection from Studies Using Gene-Deficient Mice. J. Virol. 2020, 94, e00087-20. [Google Scholar] [CrossRef]
- Sundberg, J.P.; Stearns, T.M.; Joh, J.; Proctor, M.; Ingle, A.; Silva, K.A.; Dadras, S.S.; Jenson, A.B.; Ghim, S.J. Immune status, strain background, and anatomic site of inoculation affect mouse papillomavirus (MmuPV1) induction of exophytic papillomas or endophytic trichoblastomas. PLoS ONE 2014, 9, e113582. [Google Scholar] [CrossRef]
- Beziat, V.; Rapaport, F.; Hu, J.; Titeux, M.; Bonnet des Claustres, M.; Bourgey, M.; Griffin, H.; Bandet, E.; Ma, C.S.; Sherkat, R.; et al. Humans with inherited T cell CD28 deficiency are susceptible to skin papillomaviruses but are otherwise healthy. Cell 2021, 184, 3812–3828. [Google Scholar] [CrossRef]
- Woodworth, C.D. HPV innate immunity. Front. Biosci. 2002, 7, d2058–d2071. [Google Scholar] [CrossRef]
- Moerman-Herzog, A.; Nakagawa, M. Early Defensive Mechanisms against Human Papillomavirus Infection. Clin. Vaccine Immunol. 2015, 22, 850–857. [Google Scholar] [CrossRef] [PubMed]
- Nunes, R.A.L.; Morale, M.G.; Silva, G.A.F.; Villa, L.L.; Termini, L. Innate immunity and HPV: Friends or foes. Clinics 2018, 73 (Suppl. S1), e549s. [Google Scholar] [CrossRef] [PubMed]
- Anjuere, F.; Bekri, S.; Bihl, F.; Braud, V.M.; Cuburu, N.; Czerkinsky, C.; Hervouet, C.; Luci, C. B cell and T cell immunity in the female genital tract: Potential of distinct mucosal routes of vaccination and role of tissue-associated dendritic cells and natural killer cells. Clin. Microbiol. Infect. 2012, 18 (Suppl. S5), 117–122. [Google Scholar] [CrossRef] [PubMed]
- Sistigu, A.; Di Modugno, F.; Manic, G.; Nistico, P. Deciphering the loop of epithelial-mesenchymal transition, inflammatory cytokines and cancer immunoediting. Cytokine Growth Factor Rev. 2017, 36, 67–77. [Google Scholar] [CrossRef]
- Farhood, B.; Najafi, M.; Mortezaee, K. CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: A review. J. Cell Physiol. 2019, 234, 8509–8521. [Google Scholar] [CrossRef]
- Hilligan, K.L.; Ronchese, F. Antigen presentation by dendritic cells and their instruction of CD4+ T helper cell responses. Cell. Mol. Immunol. 2020, 17, 587–599. [Google Scholar] [CrossRef]
- Gardella, B.; Dominoni, M.; Carletti, G.V.; Musacchi, V.; Amici, M.; Spinillo, A. Cytokines and chemokines levels in primary HPV infection: A pilot study. Acta Virol. 2021, 65, 75–81. [Google Scholar] [CrossRef]
- Gabriel, P.; Babiarova, K.; Zurkova, K.; Krystofova, J.; Hainz, P.; Kutinova, L.; Nemeckova, S. Chemokine binding protein vCCI attenuates vaccinia virus without affecting the cellular response elicited by immunization with a recombinant vaccinia vector carrying the HPV16 E7 gene. Viral Immunol. 2012, 25, 411–422. [Google Scholar] [CrossRef]
- Barros, M.R., Jr.; de Oliveira, T.H.A.; de Melo, C.M.L.; Venuti, A.; de Freitas, A.C. Viral Modulation of TLRs and Cytokines and the Related Immunotherapies for HPV-Associated Cancers. J. Immunol. Res. 2018, 2018, 2912671. [Google Scholar] [CrossRef]
- Karim, R.; Meyers, C.; Backendorf, C.; Ludigs, K.; Offringa, R.; van Ommen, G.J.; Melief, C.J.; van der Burg, S.H.; Boer, J.M. Human papillomavirus deregulates the response of a cellular network comprising of chemotactic and proinflammatory genes. PLoS ONE 2011, 6, e17848. [Google Scholar] [CrossRef]
- Quayle, A.J. The innate and early immune response to pathogen challenge in the female genital tract and the pivotal role of epithelial cells. J. Reprod. Immunol. 2002, 57, 61–79. [Google Scholar] [CrossRef]
- Dong, M.B.; Rahman, M.J.; Tarbell, K.V. Flow cytometric gating for spleen monocyte and DC subsets: Differences in autoimmune NOD mice and with acute inflammation. J. Immunol. Methods 2016, 432, 4–12. [Google Scholar] [CrossRef]
- Vanbervliet, B.; Homey, B.; Durand, I.; Massacrier, C.; Ait-Yahia, S.; de Bouteiller, O.; Vicari, A.; Caux, C. Sequential involvement of CCR2 and CCR6 ligands for immature dendritic cell recruitment: Possible role at inflamed epithelial surfaces. Eur. J. Immunol. 2002, 32, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Le Borgne, M.; Etchart, N.; Goubier, A.; Lira, S.A.; Sirard, J.C.; van Rooijen, N.; Caux, C.; Ait-Yahia, S.; Vicari, A.; Kaiserlian, D.; et al. Dendritic cells rapidly recruited into epithelial tissues via CCR6/CCL20 are responsible for CD8+ T cell crosspriming in vivo. Immunity 2006, 24, 191–201. [Google Scholar] [CrossRef]
- Saadeh, D.; Kurban, M.; Abbas, O. Plasmacytoid dendritic cells and type I interferon in the immunological response against warts. Clin. Exp. Dermatol. 2017, 42, 857–862. [Google Scholar] [CrossRef] [PubMed]
- Kurten, C.H.L.; Kulkarni, A.; Cillo, A.R.; Santos, P.M.; Roble, A.K.; Onkar, S.; Reeder, C.; Lang, S.; Chen, X.; Duvvuri, U.; et al. Investigating immune and non-immune cell interactions in head and neck tumors by single-cell RNA sequencing. Nat. Commun. 2021, 12, 7338. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Hua, K. Cervical Cancer: Emerging Immune Landscape and Treatment. Onco Targets Ther. 2020, 13, 8037–8047. [Google Scholar] [CrossRef]
- Wagner, S.; Bockmann, H.; Gattenlohner, S.; Klussmann, J.P.; Wittekindt, C. The innate immune system in oropharyngeal squamous cell carcinoma: Immune modulation by HPV. HNO 2018, 66, 301–307. [Google Scholar] [CrossRef]
- Faraji, F.; Fung, N.; Zaidi, M.; Gourin, C.C.; Eisele, D.W.; Rooper, L.M.; Fakhry, C. Tumor-infiltrating lymphocyte quantification stratifies early-stage human papillomavirus oropharynx cancer prognosis. Laryngoscope 2020, 130, 930–938. [Google Scholar] [CrossRef] [PubMed]
- Lechien, J.R.; Seminerio, I.; Descamps, G.; Mat, Q.; Mouawad, F.; Hans, S.; Julieron, M.; Dequanter, D.; Vanderhaegen, T.; Journe, F.; et al. Impact of HPV Infection on the Immune System in Oropharyngeal and Non-Oropharyngeal Squamous Cell Carcinoma: A Systematic Review. Cells 2019, 8, 1061. [Google Scholar] [CrossRef] [PubMed]
- Raspaglio, G.; Buttarelli, M.; Filippetti, F.; Battaglia, A.; Buzzonetti, A.; Scambia, G.; Gallo, D. Stat1 confers sensitivity to radiation in cervical cancer cells by controlling Parp1 levels: A new perspective for Parp1 inhibition. Cell Death Dis. 2021, 12, 933. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Wu, Y.; Lu, Y.; Yue, Y.; Cui, C.; Yu, M.; Wang, S.; Liu, M.; Zhao, Y.; Sun, Z. STAT1 expression and HPV16 viral load predict cervical lesion progression. Oncol. Lett. 2020, 20, 28. [Google Scholar] [CrossRef] [PubMed]
- Green, D.S.; Young, H.A.; Valencia, J.C. Current prospects of type II interferon gamma signaling and autoimmunity. J. Biol. Chem. 2017, 292, 13925–13933. [Google Scholar] [CrossRef] [PubMed]
- Welte, T.; Koch, F.; Schuler, G.; Lechner, J.; Doppler, W.; Heufler, C. Granulocyte-macrophage colony-stimulating factor induces a unique set of STAT factors in murine dendritic cells. Eur. J. Immunol. 1997, 27, 2737–2740. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Mehta, K.P.; Laimins, L.A. Suppression of STAT-1 expression by human papillomaviruses is necessary for differentiation-dependent genome amplification and plasmid maintenance. J. Virol. 2011, 85, 9486–9494. [Google Scholar] [CrossRef]
- Suay, G.; Garcia-Canaveras, J.C.; Aparisi, F.; Lahoz, A.; Juan-Vidal, O. Sex Differences in the Efficacy of Immune Checkpoint Inhibitors in Neoadjuvant Therapy of Non-Small Cell Lung Cancer: A Meta-Analysis. Cancers 2023, 15, 4433. [Google Scholar] [CrossRef]
- Salvati, L.; Biagioni, B.; Vivarelli, E.; Parronchi, P. A gendered magnifying glass on COVID-19. Clin. Mol. Allergy 2020, 18, 14. [Google Scholar] [CrossRef]
- Emran, A.A.; Gallagher, S.J.; Tiffen, J.C.; Hersey, P. Sex bias of females in survival from cancer and infections. Is X the answer? Br. J. Cancer 2021, 124, 1184–1186. [Google Scholar] [CrossRef]
- Andersen, L.; Jakobsen, K.K.; Carlander, A.L.F.; Garset-Zamani, M.; Friborg, J.; Kiss, K.; Marvig, R.L.; Olsen, C.; Nielsen, F.C.; Andersen, E.; et al. The Incidence, Survival, and HPV Impact of Second Primary Cancer following Primary Oropharyngeal Squamous Cell Carcinoma: A 20-Year Retrospective and Population-Based Study. Viruses 2023, 15, 34. [Google Scholar] [CrossRef]
- Westhoff, C. Depot-medroxyprogesterone acetate injection (Depo-Provera): A highly effective contraceptive option with proven long-term safety. Contraception 2003, 68, 75–87. [Google Scholar] [CrossRef]
- Cladel, N.M.; Jiang, P.; Li, J.J.; Peng, X.; Cooper, T.K.; Majerciak, V.; Balogh, K.K.; Meyer, T.J.; Brendle, S.A.; Budgeon, L.R.; et al. Papillomavirus can be transmitted through the blood and produce infections in blood recipients: Evidence from two animal models. Emerg. Microbes Infect. 2019, 8, 1108–1121. [Google Scholar] [CrossRef]
- Hu, J.; Cladel, N.; Peng, X.; Balogh, K.; Christensen, N.D. Protective immunity with an E1 multivalent epitope DNA vaccine against cottontail rabbit papillomavirus (CRPV) infection in an HLA-A2.1 transgenic rabbit model. Vaccine 2008, 26, 809–816. [Google Scholar] [CrossRef] [PubMed]
- Brendle, S.A.; Li, J.; Cladel, N.M.; Balogh, K.K.; Booth, J.; Shearer, D.A.; Walter, V.; Lu, S.; Christensen, N.D.; Covington, D.; et al. Passive Immunization with a Single Monoclonal Neutralizing Antibody Protects against Cutaneous and Mucosal Mouse Papillomavirus Infections. J. Virol. 2022, 96, e0070322. [Google Scholar] [CrossRef] [PubMed]
- Fein, M.R.; He, X.Y.; Almeida, A.S.; Bruzas, E.; Pommier, A.; Yan, R.; Eberhardt, A.; Fearon, D.T.; Van Aelst, L.; Wilkinson, J.E.; et al. Cancer cell CCR2 orchestrates suppression of the adaptive immune response. J. Exp. Med. 2020, 217, e20181551. [Google Scholar] [CrossRef]
- Tamaura, M.; Satoh-Takayama, N.; Tsumura, M.; Sasaki, T.; Goda, S.; Kageyama, T.; Hayakawa, S.; Kimura, S.; Asano, T.; Nakayama, M.; et al. Human gain-of-function STAT1 mutation disturbs IL-17 immunity in mice. Int. Immunol. 2020, 32, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Amador-Molina, A.; Hernandez-Valencia, J.F.; Lamoyi, E.; Contreras-Paredes, A.; Lizano, M. Role of innate immunity against human papillomavirus (HPV) infections and effect of adjuvants in promoting specific immune response. Viruses 2013, 5, 2624–2642. [Google Scholar] [CrossRef]
- Silva, M.O.; Almeida, B.S.; Sales, N.S.; Diniz, M.O.; Aps, L.R.M.M.; Rodrigues, K.B.; Silva, J.R.; Moreno, A.C.R.; Porchia, B.F.M.M.; Sulczewski, F.B.; et al. Antigen Delivery to DEC205(+) Dendritic Cells Induces Immunological Memory and Protective Therapeutic Effects against HPV-Associated Tumors at Different Anatomical Sites. Int. J. Biol. Sci. 2021, 17, 2944–2956. [Google Scholar] [CrossRef]
- Cabeza-Cabrerizo, M.; Cardoso, A.; Minutti, C.M.; Pereira da Costa, M.; Reis e Sousa, C. Dendritic Cells Revisited. Annu. Rev. Immunol. 2021, 39, 131–166. [Google Scholar] [CrossRef]
- Schneider, K.; Marbaix, E.; Bouzin, C.; Hamoir, M.; Mahy, P.; Bol, V.; Gregoire, V. Immune cell infiltration in head and neck squamous cell carcinoma and patient outcome: A retrospective study. Acta Oncol. 2018, 57, 1165–1172. [Google Scholar] [CrossRef] [PubMed]
- Lyford-Pike, S.; Peng, S.; Young, G.D.; Taube, J.M.; Westra, W.H.; Akpeng, B.; Bruno, T.C.; Richmon, J.D.; Wang, H.; Bishop, J.A.; et al. Evidence for a role of the PD-1:PD-L1 pathway in immune resistance of HPV-associated head and neck squamous cell carcinoma. Cancer Res. 2013, 73, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Jin, Y.; Qin, X. Comprehensive analysis of significant genes and immune cell infiltration in HPV-related head and neck squamous cell carcinoma. Int. Immunopharmacol. 2020, 87, 106844. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Concha-Benavente, F.; Shayan, G.; Srivastava, R.M.; Gibson, S.P.; Wang, L.; Gooding, W.E.; Ferris, R.L. STING activation enhances cetuximab-mediated NK cell activation and DC maturation and correlates with HPV(+) status in head and neck cancer. Oral. Oncol. 2018, 78, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Lisco, A.; Hsu, A.P.; Dimitrova, D.; Proctor, D.M.; Mace, E.M.; Ye, P.; Anderson, M.V.; Hicks, S.N.; Grivas, C.; Hammoud, D.A.; et al. Treatment of Relapsing HPV Diseases by Restored Function of Natural Killer Cells. N. Engl. J. Med. 2021, 385, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Blaine-Sauer, S.; Shin, M.K.; Matkowskyj, K.A.; Ward-Shaw, E.; Lambert, P.F. A Novel Model for Papillomavirus-Mediated Anal Disease and Cancer Using the Mouse Papillomavirus. mBio 2021, 12, e0161121. [Google Scholar] [CrossRef] [PubMed]
- Spurgeon, M.E.; Uberoi, A.; McGregor, S.M.; Wei, T.; Ward-Shaw, E.; Lambert, P.F. A Novel In Vivo Infection Model to Study Papillomavirus-Mediated Disease of the Female Reproductive Tract. mBio 2019, 10, e00180-19. [Google Scholar] [CrossRef]
- Uberoi, A.; Yoshida, S.; Lambert, P.F. Development of an in vivo infection model to study Mouse papillomavirus-1 (MmuPV1). J. Virol. Methods 2018, 253, 11–17. [Google Scholar] [CrossRef]
- Fischer, J.; Jung, N.; Robinson, N.; Lehmann, C. Sex differences in immune responses to infectious diseases. Infection 2015, 43, 399–403. [Google Scholar] [CrossRef]
- Hatano, T.; Sano, D.; Takahashi, H.; Oridate, N. Pathogenic Role of Immune Evasion and Integration of Human Papillomavirus in Oropharyngeal Cancer. Microorganisms 2021, 9, 891. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Brendle, S.A.; Li, J.J.; Walter, V.; Cladel, N.M.; Cooper, T.; Shearer, D.A.; Balogh, K.K.; Christensen, N.D. Depo Medroxyprogesterone (DMPA) Promotes Papillomavirus Infections but Does Not Accelerate Disease Progression in the Anogenital Tract of a Mouse Model. Viruses 2022, 14, 980. [Google Scholar] [CrossRef] [PubMed]
- Nardelli-Haefliger, D.; Roden, R.B.S.; Benyacoub, J.; Sahli, R.; Kraehenbuhl, J.P.; Schiller, J.T.; Lachat, P.; Potts, A.; De Grandi, P. Human papillomavirus type 16 virus-like particles expressed in attenuated Salmonella typhimurium elicit mucosal and systemic neutralizing antibodies in mice. Infect. Immun. 1997, 65, 3328–3336. [Google Scholar] [CrossRef] [PubMed]
- Gallichan, W.S.; Rosenthal, K.L. Effects of the estrous cycle on local humoral immune responses and protection of intranasally immunized female mice against herpes simplex virus type 2 infection in the genital tract. Virology 1996, 224, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Fisher, D.G.; Coppock, G.M.; Lopez, C.B. Virus-derived immunostimulatory RNA induces type I IFN-dependent antibodies and T-cell responses during vaccination. Vaccine 2018, 36, 4039–4045. [Google Scholar] [CrossRef] [PubMed]
- French, M.A.; Abudulai, L.N.; Fernandez, S. Isotype Diversification of IgG Antibodies to HIV Gag Proteins as a Therapeutic Vaccination Strategy for HIV Infection. Vaccines 2013, 1, 328–342. [Google Scholar] [CrossRef]
- Madera, R.F.; Libraty, D.H. The role of MyD88 signaling in heterosubtypic influenza A virus infections. Virus Res. 2013, 171, 216–221. [Google Scholar] [CrossRef]
- Liang, H.; Tang, J.; Liu, Z.; Liu, Y.; Huang, Y.; Xu, Y.; Hao, P.; Yin, Z.; Zhong, J.; Ye, L.; et al. ZIKV infection induces robust Th1-like Tfh cell and long-term protective antibody responses in immunocompetent mice. Nat. Commun. 2019, 10, 3859. [Google Scholar] [CrossRef]
- Surman, S.L.; Jones, B.G.; Penkert, R.R.; Sealy, R.E.; Marion, T.; Thomas, P.G.; Neale, G.; Xu, B.S.; Hurwitz, J.L. How Estrogen, Testosterone, and Sex Differences Influence Serum Immunoglobulin Isotype Patterns in Mice and Humans. Viruses 2023, 15, 482. [Google Scholar] [CrossRef]
- Damelang, T.; Rogerson, S.J.; Kent, S.J.; Chung, A.W. Role of IgG3 in Infectious Diseases. Trends Immunol. 2019, 40, 197–211. [Google Scholar] [CrossRef]
- Klaus, T.; Bereta, J. CH2 Domain of Mouse IgG3 Governs Antibody Oligomerization, Increases Functional Affinity to Multivalent Antigens and Enhances Hemagglutination. Front. Immunol. 2018, 9, 1096. [Google Scholar] [CrossRef]
- Armenaka, M.; Grizzanti, J.; Rosenstreich, D.L. Serum immunoglobulins and IgG subclass levels in adults with chronic sinusitis: Evidence for decreased IgG3 levels. Ann. Allergy 1994, 72, 507–514. [Google Scholar] [PubMed]
- Popa, V.; Kim, K.; Heiner, D.C. IgG deficiency in adults with recurrent respiratory infections. Ann. Allergy 1993, 70, 418–424. [Google Scholar] [PubMed]
- Olinder-Nielsen, A.M.; Granert, C.; Forsberg, P.; Friman, V.; Vietorisz, A.; Bjorkander, J. Immunoglobulin prophylaxis in 350 adults with IgG subclass deficiency and recurrent respiratory tract infections: A long-term follow-up. Scand. J. Infect. Dis. 2007, 39, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Mosley, Y.C.; Radder, J.E.; HogenEsch, H. Genetic Variation in the Magnitude and Longevity of the IgG Subclass Response to a Diphtheria-Tetanus-Acellular Pertussis (DTaP) Vaccine in Mice. Vaccines 2019, 7, 124. [Google Scholar] [CrossRef]
- Dahlstrom, K.R.; Anderson, K.S.; Field, M.S.; Chowell, D.; Ning, J.; Li, N.; Wei, Q.; Li, G.; Sturgis, E.M. Diagnostic accuracy of serum antibodies to human papillomavirus type 16 early antigens in the detection of human papillomavirus-related oropharyngeal cancer. Cancer 2017, 123, 4886–4894. [Google Scholar] [CrossRef]
- Beachler, D.C.; Pinto, L.A.; Kemp, T.J.; Nyitray, A.G.; Hildesheim, A.; Viscidi, R.; Schussler, J.; Kreimer, A.R.; Giuliano, A.R. An Examination of HPV16 Natural Immunity in Men Who Have Sex with Men (MSM) in the HPV in Men (HIM) Study. Cancer Epidemiol. Biomark. Prev. 2018, 27, 496–502. [Google Scholar] [CrossRef]
- Goncalves, A.K.; Giraldo, P.C.; Farias, K.J.; Machado, P.R.; Costa, A.P.; de Souza, L.C.; Crispim, J.C.; Eleuterio, J., Jr.; Witkin, S.S. Characterization of Immunoglobulin A/G Responses during 3 Doses of the Human Papillomavirus-16/18 ASO4-Adjuvanted Vaccine. Sex. Transm. Dis. 2016, 43, 335–339. [Google Scholar] [CrossRef]
- Wang, W.; Uberoi, A.; Spurgeon, M.; Gronski, E.; Majerciak, V.; Lobanov, A.; Hayes, M.; Loke, A.; Zheng, Z.M.; Lambert, P.F. Stress keratin 17 enhances papillomavirus infection-induced disease by downregulating T cell recruitment. PLoS Pathog. 2020, 16, e1008206. [Google Scholar] [CrossRef]
- Meyers, J.M.; Uberoi, A.; Grace, M.; Lambert, P.F.; Munger, K. Cutaneous HPV8 and MmuPV1 E6 Proteins Target the NOTCH and TGF-beta Tumor Suppressors to Inhibit Differentiation and Sustain Keratinocyte Proliferation. PLoS Pathog. 2017, 13, e1006171. [Google Scholar] [CrossRef]
- Steele, J.C.; Mann, C.H.; Rookes, S.; Rollason, T.; Murphy, D.; Freeth, M.G.; Gallimore, P.H.; Roberts, S. T-cell responses to human papillomavirus type 16 among women with different grades of cervical neoplasia. Br. J. Cancer 2005, 93, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Uberoi, A.; Yoshida, S.; Frazer, I.H.; Pitot, H.C.; Lambert, P.F. Role of Ultraviolet Radiation in Papillomavirus-Induced Disease. PLoS Pathog. 2016, 12, e1005664. [Google Scholar] [CrossRef] [PubMed]


| Strain | Sex | Viral Positive Mice /Infected Mice | Viral DNA Copies (×105) |
|---|---|---|---|
| B6 | M | 3/6 | 24, 7.2, 7.2, 0, 0, 0 |
| F | 1/6 | 0.84, 0, 0, 0, 0, 0 |
| Strain | Sex | Viral RNA Transcripts (×105) | Positive/Infected Mice (Males + Females) |
|---|---|---|---|
| B6 | M | 4, 4, 0, 0, 0, 0.1 | 7/13 |
| F | 4, 0.04, 0, 0, 0.105, 0, 0 | ||
| Rag1ko | M | 0, 0, 0.6, 6, 0.038, 0.2 | 8/13 |
| F | 0.105, 0.1075, 0, 4, 0.5, 2.25, 0 | ||
| CCR2ko | M | 0, 0, 0.009 | 2/6 |
| F | 0, 0, 0.08 | ||
| Stat1ko | M | 0, 0.003, 0.003 | 4/6 |
| F | 0.15, 0, 0.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brendle, S.A.; Li, J.J.; Walter, V.; Schell, T.D.; Kozak, M.; Balogh, K.K.; Lu, S.; Christensen, N.D.; Zhu, Y.; El-Bayoumy, K.; et al. Immune Responses in Oral Papillomavirus Clearance in the MmuPV1 Mouse Model. Pathogens 2023, 12, 1452. https://doi.org/10.3390/pathogens12121452
Brendle SA, Li JJ, Walter V, Schell TD, Kozak M, Balogh KK, Lu S, Christensen ND, Zhu Y, El-Bayoumy K, et al. Immune Responses in Oral Papillomavirus Clearance in the MmuPV1 Mouse Model. Pathogens. 2023; 12(12):1452. https://doi.org/10.3390/pathogens12121452
Chicago/Turabian StyleBrendle, Sarah A., Jingwei J. Li, Vonn Walter, Todd D. Schell, Michael Kozak, Karla K. Balogh, Song Lu, Neil D. Christensen, Yusheng Zhu, Karam El-Bayoumy, and et al. 2023. "Immune Responses in Oral Papillomavirus Clearance in the MmuPV1 Mouse Model" Pathogens 12, no. 12: 1452. https://doi.org/10.3390/pathogens12121452
APA StyleBrendle, S. A., Li, J. J., Walter, V., Schell, T. D., Kozak, M., Balogh, K. K., Lu, S., Christensen, N. D., Zhu, Y., El-Bayoumy, K., & Hu, J. (2023). Immune Responses in Oral Papillomavirus Clearance in the MmuPV1 Mouse Model. Pathogens, 12(12), 1452. https://doi.org/10.3390/pathogens12121452









