Unravelling Antigenic Cross-Reactions toward the World of Coronaviruses: Extent of the Stability of Shared Epitopes and SARS-CoV-2 Anti-Spike Cross-Neutralizing Antibodies
Abstract
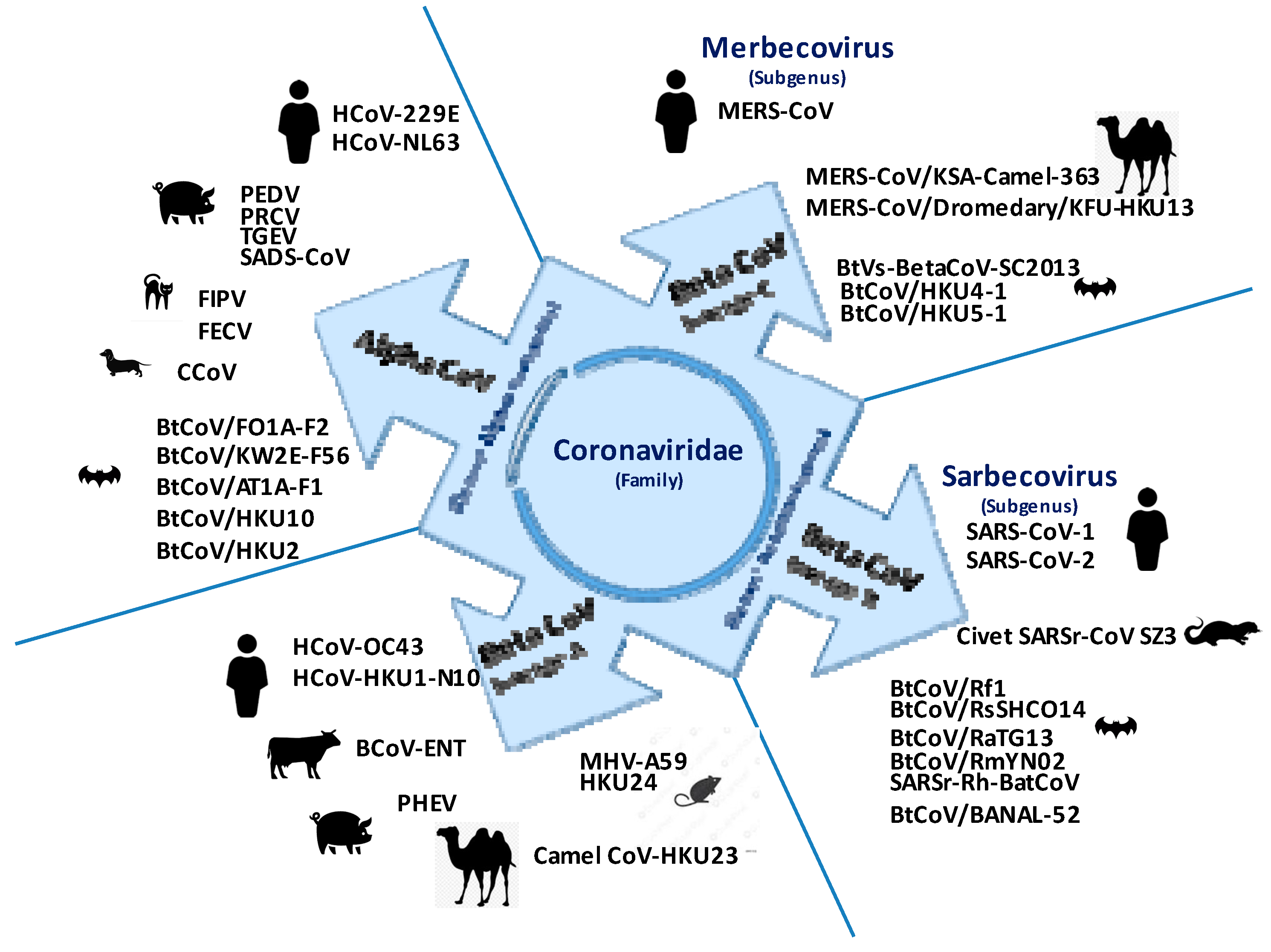
:1. Diversity and Interspecies Circulation of Coronaviruses
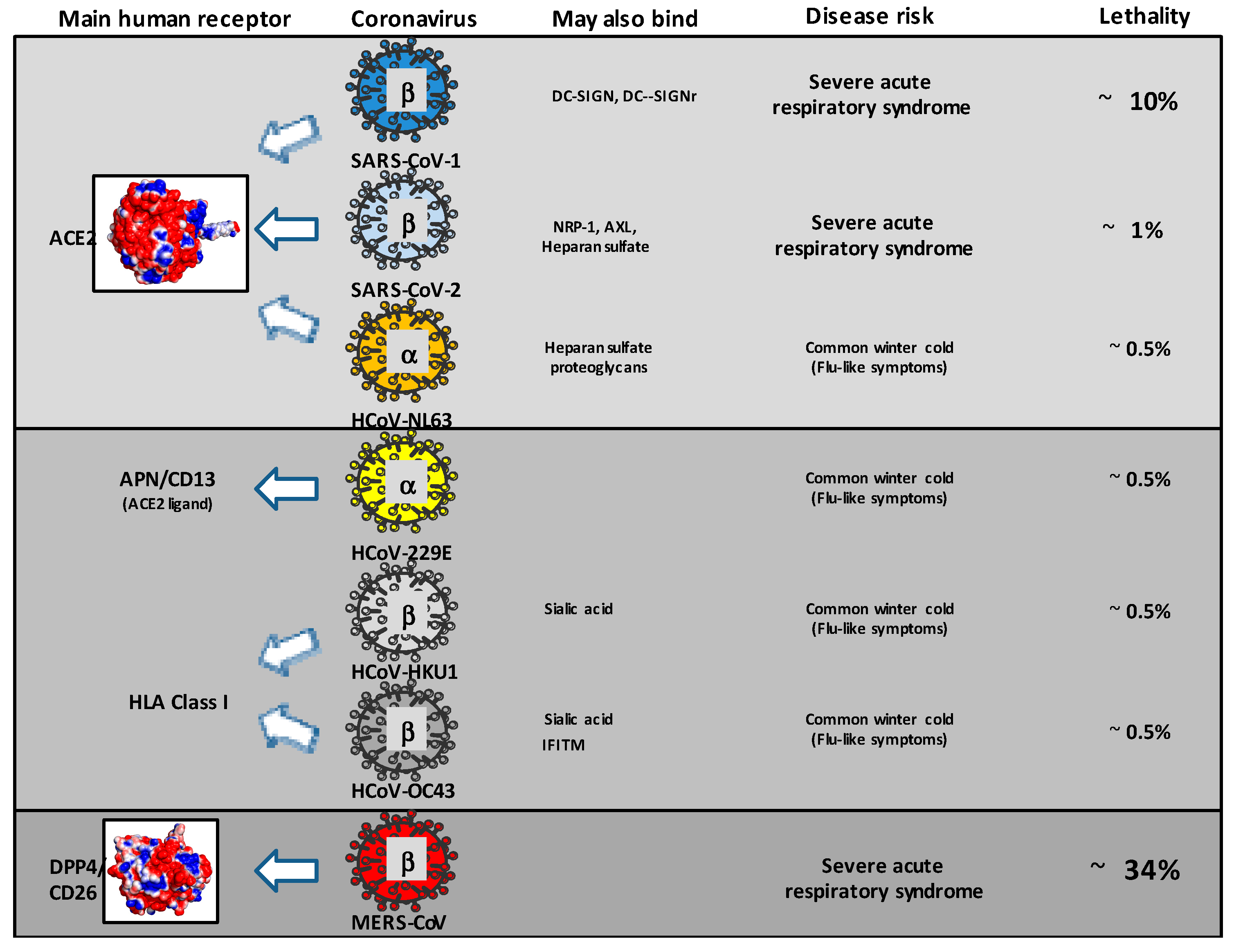
2. Evidence of Cross-Reactivity towards HCoV, MERS-CoV, SARS-CoV-1, and SARS-CoV-2
3. Evidence for Cross-Reactivity towards Animal CoVs, HCoVs, and Human CoVs including SARS-CoV-2
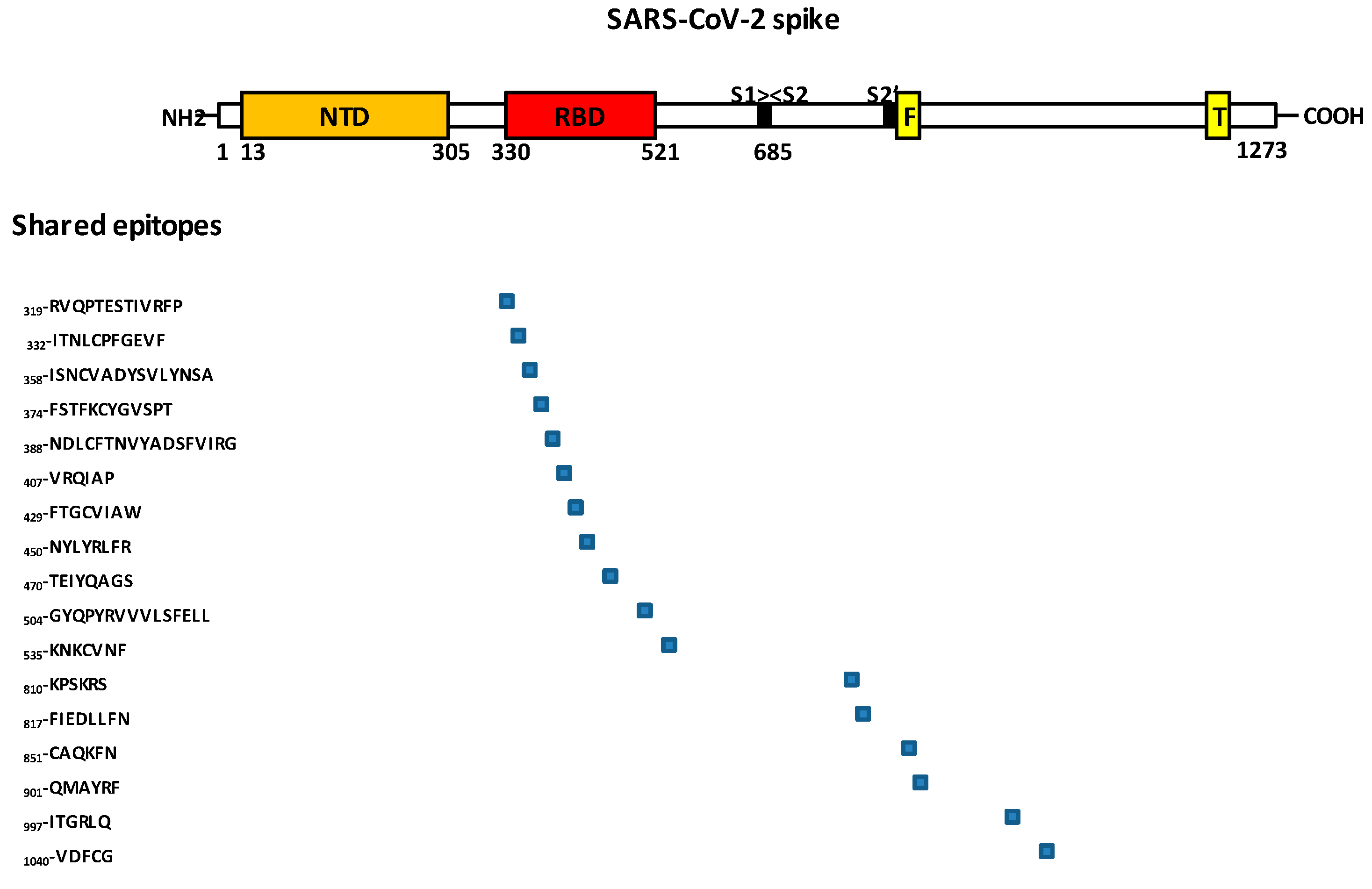
4. Epitopes in SARS-CoV-2 RBD That Binds ACE2 and Are Related to Virus Neutralization
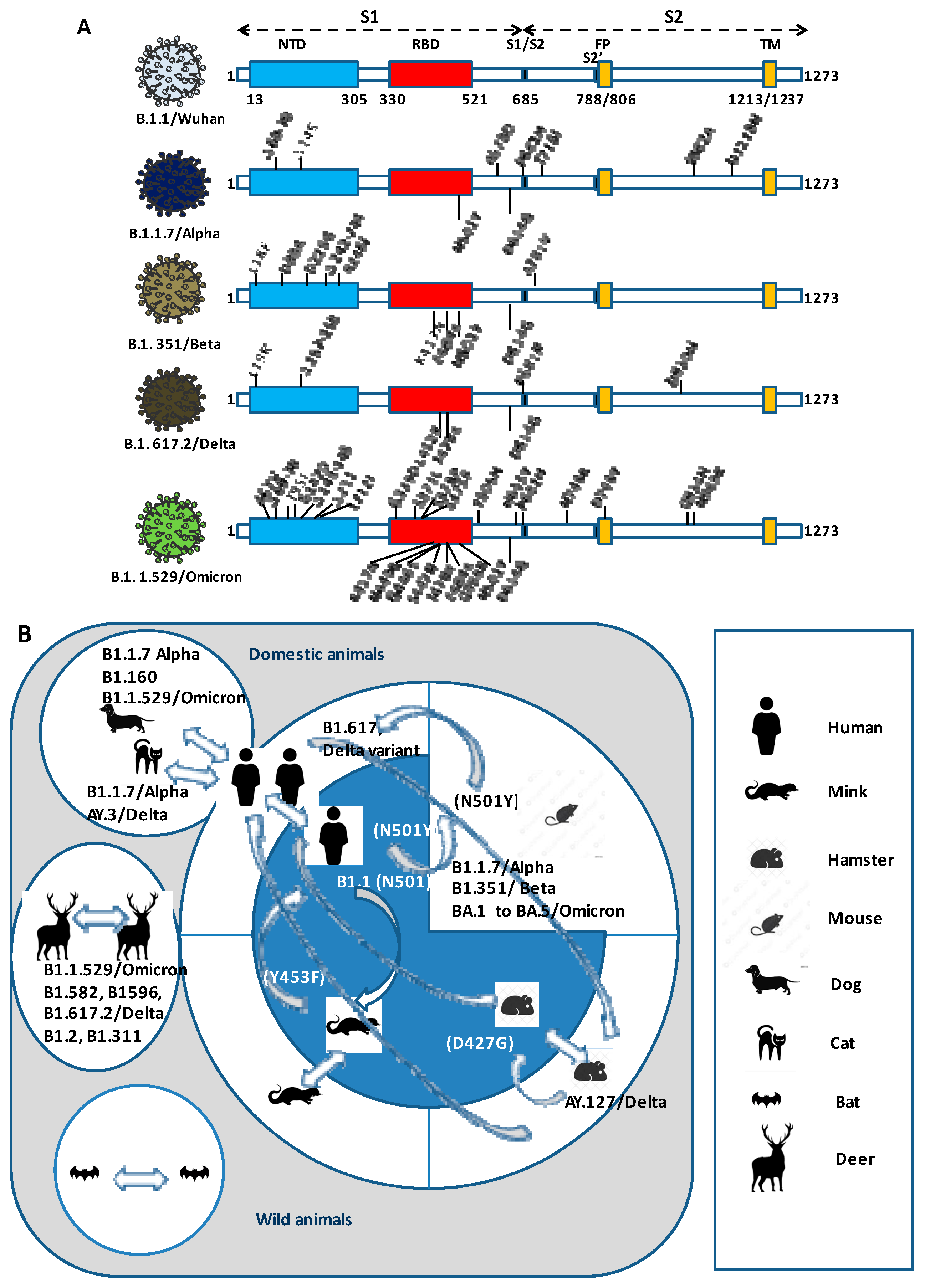
5. Repeated Intra- and Interspecies Transmission of SARS-CoV-2 and the Risk of Reintroducing to Humans Variants Which Are Less Susceptible to Neutralization
6. Prepandemic Cross Immunity to SARS-CoV-2
7. Discussion
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Woo, P.C.Y.; Lau, S.K.P.; Lam, C.S.F.; Lau, C.C.Y.; Tsang, A.K.L.; Lau, J.H.N.; Bai, R.; Teng, J.L.L.; Tsang, C.C.C.; Wang, M.; et al. Discovery of Seven Novel Mammalian and Avian Coronaviruses in the Genus Deltacoronavirus Supports Bat Coronaviruses as the Gene Source of Alphacoronavirus and Betacoronavirus and Avian Coronaviruses as the Gene Source of Gammacoronavirus and Deltacoronavirus. J. Virol. 2012, 86, 3995–4008. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.C.K.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016, 24, 490–502. [Google Scholar] [CrossRef]
- Miranda, C.; Silva, V.; Igrejas, G.; Poeta, P. Genomic evolution of the human and animal coronavirus diseases. Mol. Biol. Rep. 2021, 48, 6645–6653. [Google Scholar] [CrossRef] [PubMed]
- Wertheim, J.O.; Chu, D.K.W.; Peiris, J.S.M.; Pond, S.L.K.; Poon, L.L.M. A Case for the Ancient Origin of Coronaviruses. J. Virol. 2013, 87, 7039–7045. [Google Scholar] [CrossRef]
- Simas, P.V.M.; Barnabé, A.C.D.S.; Durães-Carvalho, R.; Neto, D.F.D.L.; Caserta, L.C.; Artacho, L.; Jacomassa, F.A.F.; Martini, M.C.; Dos Santos, M.M.A.B.; Felippe, P.A.N.; et al. Bat Coronavirus in Brazil Related to Appalachian Ridge and Porcine Epidemic Diarrhea Viruses. Emerg. Infect. Dis. 2015, 21, 729–731. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Fan, H.; Lan, T.; Yáng, X.-L.; Shi, W.-F.; Zhang, W.; Zhu, Y.; Zhang, Y.-W.; Xie, Q.-M.; Mani, S.; et al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 2018, 556, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Garbino, J.; Crespo, S.; Aubert, J.; Rochat, T.; Ninet, B.; Deffernez, C.; Wunderli, W.; Pache, J.; Soccal, P.M.; Kaiser, L. A Prospective Hospital-Based Study of the Clinical Impact of Non–Severe Acute Respiratory Syndrome (Non-SARS)–Related Human Coronavirus Infection. Clin. Infect. Dis. 2006, 43, 1009–1015. [Google Scholar] [CrossRef]
- Gaunt, E.R.; Hardie, A.; Claas, E.C.J.; Simmonds, P.; Templeton, K.E. Epidemiology and Clinical Presentations of the Four Human Coronaviruses 229E, HKU1, NL63, and OC43 Detected over 3 Years Using a Novel Multiplex Real-Time PCR Method. J. Clin. Microbiol. 2010, 48, 2940–2947. [Google Scholar] [CrossRef]
- Zaki, A.M.; Van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef]
- Marra, M.A.; Jones, S.J.M.; Astell, C.R.; Holt, R.A.; Brooks-Wilson, A.; Butterfield, Y.S.N.; Khattra, J.; Asano, J.K.; Barber, S.A.; Chan, S.Y.; et al. The Genome Sequence of the SARS-Associated Coronavirus. Science 2003, 300, 1399–1404. [Google Scholar] [CrossRef]
- Xu, R.H.; He, J.F.; Evans, M.R.; Peng, G.W.; Field, H.E.; Yu, D.W.; Lee, C.K.; Luo, H.M.; Lin, W.S.; Lin, P.; et al. Epidemiologic Clues to SARS Origin in China. Emerg. Infect. Dis. 2004, 10, 1030–1037. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Forni, D.; Cagliani, R.; Clerici, M.; Sironi, M. Molecular Evolution of Human Coronavirus Genomes. Trends Microbiol. 2017, 25, 35–48. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef]
- Graham, R.L.; Donaldson, E.F.; Baric, R.S. A decade after SARS: Strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013, 11, 836–848. [Google Scholar] [CrossRef]
- Lan, J.; Chen, P.; Liu, W.; Ren, W.; Zhang, L.; Ding, Q.; Zhang, Q.; Wang, X.; Ge, J. Structural insights into the binding of SARS-CoV-2, SARS-CoV, and hCoV-NL63 spike receptor-binding domain to horse ACE2. Structure 2022, 30, 1432–1442.e4. [Google Scholar] [CrossRef]
- Bakkers, M.J.; Lang, Y.; Feitsma, L.J.; Hulswit, R.J.; de Poot, S.A.; van Vliet, A.L.; Margine, I.; de Groot-Mijnes, J.D.; van Kuppeveld, F.J.; Langereis, M.A.; et al. Betacoronavirus Adaptation to Humans Involved Progressive Loss of Hemagglutinin-Esterase Lectin Activity. Cell Host Microbe 2017, 21, 356–366. [Google Scholar] [CrossRef]
- Pyrc, K.; Dijkman, R.; Deng, L.; Jebbink, M.F.; Ross, H.A.; Berkhout, B.; van der Hoek, L. Mosaic Structure of Human Coronavirus NL63, One Thousand Years of Evolution. J. Mol. Biol. 2006, 364, 964–973. [Google Scholar] [CrossRef]
- Tao, Y.; Shi, M.; Chommanard, C.; Queen, K.; Zhang, J.; Markotter, W.; Kuzmin, I.V.; Holmes, E.C.; Tong, S. Surveillance of Bat Coronaviruses in Kenya Identifies Relatives of Human Coronaviruses NL63 and 229E and Their Recombination History. J. Virol. 2017, 91, e01953-16. [Google Scholar] [CrossRef]
- Kiyuka, P.K.; Agoti, C.N.; Munywoki, P.K.; Njeru, R.; Bett, A.; Otieno, J.R.; Otieno, G.P.; Kamau, E.; Clark, T.G.; Van Der Hoek, L.; et al. Human Coronavirus NL63 Molecular Epidemiology and Evolutionary Patterns in Rural Coastal Kenya. J. Infect. Dis. 2018, 217, 1728–1739. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.-F.; Tuo, J.-L.; Huang, X.-B.; Zhu, X.; Zhang, D.-M.; Zhou, K.; Yuan, L.; Luo, H.-J.; Zheng, B.-J.; Yuen, K.-Y.; et al. Epidemiology characteristics of human coronaviruses in patients with respiratory infection symptoms and phylogenetic analysis of HCoV-OC43 during 2010–2015 in Guangzhou. PLoS ONE 2018, 13, e0191789. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.-I.; Kim, I.B.; Park, S.J.; Ha, E.-H.; Lee, C.W. Comparison of the clinical characteristics and mortality of adults infected with human coronaviruses 229E and OC43. Sci. Rep. 2021, 11, 4499. [Google Scholar] [CrossRef] [PubMed]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.; Müller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.; Zaki, A.; Fouchier, R.A.; et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef]
- Wang, N.; Shi, X.; Jiang, L.; Zhang, S.; Wang, D.; Tong, P.; Guo, D.; Fu, L.; Cui, Y.; Liu, X.; et al. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013, 23, 986–993. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Frutos, R.; Roig, M.L.; Serra-Cobo, J.; Devaux, C.A. COVID-19: The Conjunction of Events Leading to the Coronavirus Pandemic and Lessons to Learn for Future Threats. Front. Med. 2020, 7, 223. [Google Scholar] [CrossRef]
- Dearlove, B.; Lewitus, E.; Bai, H.; Li, Y.; Reeves, D.B.; Joyce, M.G.; Scott, P.T.; Amare, M.F.; Vasan, S.; Michael, N.L.; et al. A SARS-CoV-2 vaccine candidate would likely match all currently circulating variants. Proc. Natl. Acad. Sci. USA 2020, 117, 23652–23662. [Google Scholar] [CrossRef]
- Rausch, J.W.; Capoferri, A.A.; Katusiime, M.G.; Patro, S.C.; Kearney, M.F. Low genetic diversity may be an Achilles heel of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 24614–24616. [Google Scholar] [CrossRef] [PubMed]
- Pachetti, M.; Marini, B.; Benedetti, F.; Giudici, F.; Mauro, E.; Storici, P.; Masciovecchio, C.; Angeletti, S.; Ciccozzi, M.; Gallo, R.C.; et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020, 18, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Takada, K.; Ueda, M.T.; Shichinohe, S.; Kida, Y.; Ono, C.; Matsuura, Y.; Watanabe, T.; Nakagawa, S. Genomic diversity of SARS-CoV-2 can be accelerated by mutations in the nsp14 gene. iScience 2023, 26, 106210. [Google Scholar] [CrossRef]
- Yin, X.; Popa, H.; Stapon, A.; Bouda, E.; Garcia-Diaz, M. Fidelity of Ribonucleotide Incorporation by the SARS-CoV-2 Replication Complex. J. Mol. Biol. 2023, 435, 167973. [Google Scholar] [CrossRef] [PubMed]
- Oude Munnink, B.B.; Sikkema, R.S.; Nieuwenhuijse, D.F.; Molenaar, R.J.; Munger, E.; Molenkamp, R.; van der Spek, A.; Tolsma, P.; Rietveld, A.; Brouwer, M.; et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 2021, 371, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Li, P.; Ji, Y.; Ikram, A.; Pan, Q. Cross-reactivity towards SARS-CoV-2: The potential role of low-pathogenic human coronaviruses. Lancet Microbe 2020, 1, e151. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Ma, Z.; Peppelenbosch, M.P.; Pan, Q. Potential association between COVID-19 mortality and health-care resource availability. Lancet Glob. Health 2020, 8, e480. [Google Scholar] [CrossRef]
- Lewitus, E.; Bai, H.; Rolland, M. Design of a pan-betacoronavirus vaccine candidate through a phylogenetically informed approach. Sci. Adv. 2023, 9, eabq4149. [Google Scholar] [CrossRef]
- Sun, Z.F.; Meng, X.J. Antigenic Cross-Reactivity between the Nucleocapsid Protein of Severe Acute Respiratory Syndrome (SARS) Coronavirus and Polyclonal Antisera of Antigenic Group I Animal Coronaviruses: Implication for SARS Diagnosis. J. Clin. Microbiol. 2004, 42, 2351–2352. [Google Scholar] [CrossRef]
- Rockstroh, A.; Wolf, J.; Fertey, J.; Kalbitz, S.; Schroth, S.; Lübbert, C.; Ulbert, S.; Borte, S. Correlation of humoral immune responses to different SARS-CoV-2 antigens with virus neutralizing antibodies and symptomatic severity in a German COVID-19 cohort. Emerg. Microbes Infect. 2021, 10, 774–781. [Google Scholar] [CrossRef]
- Guo, L.; Ren, L.; Yang, S.; Xiao, M.; Chang, D.; Yang, F.; Cruz, C.S.D.; Wang, Y.; Wu, C.; Xiao, Y.; et al. Profiling Early Humoral Response to Diagnose Novel Coronavirus Disease (COVID-19). Clin. Infect. Dis. 2020, 71, 778–785. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.K.; Khan, M.S.; Choudhury, S.; Mukhopadhyay, A.; Rastogi, A.; Thakur, A.; Kumari, P.; Kaur, M.; Saini, C.; Sapehia, V.; et al. CoronaVR: A Computational Resource and Analysis of Epitopes and Therapeutics for Severe Acute Respiratory Syndrome Coronavirus-2. Front. Microbiol. 2020, 11, 1858. [Google Scholar] [CrossRef] [PubMed]
- Cantoni, D.; Siracusano, G.; Mayora-Neto, M.; Pastori, C.; Fantoni, T.; Lytras, S. Analysis of antibody neutralisation activity against SARS-CoV-2 variants and seasonal Human Coronaviruses NL63, HKU1, and 229E induced by three different COVID-19 vaccine platforms. Vaccines 2023, 11, 58. [Google Scholar] [CrossRef] [PubMed]
- Ksiazek, T.G.; Erdman, D.; Goldsmith, C.S.; Zaki, S.R.; Peret, T.; Emery, S.; Tong, S.; Urbani, C.; Comer, J.A.; Lim, W.; et al. A Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003, 348, 1953–1966. [Google Scholar] [CrossRef] [PubMed]
- Che, X.Y.; Qiu, L.W.; Liao, Z.Y.; Wang, Y.D.; Wen, K.; Pan, Y.X.; Hao, W.; Mei, Y.B.; Cheng, V.C.; Yuen, K.Y. Antigenic Cross-Reactivity between Severe Acute Respiratory Syndrome–Associated Coronavirus and Human Coronaviruses 229E and OC43. J. Infect. Dis. 2005, 191, 2033–2037. [Google Scholar] [CrossRef]
- Mu, F.; Niu, D.; Mu, J.; He, B.; Han, W.; Fan, B.; Huang, S.; Qiu, Y.; You, B.; Chen, W. The expression and antigenicity of a truncated spike-nucleocapsid fusion protein of severe acute respiratory syndrome-associated coronavirus. BMC Microbiol. 2008, 8, 207. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Wang, Z.; Ren, L.; Hao, Y.; Zhu, M.; Jiang, H.; Wang, S.; Li, D.; Shao, Y. Pre-existing anti-HCoV-OC43 immunity influences the durability and cross-reactivity of humoral response to SARS-CoV-2 vaccination. Front. Cell. Infect. Microbiol. 2022, 12, 978440. [Google Scholar] [CrossRef]
- Ahmed, S.F.; Quadeer, A.A.; McKay, M.R. Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies. Viruses 2020, 12, 254. [Google Scholar] [CrossRef]
- Shrock, E.; Fujimura, E.; Kula, T.; Timms, R.T.; Lee, I.-H.; Leng, Y.; Robinson, M.L.; Sie, B.M.; Li, M.Z.; Chen, Y.; et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science 2020, 370, eabd4250. [Google Scholar] [CrossRef]
- Ng, K.W.; Faulkner, N.; Cornish, G.H.; Rosa, A.; Harvey, R.; Hussain, S.; Ulferts, R.; Earl, C.; Wrobel, A.G.; Benton, D.J.; et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science 2020, 370, 1339–1343. [Google Scholar] [CrossRef]
- Rak, A.; Donina, S.; Zabrodskaya, Y.; Rudenko, L.; Isakova-Sivak, I. Cross-Reactivity of SARS-CoV-2 Nucleocapsid-Binding Antibodies and Its Implication for COVID-19 Serology Tests. Viruses 2022, 14, 2041. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.-W.; Cheng, V.C.-C.; Cai, J.-P.; Chan, K.-H.; Chen, L.-L.; Wong, L.-H.; Choi, C.Y.-K.; Fong, C.H.-Y.; Ng, A.C.-K.; Lu, L.; et al. Seroprevalence of SARS-CoV-2 in Hong Kong and in residents evacuated from Hubei province, China: A multicohort study. Lancet Microbe 2020, 1, e111–e118. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.A.; Alahmari, A.A.; Almuzaini, Y.; Alamri, F.; Alsofayan, Y.M.; Aburas, A.; Al-Muhsen, S.; Van Kerkhove, M.; Yezli, S.; Ciottone, G.R.; et al. Potential Cross-Reactive Immunity to COVID-19 Infection in Individuals With Laboratory-Confirmed MERS-CoV Infection: A National Retrospective Cohort Study From Saudi Arabia. Front. Immunol. 2021, 12, 727989. [Google Scholar] [CrossRef] [PubMed]
- AlKhalifah, J.M.; Seddiq, W.; Alshehri, M.A.; Alhetheel, A.; Albarrag, A.; Meo, S.A.; Al-Tawfiq, J.A.; Barry, M. Impact of MERS-CoV and SARS-CoV-2 Viral Infection on Immunoglobulin-IgG Cross-Reactivity. Vaccines 2023, 11, 552. [Google Scholar] [CrossRef]
- Al Maani, A.; Al-Jardani, A.; Karrar, H.; Petersen, E.; Al Abri, S. COVID-19 in a case previously infected with MERS-CoV: No cross immunity. J. Infect. 2020, 82, e28–e29. [Google Scholar] [CrossRef]
- Hemida, M.G.; Perera, R.A.; Al Jassim, R.A.; Kayali, G.; Siu, L.Y.; Wang, P.; Chu, K.W.; Perlman, S.; Ali, M.A.; Alnaeem, A.; et al. Seroepidemiology of Middle East respiratory syndrome (MERS) coronavirus in Saudi Arabia (1993) and Australia (2014) and characterisation of assay specificity. Eurosurveillance 2014, 19, 20828. [Google Scholar] [CrossRef]
- Klompus, S.; Leviatan, S.; Vogl, T.; Mazor, R.D.; Kalka, I.N.; Stoler-Barak, L.; Nathan, N.; Peres, A.; Moss, L.; Godneva, A.; et al. Cross-reactive antibodies against human coronaviruses and the animal coronavirome suggest diagnostics for future zoonotic spillovers. Sci. Immunol. 2021, 6, eabe9950. [Google Scholar] [CrossRef]
- Song, G.; He, W.-T.; Callaghan, S.; Anzanello, F.; Huang, D.; Ricketts, J.; Torres, J.L.; Beutler, N.; Peng, L.; Vargas, S.; et al. Cross-reactive serum and memory B-cell responses to spike protein in SARS-CoV-2 and endemic coronavirus infection. Nat. Commun. 2021, 12, 2938. [Google Scholar] [CrossRef]
- Jaago, M.; Rähni, A.; Pupina, N.; Pihlak, A.; Sadam, H.; Tuvikene, J.; Avarlaid, A.; Planken, A.; Planken, M.; Haring, L.; et al. Differential patterns of cross-reactive antibody response against SARS-CoV-2 spike protein detected for chronically ill and healthy COVID-19 naïve individuals. Sci. Rep. 2022, 12, 168–179. [Google Scholar] [CrossRef]
- Amanat, F.; Thapa, M.; Lei, T.; Ahmed, S.M.S.; Adelsberg, D.C.; Carreño, J.M.; Strohmeier, S.; Schmitz, A.J.; Zafar, S.; Zhou, J.Q.; et al. SARS-CoV-2 mRNA vaccination induces functionally diverse antibodies to NTD, RBD, and S2. Cell 2021, 184, 3936–3948.e10. [Google Scholar] [CrossRef]
- Angyal, A.; Longet, S.; Moore, S.C.; Payne, R.P.; Harding, A.; Tipton, T.; Rongkard, P.; Ali, M.; Hering, L.M.; Meardon, N.; et al. T-cell and antibody responses to first BNT162b2 vaccine dose in previously infected and SARS-CoV-2-naive UK health-care workers: A multicentre prospective cohort study. Lancet Microbe 2021, 3, e21–e31. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.M.; Goodwin, E.C.; Verma, A.; Arevalo, C.P.; Bolton, M.J.; Weirick, M.E.; Gouma, S.; McAllister, C.M.; Christensen, S.R.; Weaver, J.; et al. Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell 2021, 184, 1858–1864.e10. [Google Scholar] [CrossRef] [PubMed]
- Gombar, S.; Bergquist, T.; Pejaver, V.; Hammarlund, N.E.; Murugesan, K.; Mooney, S.; Shah, N.; Pinsky, B.A.; Banaei, N. SARS-CoV-2 infection and COVID-19 severity in individuals with prior seasonal coronavirus infection. Diagn. Microbiol. Infect. Dis. 2021, 100, 115338. [Google Scholar] [CrossRef]
- Silva, C.; Mullis, L.B.; Pereira, O.; Saif, L.J.; Vlasova, A.; Zhang, X.; Owens, R.J.; Paulson, D.; Taylor, D.; Haynes, L.M.; et al. Human Respiratory Coronaviruses Detected in Patients with Influenza-Like Illness in Arkansas, USA. Virol. Mycol. 2014, 2014 (Suppl. 2), 4. [Google Scholar] [CrossRef]
- Lednicky, J.A.; Tagliamonte, M.S.; White, S.K.; Blohm, G.M.; Alam, M.; Iovine, N.M.; Salemi, M.; Mavian, C.; Morris, J.G. Isolation of a Novel Recombinant Canine Coronavirus from a Visitor to Haiti: Further Evidence of Transmission of Coronaviruses of Zoonotic Origin to Humans. Clin. Infect. Dis. 2022, 75, e1184–e1187. [Google Scholar] [CrossRef] [PubMed]
- Vlasova, A.N.; Diaz, A.; Damtie, D.; Xiu, L.; Toh, T.-H.; Lee, J.S.-Y.; Saif, L.J.; Gray, G.C. Novel Canine Coronavirus Isolated from a Hospitalized Patient with Pneumonia in East Malaysia. Clin. Infect. Dis. 2022, 74, 446–454. [Google Scholar] [CrossRef]
- Nguyen, T.T.K.; Ngo, T.T.; Tran, P.M.; Pham, T.T.T.; Vu, H.T.T.; Nguyen, N.T.H.; Thwaites, G.; Virtala, A.K.; Vapalahti, O.; Baker, S.; et al. Respiratory viruses in individuals with a high frequency of animal exposure in southern and highland Vietnam. J. Med. Virol. 2020, 92, 971–981. [Google Scholar] [CrossRef]
- Kong, F.; Wang, Q.; Kenney, S.P.; Jung, K.; Vlasova, A.N.; Saif, L.J. Porcine Deltacoronaviruses: Origin, Evolution, Cross-Species Transmission and Zoonotic Potential. Pathogens 2022, 11, 79. [Google Scholar] [CrossRef] [PubMed]
- Zhai, S.-L.; Sun, M.-F.; Zhang, J.-F.; Zheng, C.; Liao, M. Spillover infection of common animal coronaviruses to humans. Lancet Microbe 2022, 3, e808. [Google Scholar] [CrossRef]
- Fouchier, R.A.M.; Hartwig, N.G.; Bestebroer, T.M.; Niemeyer, B.; de Jong, J.C.; Simon, J.H.; Osterhaus, A.D.M.E. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. USA 2004, 101, 6212–6216. [Google Scholar] [CrossRef]
- Dee, S.; Neill, C.; Singrey, A.; Clement, T.; Cochrane, R.; Jones, C.; Patterson, G.; Spronk, G.; Christopher-Hennings, J.; Nelson, E. Modeling the transboundary risk of feed ingredients contaminated with porcine epidemic diarrhea virus. BMC Veter-Res. 2016, 12, 51. [Google Scholar] [CrossRef] [PubMed]
- Edwards, C.E.; Yount, B.L.; Graham, R.L.; Leist, S.R.; Hou, Y.J.; Dinnon, K.H.; Sims, A.C.; Swanstrom, J.; Gully, K.; Scobey, T.D.; et al. Swine acute diarrhea syndrome coronavirus replication in primary human cells reveals potential susceptibility to infection. Proc. Natl. Acad. Sci. USA 2020, 117, 26915–26925. [Google Scholar] [CrossRef] [PubMed]
- Mihindukulasuriya, K.; Wu, G.; Leger, J.S.; Nordhausen, R.W.; Wang, D. Identification of a Novel Coronavirus from a Beluga Whale by Using a Panviral Microarray. J. Virol. 2008, 82, 5084–5088. [Google Scholar] [CrossRef] [PubMed]
- Schütze, H. Coronaviruses in aquatic organisms. In Aquaculture Virology; Kibenge, F.S.B., Godoy, M.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 323–332. [Google Scholar]
- Rathod, N.B.; Elabed, N.; Özogul, F.; Regenstein, J.M.; Galanakis, C.M.; Aljaloud, S.O.; Ibrahim, S.A. The Impact of COVID-19 Pandemic on Seafood Safety and Human Health. Front. Microbiol. 2022, 13, 875164. [Google Scholar] [CrossRef]
- Pedersen, N.C.; Ward, J.; Mengeling, W.L. Antigenic relationship of the feline infectious peritonitis virus to coronaviruses of other species. Arch. Virol. 1978, 58, 45–53. [Google Scholar] [CrossRef]
- Horzinek, M.C.; Lutz, H.; Pedersen, N.C. Antigenic relationships among homologous structural polypeptides of porcine, feline, and canine coronaviruses. Infect. Immun. 1982, 37, 1148–1155. [Google Scholar] [CrossRef]
- Hogue, B.G.; King, B.; Brian, D.A. Antigenic relationships among proteins of bovine coronavirus, human respiratory coronavirus OC43, and mouse hepatitis coronavirus A59. J. Virol. 1984, 51, 384–388. [Google Scholar] [CrossRef]
- Vlasova, A.N.; Zhang, X.; Hasoksuz, M.; Nagesha, H.S.; Haynes, L.M.; Fang, Y.; Lu, S.; Saif, L.J. Two-Way Antigenic Cross-Reactivity between Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and Group 1 Animal CoVs Is Mediated through an Antigenic Site in the N-Terminal Region of the SARS-CoV Nucleoprotein. J. Virol. 2007, 81, 13365–13377. [Google Scholar] [CrossRef]
- Agnihothram, S.; Gopal, R.; Yount, B.L., Jr.; Donaldson, E.F.; Menachery, V.D.; Graham, R.L.; Scobey, T.D.; Gralinski, L.E.; Denison, M.R.; Zambon, M.; et al. Evaluation of Serologic and Antigenic Relationships Between Middle Eastern Respiratory Syndrome Coronavirus and Other Coronaviruses to Develop Vaccine Platforms for the Rapid Response to Emerging Coronaviruses. J. Infect. Dis. 2014, 209, 995–1006. [Google Scholar] [CrossRef]
- Han, Y.; Du, J.; Su, H.; Zhang, J.; Zhu, G.; Zhang, S.; Wu, Z.; Jin, Q. Identification of Diverse Bat Alphacoronaviruses and Betacoronaviruses in China Provides New Insights Into the Evolution and Origin of Coronavirus-Related Diseases. Front. Microbiol. 2019, 10, 1900. [Google Scholar] [CrossRef]
- Lavi, E.; Haluskey, J.A.; Masters, P.S. The pathogenesis of MHV nucleocapsid gene chimeric viruses. Adv. Exp. Med. Biol. 1998, 440, 537–541. [Google Scholar]
- Herrewegh, A.A.P.M.; Smeenk, I.; Horzinek, M.C.; Rottier, P.J.M.; de Groot, R.J. Feline Coronavirus Type II Strains 79-1683 and 79-1146 Originate from a Double Recombination between Feline Coronavirus Type I and Canine Coronavirus. J. Virol. 1998, 72, 4508–4514. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.P.; Li, K.S.M.; Huang, Y.; Shek, C.-T.; Tse, H.; Wang, M.; Choi, G.K.Y.; Xu, H.; Lam, C.S.F.; Guo, R.; et al. Ecoepidemiology and Complete Genome Comparison of Different Strains of Severe Acute Respiratory Syndrome-Related Rhinolophus Bat Coronavirus in China Reveal Bats as a Reservoir for Acute, Self-Limiting Infection That Allows Recombination Events. J. Virol. 2010, 84, 2808–2819. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Yip, C.C.Y.; Huang, Y.; Tsoi, H.-W.; Chan, K.-H.; Yuen, K.-Y. Comparative Analysis of 22 Coronavirus HKU1 Genomes Reveals a Novel Genotype and Evidence of Natural Recombination in Coronavirus HKU1. J. Virol. 2006, 80, 7136–7145. [Google Scholar] [CrossRef]
- Kozlakidis, Z. Evidence for Recombination as an Evolutionary Mechanism in Coronaviruses: Is SARS-CoV-2 an Exception? Front. Public Health 2022, 10, 859900. [Google Scholar] [CrossRef]
- Colson, P.; Fournier, P.E.; Delerce, J.; Million, M.; Bedotto, M.; Houhamdi, L.; Yahi, N.; Bayette, J.; Levasseur, A.; Fantini, J.; et al. Culture and identification of a “Deltamicron” SARS-CoV-2 in a three cases cluster in southern France. J. Med. Virol. 2022, 94, 3739–3749. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.-Y.; Li, J.-L.; Yang, X.-L.; Chmura, A.A.; Zhu, G.; Epstein, J.H.; Mazet, J.K.; Hu, B.; Zhang, W.; Peng, C.; et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 2013, 503, 535–538. [Google Scholar] [CrossRef]
- Ben Hu, B.; Ge, X.; Wang, L.-F.; Shi, Z. Bat origin of human coronaviruses. Virol. J. 2015, 12, 221. [Google Scholar] [CrossRef]
- Afelt, A.; Frutos, R.; Devaux, C. Bats, Coronaviruses, and Deforestation: Toward the Emergence of Novel Infectious Diseases? Front. Microbiol. 2018, 9, 702. [Google Scholar] [CrossRef]
- Frutos, R.; Serra-Cobo, J.; Pinault, L.; Roig, M.L.; Devaux, C.A. Emergence of Bat-Related Betacoronaviruses: Hazard and Risks. Front. Microbiol. 2021, 12, 591535. [Google Scholar] [CrossRef]
- Frutos, R.; Serra-Cobo, J.; Chen, T.; Devaux, C.A. COVID-19: Time to exonerate the pangolin from the transmission of SARS-CoV-2 to humans. Infect. Genet. Evol. 2020, 84, 104493. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Chen, X.; Hu, T.; Li, J.; Song, H.; Liu, Y.; Wang, P.; Liu, D.; Yang, J.; Holmes, E.C.; et al. A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein. Curr. Biol. 2020, 30, 2196–2203.e3. [Google Scholar] [CrossRef] [PubMed]
- Wacharapluesadee, S.; Tan, C.W.; Maneeorn, P.; Duengkae, P.; Zhu, F.; Joyjinda, Y.; Kaewpom, T.; Ni Chia, W.; Ampoot, W.; Lim, B.L.; et al. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat. Commun. 2021, 12, 972. [Google Scholar] [CrossRef] [PubMed]
- Delaune, D.; Hul, V.; Karlsson, E.A.; Hassanin, A.; Ou, T.P.; Baidaliuk, A.; Gámbaro, F.; Prot, M.; Tu, V.T.; Chea, S.; et al. A novel SARS-CoV-2 related coronavirus in bats from Cambodia. Nat. Commun. 2021, 12, 6563. [Google Scholar] [CrossRef]
- Temmam, S.; Vongphayloth, K.; Baquero, E.; Munier, S.; Bonomi, M.; Regnault, B.; Douangboubpha, B.; Karami, Y.; Chrétien, D.; Sanamxay, D.; et al. Bat coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature 2022, 604, 330–336. [Google Scholar] [CrossRef]
- Zhu, Z.; Chakraborti, S.; He, Y.; Roberts, A.; Sheahan, T.; Xiao, X.; Hensley, L.E.; Prabakaran, P.; Rockx, B.; Sidorov, I.A.; et al. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc. Natl. Acad. Sci. USA 2007, 104, 12123–12128. [Google Scholar] [CrossRef]
- Tian, X.; Li, C.; Huang, A.; Xia, S.; Lu, S.; Shi, Z.; Lu, L.; Jiang, S.; Yang, Z.; Wu, Y.; et al. Potent Binding of 2019 Novel Coronavirus Spike Protein by a SARS Coronavirus-Specific Human Monoclonal Antibody. Emerg. Microbes Infect. 2020, 9, 382–385. [Google Scholar] [CrossRef]
- Jiang, S.; Hillyer, C.; Du, L. Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends Immunol. 2020, 41, 355–359. [Google Scholar] [CrossRef]
- Yarmarkovich, M.; Warrington, J.M.; Farrel, A.; Maris, J.M. Identification of SARS-CoV-2 Vaccine Epitopes Predicted to Induce Long-Term Population-Scale Immunity. Cell Rep. Med. 2020, 1, 100036. [Google Scholar] [CrossRef]
- Li, W.; Zhang, C.; Sui, J.; Kuhn, J.H.; Moore, M.J.; Luo, S.; Wong, S.-K.; Huang, I.-C.; Xu, K.; Vasilieva, N.; et al. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005, 24, 1634–1643. [Google Scholar] [CrossRef]
- Glowacka, I.; Bertram, S.; Herzog, P.; Pfefferle, S.; Steffen, I.; Muench, M.O.; Simmons, G.; Hofmann, H.; Kuri, T.; Weber, F.; et al. Differential Downregulation of ACE2 by the Spike Proteins of Severe Acute Respiratory Syndrome Coronavirus and Human Coronavirus NL63. J. Virol. 2010, 84, 1198–1205. [Google Scholar] [CrossRef] [PubMed]
- Kucharski, A.J.; Russell, T.W.; Diamond, C.; Liu, Y.; Edmunds, J.; Funk, S.; Eggo, R.M.; Sun, F.; Jit, M.; Munday, J.D.; et al. Early dynamics of transmission and control of COVID-19: A mathematical modelling study. Lancet Infect. Dis. 2020, 20, 553–558. [Google Scholar] [CrossRef]
- Daszak, P.; Cunningham, A.A.; Hyatt, A.D. Emerging Infectious Diseases of Wildlife—Threats to Biodiversity and Human Health. Science 2000, 287, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Power, A.G.; Mitchell, C.E. Pathogen Spillover in Disease Epidemics. Am. Nat. 2004, 164, S79–S89. [Google Scholar] [CrossRef] [PubMed]
- Frutos, R.; Gavotte, L.; Devaux, C.A. Understanding the origin of COVID-19 requires to change the paradigm on zoonotic emergence from the spillover to the circulation model. Infect. Genet. Evol. 2021, 95, 104812. [Google Scholar] [CrossRef]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 2020, 368, 1016–1020. [Google Scholar] [CrossRef]
- McAloose, D.; Laverack, M.; Wang, L.; Killian, M.L.; Caserta, L.C.; Yuan, F.; Mitchell, P.K.; Queen, K.; Mauldin, M.R.; Cronk, B.D.; et al. From People to Panthera: Natural SARS-CoV-2 Infection in Tigers and Lions at the Bronx Zoo. mBio 2021, 11, e02220-20. [Google Scholar] [CrossRef]
- Hale, V.L.; Dennis, P.M.; McBride, D.S.; Nolting, J.M.; Madden, C.; Huey, D.; Ehrlich, M.; Grieser, J.; Winston, J.; Lombardi, D.; et al. SARS-CoV-2 infection in free-ranging white-tailed deer. Nature 2022, 602, 481–486. [Google Scholar] [CrossRef]
- Halfmann, P.J.; Iida, S.; Iwatsuki-Horimoto, K.; Maemura, T.; Kiso, M.; Scheaffer, S.M.; Darling, T.L.; Joshi, A.; Loeber, S.; Singh, G.; et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature 2022, 603, 687–695. [Google Scholar] [CrossRef]
- Woo, H.-J.; Reifman, J. A quantitative quasispecies theory-based model of virus escape mutation under immune selection. Proc. Natl. Acad. Sci. USA 2012, 109, 12980–12985. [Google Scholar] [CrossRef]
- Domingo, E.; Perales, C. Viral quasispecies. PLoS Genet. 2019, 15, e1008271. [Google Scholar] [CrossRef] [PubMed]
- Karamitros, T.; Papadopoulou, G.; Bousali, M.; Mexias, A.; Tsiodras, S.; Mentis, A. SARS-CoV-2 exhibits intra-host genomic plasticity and low-frequency polymorphic quasispecies. J. Clin. Virol. 2020, 131, 104585. [Google Scholar] [CrossRef] [PubMed]
- Jary, A.; Leducq, V.; Malet, I.; Marot, S.; Klement-Frutos, E.; Teyssou, E.; Soulié, C.; Abdi, B.; Wirden, M.; Pourcher, V.; et al. Evolution of viral quasispecies during SARS-CoV-2 infection. Clin. Microbiol. Infect. 2020, 26, 1560.e1–1560.e4. [Google Scholar] [CrossRef] [PubMed]
- Bashor, L.; Gagne, R.B.; Bosco-Lauth, A.M.; Bowen, R.A.; Stenglein, M.; VandeWoude, S. SARS-CoV-2 evolution in animals suggests mechanisms for rapid variant selection. Proc. Natl. Acad. Sci. USA 2021, 118, e2105253118. [Google Scholar] [CrossRef] [PubMed]
- Devaux, C.A.; Fantini, J. ACE2 receptor polymorphism in humans and animals increases the risk of the emergence of SARS-CoV-2 variants during repeated intra- and inter-species host-switching of the virus. Front. Microbiol. 2023; manuscript submitted for publication. [Google Scholar]
- Hammer, A.S.; Quaade, M.L.; Rasmussen, T.B.; Fonager, J.; Rasmussen, M.; Mundbjerg, K.; Lohse, L.; Strandbygaard, B.; Jørgensen, C.S.; Alfaro-Núñez, A.; et al. SARS-CoV-2 Transmission between Mink (Neovison vison) and Humans, Denmark. Emerg. Infect. Dis. 2021, 27, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Frutos, R.; Devaux, C. Mass culling of minks to protect the COVID-19 vaccines: Is it rational? New Microbes New Infect. 2020, 38, 100816. [Google Scholar] [CrossRef]
- Kok, K.-H.; Wong, S.-C.; Chan, W.-M.; Wen, L.; Chu, A.W.-H.; Ip, J.D.; Lee, L.-K.; Wong, I.T.-F.; Lo, H.W.-H.; Cheng, V.C.-C.; et al. Co-circulation of two SARS-CoV-2 variant strains within imported pet hamsters in Hong Kong. Emerg. Microbes Infect. 2022, 11, 689–698. [Google Scholar] [CrossRef]
- Frutos, R.; Yahi, N.; Gavotte, L.; Fantini, J.; Devaux, C.A. Role of spike compensatory mutations in the interspecies transmission of SARS-CoV-2. One Health 2022, 15, 100429. [Google Scholar] [CrossRef]
- Fantini, J.; Devaux, C.A.; Yahi, N.; Frutos, R. The novel hamster-adapted SARS-CoV-2 Delta variant may be selectively advantaged in humans. J. Infect. 2022, 84, e53–e54. [Google Scholar] [CrossRef]
- Palmer, M.V.; Martins, M.; Falkenberg, S.; Buckley, A.; Caserta, L.C.; Mitchell, P.K.; Cassmann, E.D.; Rollins, A.; Zylich, N.C.; Renshaw, R.W.; et al. Susceptibility of White-Tailed Deer (Odocoileus virginianus) to SARS-CoV-2. J. Virol. 2021, 95, e00083-21. [Google Scholar] [CrossRef]
- Chandler, J.C.; Bevins, S.N.; Ellis, J.W.; Linder, T.J.; Tell, R.M.; Jenkins-Moore, M.; Root, J.J.; Lenoch, J.B.; Robbe-Austerman, S.; DeLiberto, T.J.; et al. SARS-CoV-2 exposure in wild white-tailed deer (Odocoileus virginianus). Proc. Natl. Acad. Sci. USA 2021, 118, e2114828118. [Google Scholar] [CrossRef] [PubMed]
- Palermo, P.M.; Orbegozo, J.; Watts, D.M.; Morrill, J.C. SARS-CoV-2 Neutralizing Antibodies in White-Tailed Deer from Texas. Vector-Borne Zoonotic Dis. 2022, 22, 62–64. [Google Scholar] [CrossRef] [PubMed]
- Kuchipudi, S.V.; Surendran-Nair, M.; Ruden, R.M.; Yon, M.; Nissly, R.H.; Vandegrift, K.J.; Nelli, R.K.; Li, L.; Jayarao, B.M.; Maranas, C.D.; et al. Multiple spillovers from humans and onward transmission of SARS-CoV-2 in white-tailed deer. Proc. Natl. Acad. Sci. USA 2022, 119, e2121644119. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.; Boggiatto, P.M.; Buckley, A.; Cassmann, E.D.; Falkenberg, S.; Caserta, L.C.; Fernandes, M.H.V.; Kanipe, C.; Lager, K.; Palmer, M.V.; et al. From Deer-to-Deer: SARS-CoV-2 is efficiently transmitted and presents broad tissue tropism and replication sites in white-tailed deer. PLoS Pathog. 2022, 18, e1010197. [Google Scholar] [CrossRef] [PubMed]
- Pickering, B.; Lung, O.; Maguire, F.; Kruczkiewicz, P.; Kotwa, J.D.; Buchanan, T.; Gagnier, M.; Guthrie, J.L.; Jardine, C.M.; Marchand-Austin, A.; et al. Divergent SARS-CoV-2 variant emerges in white-tailed deer with deer-to-human transmission. Nat. Microbiol. 2022, 7, 2011–2024. [Google Scholar] [CrossRef]
- Wei, C.; Shan, K.-J.; Wang, W.; Zhang, S.; Huan, Q.; Qian, W. Evidence for a mouse origin of the SARS-CoV-2 Omicron variant. J. Genet. Genom. 2021, 48, 1111–1121. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, Y.; Iketani, S.; Nair, M.S.; Li, Z.; Mohri, H.; Wang, M.; Yu, J.; Bowen, A.D.; Chang, J.Y.; et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature 2022, 608, 603–608. [Google Scholar] [CrossRef]
- Yuan, M.; Zhu, X.; He, W.-T.; Zhou, P.; Kaku, C.I.; Capozzola, T.; Zhu, C.Y.; Yu, X.; Liu, H.; Yu, W.; et al. A broad and potent neutralization epitope in SARS-related coronaviruses. Proc. Natl. Acad. Sci. USA 2022, 119, e2205784119. [Google Scholar] [CrossRef]
- Cao, Y.; Jian, F.; Wang, J.; Yu, Y.; Song, W.; Yisimayi, A.; An, R.; Chen, X.; Na Zhang, N.; Wang, Y.; et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature 2023, 614, 521–529. [Google Scholar] [CrossRef]
- Wang, J.; Guo, C.; Cai, L.; Liao, C.; Yi, H.; Li, Q.; Hu, H.; Deng, Q.; Lu, Y.; Guo, Z.; et al. Pre-Existing Cross-Reactive Antibody Responses Do Not Significantly Impact Inactivated COVID-19 Vaccine-Induced Neutralization. Front. Immunol. 2021, 12, 772511. [Google Scholar] [CrossRef]
- Majdoubi, A.; Michalski, C.; O’Connell, S.E.; Dada, S.; Narpala, S.; Gelinas, J.; Mehta, D.; Cheung, C.; Winkler, D.F.; Basappa, M.; et al. A majority of uninfected adults show preexisting antibody reactivity against SARS-CoV-2. JCI Insight 2021, 6, e146316. [Google Scholar] [CrossRef] [PubMed]
- Souris, M.; Tshilolo, L.; Parzy, D.; Ingoba, L.L.; Ntoumi, F.; Kamgaing, R.; Ndour, M.; Mbongi, D.; Phoba, B.; Tshilolo, M.-A.; et al. Pre-Pandemic Cross-Reactive Immunity against SARS-CoV-2 among Central and West African Populations. Viruses 2022, 14, 2259. [Google Scholar] [CrossRef] [PubMed]
- Nzoghe, A.M.; Essone, P.N.; Leboueny, M.; Siawaya, A.C.M.; Bongho, E.C.; Ndjindji, O.M.; Houechenou, R.M.A.; Agnandji, S.T.; Siawaya, J.F.D. Evidence and implications of pre-existing humoral cross-reactive immunity to SARS-CoV-2. Immun. Inflamm. Dis. 2021, 9, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Aissatou, A.; Fokam, J.; Semengue, E.N.J.; Takou, D.; Ka’e, A.C.; Ambe, C.C.; Nka, A.D.; Djupsa, S.C.; Beloumou, G.; Ciaffi, L.; et al. Pre-existing immunity to SARS-CoV-2 before the COVID-19 pandemic era in Cameroon: A comparative analysis according to HIV-status. Front. Immunol. 2023, 14, 1155855. [Google Scholar] [CrossRef] [PubMed]
- Goel, R.R.; Apostolidis, S.A.; Painter, M.M.; Mathew, D.; Pattekar, A.; Kuthuru, O.; Gouma, S.; Hicks, P.; Meng, W.; Rosenfeld, A.M.; et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naïve and recovered individuals after mRNA vaccination. Sci. Immunol. 2021, 6, eabi6950. [Google Scholar] [CrossRef] [PubMed]
- Wolf, C.; Köppert, S.; Becza, N.; Kuerten, S.; Kirchenbaum, G.A.; Lehmann, P.V. Antibody Levels Poorly Reflect on the Frequency of Memory B Cells Generated following SARS-CoV-2, Seasonal Influenza, or EBV Infection. Cells 2022, 11, 3662. [Google Scholar] [CrossRef] [PubMed]
- Namuniina, A.; Muyanja, E.S.; Biribawa, V.M.; Okech, B.A.; Ssemaganda, A.; Price, M.A.; Hills, N.; Nanteza, A.; Bagaya, B.S.; Weiskopf, D.; et al. High proportion of Ugandans with pre-pandemic SARS-CoV-2 cross-reactive CD4+ and CD8+ T-cell 2 responses. MedRxiv, 2023; not peer reviewed. [Google Scholar] [CrossRef]
- Eguia, R.T.; Crawford, K.H.D.; Stevens-Ayers, T.; Kelnhofer-Millevolte, L.; Greninger, A.L.; Englund, J.A.; Boeckh, M.J.; Bloom, J.D. A human coronavirus evolves antigenically to escape antibody immunity. PLoS Pathog. 2021, 17, e1009453. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; van Haperen, R.; Gutiérrez-Álvarez, J.; Li, W.; Okba, N.M.A.; Albulescu, I.; Widjaja, I.; van Dieren, B.; Fernandez-Delgado, R.; Sola, I.; et al. A conserved immunogenic and vulnerable site on the coronavirus spike protein delineated by cross-reactive monoclonal antibodies. Nat. Commun. 2021, 12, 1715. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Yi, C.; Zhu, Y.; Ding, L.; Xia, S.; Chen, X.; Liu, M.; Gu, C.; Lu, X.; Fu, Y.; et al. Neutralization mechanism of a human antibody with pan-coronavirus reactivity including SARS-CoV-2. Nat. Microbiol. 2022, 7, 1063–1074. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.E.; Brown-Augsburger, P.L.; Corbett, K.S.; Westendorf, K.; Davies, J.; Cujec, T.P.; Wiethoff, C.M.; Blackbourne, J.L.; Heinz, B.A.; Foster, D.; et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 2021, 13, eabf1906. [Google Scholar] [CrossRef] [PubMed]
- Jacob-Dolan, C.; Feldman, J.; McMahan, K.; Yu, J.; Zahn, R.; Wegmann, F.; Schuitemaker, H.; Schmidt, A.G.; Barouch, D.H. Coronavirus-Specific Antibody Cross Reactivity in Rhesus Macaques following SARS-CoV-2 Vaccination and Infection. J. Virol. 2021, 95, e00117-21. [Google Scholar] [CrossRef]
- Sagar, M.; Reifler, K.; Rossi, M.; Miller, N.S.; Sinha, P.; White, L.F.; Mizgerd, J.P. Recent endemic coronavirus infection is associated with less-severe COVID-19. J. Clin. Investig. 2021, 131, e143380. [Google Scholar] [CrossRef] [PubMed]
- Beretta, A.; Cranage, M.; Zipeto, D. Is Cross-Reactive Immunity Triggering COVID-19 Immunopathogenesis? Front. Immunol. 2020, 11, 567710. [Google Scholar] [CrossRef] [PubMed]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501.e1415. [Google Scholar] [CrossRef]
- Sette, A.; Crotty, S. Pre-existing immunity to SARS-CoV-2: The knowns and unknowns. Nat. Rev. Immunol. 2020, 20, 457–458. [Google Scholar] [CrossRef]
- Nelde, A.; Bilich, T.; Heitmann, J.S.; Maringer, Y.; Salih, H.R.; Roerden, M.; Lübke, M.; Bauer, J.; Rieth, J.; Wacker, M.; et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Microbiol. 2021, 22, 74–85. [Google Scholar] [CrossRef]
- Kundu, R.; Narean, J.S.; Wang, L.; Fenn, J.; Pillay, T.; Fernandez, N.D.; Conibear, E.; Koycheva, A.; Davies, M.; Tolosa-Wright, M.; et al. Cross-reactive memory T cells associate with protection against SARS-CoV-2 infection in COVID-19 contacts. Nat. Commun. 2022, 13, 80. [Google Scholar] [CrossRef] [PubMed]
- Monto, A.S.; Lim, S.K. The Tecumseh Study of Respiratory Illness. VI. Frequency of and Relationship between Outbreaks of Coronavims Infection. J. Infect. Dis. 1974, 129, 271–276. [Google Scholar] [CrossRef]
- Hamre, D.; Beem, M. Virologic studies of acute respiratory disease in young adults. V. Coronavirus 229E infections during six years of surveillance. Am. J. Epidemiol. 1972, 96, 94–106. [Google Scholar] [CrossRef] [PubMed]
- Van Der Hoek, L.; Pyrc, K.; Berkhout, B. Human coronavirus NL63, a new respiratory virus. FEMS Microbiol. Rev. 2006, 30, 760–773. [Google Scholar] [CrossRef]
- Talbot, H.K.; Shepherd, B.E.; Crowe, J.; Griffin, M.R.; Edwards, K.M.; Podsiad, A.B.; Tollefson, S.J.; Wright, P.F.; Williams, J. The Pediatric Burden of Human Coronaviruses Evaluated for Twenty Years. Pediatr. Infect. Dis. J. 2009, 28, 682–687. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, P.; Curtis, N. Coronavirus Infections in Children Including COVID-19. Pediatr. Infect. Dis. J. 2020, 39, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, S.B. Update on Human Rhinovirus and Coronavirus Infections. Semin. Respir. Crit. Care Med. 2016, 37, 555–571. [Google Scholar] [CrossRef]
- Walsh, E.E.; Shin, J.H.; Falsey, A.R. Clinical Impact of Human Coronaviruses 229E and OC43 Infection in Diverse Adult Populations. J. Infect. Dis. 2013, 208, 1634–1642. [Google Scholar] [CrossRef] [PubMed]
- Edridge, A.W.D.; Kaczorowska, J.; Hoste, A.C.R.; Bakker, M.; Klein, M.; Loens, K.; Jebbink, M.F.; Matser, A.; Kinsella, C.M.; Rueda, P.; et al. Seasonal coronavirus protective immunity is short-lasting. Nat. Med. 2020, 26, 1691–1693. [Google Scholar] [CrossRef]
- Waterlow, N.R.; van Leeuwen, E.; Davies, N.G.; Flasche, S.; Eggo, R.M.; CMMID COVID-19 Working Group. How immunity from and interaction with seasonal coronaviruses can shape SARS-CoV-2 epidemiology. Proc. Natl. Acad. Sci. USA 2021, 118, e2108395118. [Google Scholar] [CrossRef] [PubMed]
- Nyaguthii, D.M.; Otieno, G.P.; Kombe, I.K.; Koech, D.; Mutunga, M.; Medley, G.F.; Nokes, D.J.; Munywoki, P.K. Infection patterns of endemic human coronaviruses in rural households in coastal Kenya [version 1; peer review: 1 approved, 2 approved with reservations]. Wellcome Open Res. 2021, 6, 27. [Google Scholar] [CrossRef]
- Monto, A.S.; Dejonge, P.M.; Callear, A.P.; Bazzi, L.A.; Capriola, S.B.; Malosh, R.E.; Martin, E.T.; Petrie, J.G. Coronavirus Occurrence and Transmission Over 8 Years in the HIVE Cohort of Households in Michigan. J. Infect. Dis. 2020, 222, 9–16. [Google Scholar] [CrossRef]
- Nickbakhsh, S.; Ho, A.; Marques, D.F.P.; McMenamin, J.; Gunson, R.N.; Murcia, P.R. Epidemiology of Seasonal Coronaviruses: Establishing the Context for the Emergence of Coronavirus Disease 2019. J. Infect. Dis. 2020, 222, 17–25. [Google Scholar] [CrossRef]
- Lalaoui, R.; Bakour, S.; Raoult, D.; Verger, P.; Sokhna, C.; Devaux, C.; Pradines, B.; Rolain, J.-M. What could explain the late emergence of COVID-19 in Africa? New Microbes New Infect. 2020, 38, 100760. [Google Scholar] [CrossRef]
- Lone, S.A.; Ahmad, A. COVID-19 pandemic—An African perspective. Emerg. Microbes Infect. 2020, 9, 1300–1308. [Google Scholar] [CrossRef] [PubMed]
- Osman, I.O.; Levasseur, A.; Brechard, L.; Hassan, I.A.; Abdillahi, I.S.; Waberi, Z.A.; Delerce, J.; Bedotto, M.; Houhamdi, L.; Fournier, P.-E.; et al. Whole Genome Sequencing of SARS-CoV-2 Strains in COVID-19 Patients From Djibouti Shows Novel Mutations and Clades Replacing Over Time. Front. Med. 2021, 8, 737602. [Google Scholar] [CrossRef] [PubMed]
- Lekana-Douki, S.E.; N’Dilimabaka, N.; Levasseur, A.; Colson, P.; Andeko, J.C.; Minko, O.Z.; Mve-Ella, O.B.; Fournier, P.-E.; Devaux, C.; Ondo, B.M.; et al. Screening and Whole Genome Sequencing of SARS-CoV-2 Circulating During the First Three Waves of the COVID-19 Pandemic in Libreville and the Haut-Ogooué Province in Gabon. Front. Med. 2022, 9, 877391. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Liu, J.; Ma, Z.; Bramer, W.M.; Peppelenbosch, M.P.; Pan, Q. Estimating Global Epidemiology of Low-Pathogenic Human Coronaviruses in Relation to the COVID-19 Context. J. Infect. Dis. 2020, 222, 695–696. [Google Scholar] [CrossRef] [PubMed]
- Van Cakenberghe, V.; Tungaluna, G.-C.G.; Akawa, P.M.; Seamark, E.; Verheyen, E. The bats of the Congo and of Rwanda and Burundi revisited (Mammalia: Chiroptera). Eur. J. Taxon. 2017, 382, 1–327. [Google Scholar] [CrossRef]
- Nziza, J.; Goldstein, T.; Cranfield, M.; Webala, P.; Nsengimana, O.; Nyatanyi, T.; Mudakikwa, A.; Tremeau-Bravard, A.; Byarugaba, D.; Tumushime, J.C.; et al. Coronaviruses Detected in Bats in Close Contact with Humans in Rwanda. EcoHealth 2020, 17, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Devaux, C.A.; Mediannikov, O.; Medkour, H.; Raoult, D. Infectious Disease Risk Across the Growing Human-Non Human Primate Interface: A Review of the Evidence. Front. Public Health 2019, 7, 305. [Google Scholar] [CrossRef]
- Dolgin, E. Pan-coronavirus vaccine pipeline takes form. Nat. Rev. Drug Discov. 2022, 21, 324–326. [Google Scholar] [CrossRef]
- Murray, S.M.; Ansari, A.M.; Frater, J.; Klenerman, P.; Dunachie, S.; Barnes, E.; Ogbe, A. The impact of pre-existing cross-reactive immunity on SARS-CoV-2 infection and vaccine responses. Nat. Rev. Immunol. 2022, 23, 304–316. [Google Scholar] [CrossRef]
- Zhao, Y.; Ni, W.; Liang, S.; Dong, L.; Xiang, M.; Cai, Z.; Niu, D.; Zhang, Q.; Wang, D.; Zheng, Y.; et al. Vaccination with Span, an antigen guided by SARS-CoV-2 S protein evolution, protects against challenge with viral variants in mice. Sci. Transl. Med. 2023, 15, eabo3332. [Google Scholar] [CrossRef]
- Dutta, D.; Ghosh, A.; Dutta, C.; Sukla, S.; Biswas, S. Cross-reactivity of SARS-CoV-2 with other pathogens, especially dengue virus: A historical perspective. J. Med. Virol. 2023, 95, e28557. [Google Scholar] [CrossRef] [PubMed]
- Brouqui, P.; Colson, P.; Melenotte, C.; Houhamdi, L.; Bedotto, M.; Devaux, C.; Gautret, P.; Million, M.; Parola, P.; Stoupan, D.; et al. COVID-19 re-infection. Eur. J. Clin. Investig. 2021, 51, e13537. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Russell, R.M.; Bibollet-Ruche, F.; Skelly, A.N.; Sherrill-Mix, S.; Freeman, D.A.; Stoltz, R.; Lindemuth, E.; Lee, F.H.; Sterrett, S.; et al. Predictors of nonseroconversion after SARS-CoV-2 infection. Emerg. Infect. Dis. 2021, 27, 2454–2458. [Google Scholar] [CrossRef]
- Alejo, J.L.; Mitchell, J.; Chang, A.; Chiang, T.P.Y.; Massie, A.B.; Segev, D.L.; Makary, M.A. Prevalence and Durability of SARS-CoV-2 Antibodies Among Unvaccinated US Adults by History of COVID-19. JAMA 2022, 327, 1085–1087. [Google Scholar] [CrossRef] [PubMed]
- Townsend, J.P.; Hassler, H.B.; Sah, P.; Galvani, A.P.; Dornburg, A. The durability of natural infection and vaccine-induced immunity against future infection by SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2021, 119, e666–e675. [Google Scholar] [CrossRef]
- Hachmann, N.P.; Miller, J.; Collier, A.-R.Y.; Ventura, J.D.; Yu, J.; Rowe, M.; Bondzie, E.A.; Powers, O.; Surve, N.; Hall, K.; et al. Neutralization Escape by SARS-CoV-2 Omicron Subvariants BA.2.12.1, BA.4, and BA.5. N. Engl. J. Med. 2022, 387, 86–88. [Google Scholar] [CrossRef]
- Muecksch, F.; Wang, Z.; Cho, A.; Gaebler, C.; Ben Tanfous, T.; DaSilva, J.; Bednarski, E.; Ramos, V.; Zong, S.; Johnson, B.; et al. Increased memory B cell potency and breadth after a SARS-CoV-2 mRNA boost. Nature 2022, 607, 128–134. [Google Scholar] [CrossRef]
- Tan, S.T.; Kwan, A.T.; Rodríguez-Barraquer, I.; Singer, B.J.; Park, H.J.; Lewnard, J.A.; Sears, D.; Lo, N.C. Infectiousness of SARS-CoV-2 breakthrough infections and reinfections during the Omicron wave. Nat. Med. 2023, 29, 358–365. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Devaux, C.A.; Fantini, J. Unravelling Antigenic Cross-Reactions toward the World of Coronaviruses: Extent of the Stability of Shared Epitopes and SARS-CoV-2 Anti-Spike Cross-Neutralizing Antibodies. Pathogens 2023, 12, 713. https://doi.org/10.3390/pathogens12050713
Devaux CA, Fantini J. Unravelling Antigenic Cross-Reactions toward the World of Coronaviruses: Extent of the Stability of Shared Epitopes and SARS-CoV-2 Anti-Spike Cross-Neutralizing Antibodies. Pathogens. 2023; 12(5):713. https://doi.org/10.3390/pathogens12050713
Chicago/Turabian StyleDevaux, Christian A., and Jacques Fantini. 2023. "Unravelling Antigenic Cross-Reactions toward the World of Coronaviruses: Extent of the Stability of Shared Epitopes and SARS-CoV-2 Anti-Spike Cross-Neutralizing Antibodies" Pathogens 12, no. 5: 713. https://doi.org/10.3390/pathogens12050713




