Molecular Confirmation of Accipiter Birds of Prey as Definitive Hosts of Numerous Sarcocystis Species, including Sarcocystis sp., Closely Related to Pathogenic S. calchasi
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Isolation of Oocysts/Sporocysts
2.3. Molecular Identification and Phylogenetic Analysis
2.4. Analysis of DNA Sequence and Statistical Data
3. Results
3.1. Microscopical Examination of Sarcocystis spp. Oocysts/Sporocysts Isolated from Accipiter Hawks
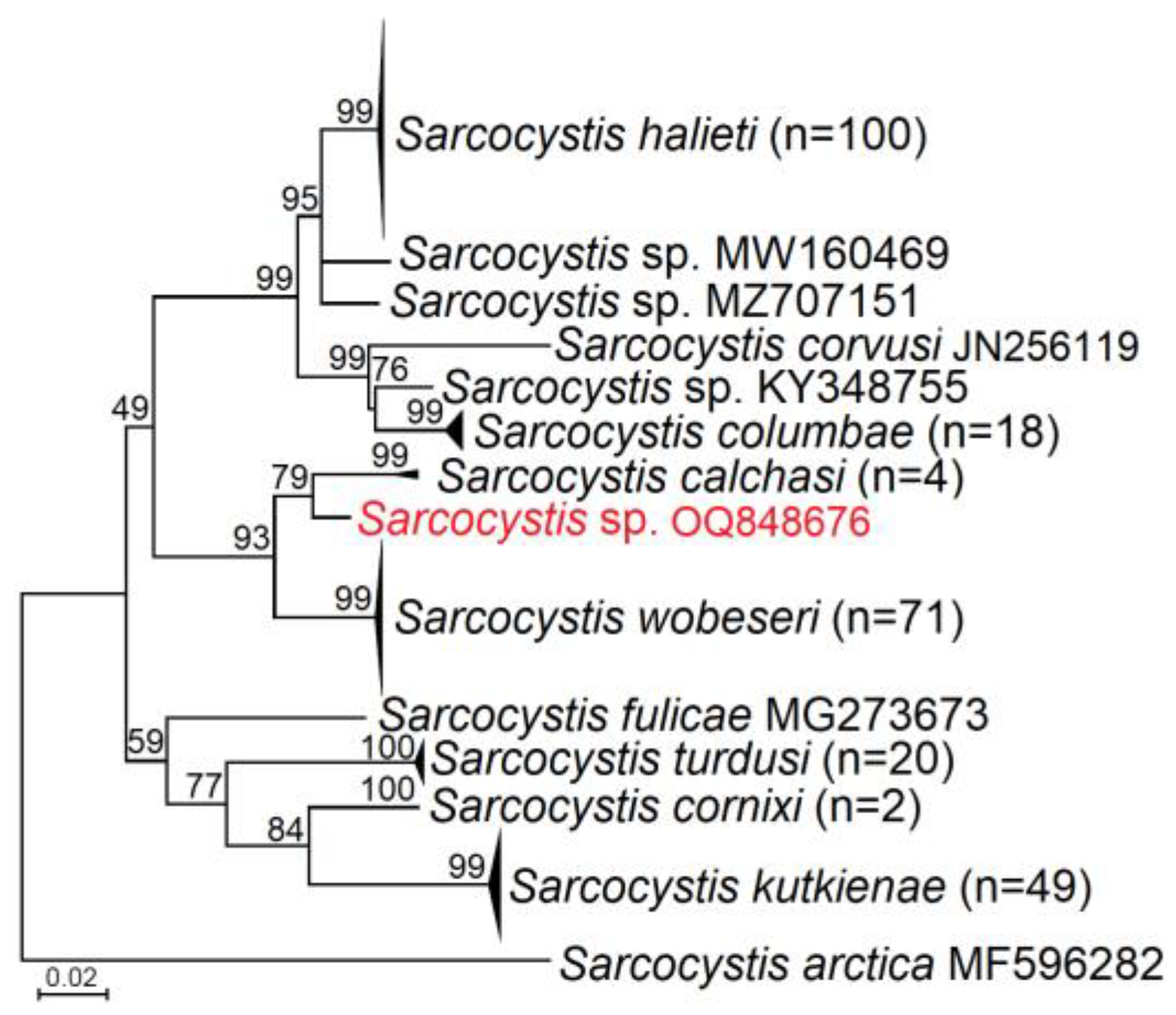
3.2. Molecular Identification and Phylogenetic Analysis of Sarcocystis spp. Closely Related to S. calchasi
3.3. The Identification and Distribution of Sarcocystis spp. in Intestines of Accipiter spp.
4. Discussion
4.1. Determination of the Definitive Hosts of Sarcocystis spp.
4.2. Distribution of Detected Sarcocystis spp. in the Definitive Hosts
| Sarcocystis spp. | Intermediate Hosts | Definitive Hosts Defined by Molecular Methods |
|---|---|---|
| S. calchasi | Cacatuidae [44]; Columbidae [4,12,15,16,41,42,43,44,45,46,47,48]; Numididae [20]; Phalacrocoracidae [17]; Picidae [14]; Psittacidae [19]; Psittaculidae [23] | Accipitridae [4,18,46, PS] |
| S. columbae | Columbidae [5,82]; Laridae [54] | Accipitridae [5,18, PS]; Corvidae [65] |
| S. cornixi | Corvidae [55,69] | Accipitridae [51, PS]; Corvidae [65] |
| S. halieti | Accipitridae [87]; Corvidae [69]; Laridae [54]; Phalacrocoracidae [53]; Procellariidae [22]; Strigidae [49]; Sturnidae [52] | Accipitridae [18,50,51,52, PS]; Corvidae [65] |
| S. kutkienae | Corvidae [55,62] | Accipitridae [PS]; Corvidae [65] |
| S. lari | Laridae [7,54] | Accipitridae [51, PS]; Corvidae [65] |
| S. turdusi | Turdidae [71]; Muscicapidae * | Accipitridae [18,50, PS]; Corvidae [65] |
| S. wobeseri | Accipitridae [83]; Anatidae [70]; Laridae [54] | Accipitridae [PS]; Corvidae [65] |
| Sarcocystis spp. | Not determined | Accipitridae [PS] |
4.3. Sarcocystis spp. Closely Related to S. calchasi
4.4. Identification of S. calchasi in Lithuania
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dubey, J.P.; Calero-Bernal, R.; Rosenthal, B.M.; Speer, C.A.; Fayer, R. Sarcocystosis of Animals and Humans, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Fayer, R. Sarcocystis spp. in Human Infections. Clin. Microbiol. Rev. 2004, 17, 894–902. [Google Scholar] [CrossRef] [PubMed]
- Box, E.D.; Meier, J.L.; Smith, J.H. Description of Sarcocystis falcatula Stiles, 1983, a Parasite of Birds and Opossum. J. Protozool. 1984, 31, 521–524. [Google Scholar] [CrossRef] [PubMed]
- Olias, P.; Gruber, A.D.; Hafez, H.M.; Heydorn, A.O.; Mehlhorn, H.; Lierz, M. Sarcocystis calchasi sp. nov. of the domestic pigeon (Columba livia f. domestica) and the Northern goshawk (Accipiter gentilis): Light and electron microscopical characteristics. Parasitol. Res. 2010, 106, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Olias, P.; Olias, L.; Lierz, M.; Mehlhorn, H.; Gruber, A.D. Sarcocystis calchasi is Distinct to Sarcocystis columbae sp. nov. from the Wood Pigeon (Columba palumbus) and Sarcocystis sp. from the Sparrowhawk (Accipiter nisus). Vet. Parasitol. 2010, 171, 7–14. [Google Scholar] [CrossRef]
- Kutkienė, L.; Prakas, P.; Sruoga, A.; Butkauskas, D. Description of Sarcocystis anasi sp. nov. and Sarcocystis albifronsi sp. nov. in Birds of the Order Anseriformes. Parasitol. Res. 2012, 110, 1043–1046. [Google Scholar] [CrossRef]
- Prakas, P.; Kutkienė, L.; Butkauskas, D.; Sruoga, A.; Žalakevičius, M. Description of Sarcocystis lari sp. n. (Apicomplexa: Sarcocystidae) from the Great Black-Backed Gull, Larus marinus (Charadriiformes: Laridae), on the Basis of Cyst Morphology and Molecular Data. Folia Parasitol. 2014, 61, 11–17. [Google Scholar] [CrossRef]
- Prakas, P.; Butkauskas, D.; Švažas, S.; Juozaitytė-Ngugu, E.; Stanevičius, V. Morphologic and Genetic Identification of Sarcocystis fulicae n. sp. (Apicomplexa: Sarcocystidae) from the Eurasian coot (Fulica atra). J. Wildl. Dis. 2018, 54, 765–771. [Google Scholar] [CrossRef]
- Máca, O.; González-Solís, D. Sarcocystis cristata sp. nov. (Apicomplexa, Sarcocystidae) in the Imported Great Blue Turaco Corythaeola cristata (Aves, Musophagidae). Parasit. Vectors 2021, 14, 56. [Google Scholar] [CrossRef]
- Hu, J.; Zhang, M.; Wu, Z.; Zeng, H.; Tao, J. Description of Sarcocystis platyrhynchosi n. sp. (Apicomplexa: Sarcocystidae) from Domestic Ducks Anas platyrhynchos (Anseriformes: Anatidae) in China. Parasit. Vectors 2023, 16, 50. [Google Scholar] [CrossRef]
- Wünschmann, A.; Rejmanek, D.; Conrad, P.A.; Hall, N.; Cruz-Martinez, L.; Vaughn, S.B.; Barr, B.C. Natural Fatal Sarcocystis falcatula Infections in Free-Ranging Eagles in North America. J. Vet. Diagn. Investig. 2010, 22, 282–289. [Google Scholar] [CrossRef]
- Hodo, C.L.; Whitley, D.B.; Hamer, S.A.; Corapi, W.V.; Snowden, K.; Heatley, J.J.; Hoffmann, A.R. Histopathologic and Molecular Characterization of Sarcocystis calchasi Encephalitis in White-winged Doves (Zenaida asiatica) and Eurasian Collared Doves (Streptopelia decaocto), East-central Texas, USA, 2010–2013. J. Wildl. Dis. 2016, 52, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Konradt, G.; Bianchi, M.V.; Leite-Filho, R.V.; da Silva, B.Z.; Soares, R.M.; Pavarini, S.P.; Driemeier, D. Necrotizing Meningoencephalitis Caused by Sarcocystis falcatula in Bare-Faced Ibis (Phimosus infuscatus). Parasitol. Res. 2017, 116, 809–812. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, L.; Parmentier, S.L.; Fischer, D.; Heckmann, J.; Klopfleisch, R.; Kershaw, O.; Ziegler, U.; Neurath, H.; Schmidt, V.; Lierz, M. Investigations into Causes of Neurologic Signs and Mortality and the First Identification of Sarcocystis calchasi in Free-ranging Woodpeckers in Germany. J. Zoo. Wildl. Med. 2018, 49, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Mete, A.; Rogers, K.H.; Wolking, R.; Bradway, D.S.; Kelly, T.; Piazza, M.; Crossley, B. Sarcocystis calchasi Outbreak in Feral Rock Pigeons (Columba livia) in California. Vet. Pathol. 2019, 56, 317–321. [Google Scholar] [CrossRef]
- Parmentier, S.L.; Maier-Sam, K.; Failing, K.; Gruber, A.D.; Lierz, M. High Prevalence of Sarcocystis calchasi in Racing Pigeon Flocks in Germany. PLoS ONE 2019, 14, e0215241. [Google Scholar] [CrossRef]
- Bamac, O.E.; Rogers, K.H.; Arranz-Solís, D.; Saeij, J.P.J.; Lewis, S.; Duerr, R.; Skoglund, J.; Peronne, L.; Mete, A. Protozoal Encephalitis Associated with Sarcocystis calchasi and S. falcatula during an Epizootic Involving Brandt’s Cormorants (Phalacrocorax penicillatus) in Coastal Southern California, USA. Int. J. Parasitol. Parasites Wildl. 2020, 12, 185–191. [Google Scholar] [CrossRef]
- Rogers, K.H.; Arranz-Solís, D.; Saeij, J.P.J.; Lewis, S.; Mete, A. Sarcocystis calchasi and other Sarcocystidae Detected in Predatory Birds in California, USA. Int. J. Parasitol. Parasites Wildl. 2021, 17, 91–99. [Google Scholar] [CrossRef]
- Söderström, M.; Malkamäki, S.; Sukura, A.; Sainmaa, S.; Airas, N. Sarcocystis calchasi in a Captive Patagonian Conure (Cyanoliseus patagonus) in Finland. J. Comp. Pathol. 2021, 189, 135–140. [Google Scholar] [CrossRef]
- Gadsby, S.; Garner, M.M.; Bolin, S.R.; Sanchez, C.R.; Flaminio, K.P.; Sim, R.R. Fatal Sarcocystis calchasi-Associated Meningoencephalitis in 2 Captive Vulturine Guineafowl. J. Vet. Diagn. Investig. 2022, 34, 543–546. [Google Scholar] [CrossRef]
- Llano, H.A.B.; Zavatieri Polato, H.; Borges Keid, L.; Ferreira de Souza Oliveira, T.M.; Zwarg, T.; de Oliveira, A.S.; Sanches, T.C.; Joppert, A.M.; Gondim, L.F.P.; Martins Soares, R. Molecular Screening for Sarcocystidae in Muscles of Wild Birds from Brazil suggests a Plethora of Intermediate Hosts for Sarcocystis falcatula. Int. J. Parasitol. Parasites Wildl. 2022, 17, 230–238. [Google Scholar] [CrossRef]
- Sato, A.P.; da Silva, T.C.E.; de Pontes, T.P.; Sanches, A.W.D.; Prakas, P.; Locatelli-Dittrich, R. Molecular Characterization of Sarcocystis spp. in Seabirds from Southern Brazil. Parasitol. Int. 2022, 90, 102595. [Google Scholar] [CrossRef] [PubMed]
- Gonzales-Viera, O.; Arranz-Solís, D.; Smith, J.; Saeij, J.P.J.; Mete, A. Fatal Sarcocystis calchasi Hepatitis in a Captive Indian Ringneck Parakeet (Psittacula krameri manillensis). Vet. Parasitol. Reg. Stud. Rep. 2023, 39, 100841. [Google Scholar] [CrossRef]
- Dubey, J.P.; Venturini, L.; Venturini, C.; Basso, W.; Unzaga, J. Isolation of Sarcocystis falcatula from the South American Opossum (Didelphis albiventris) from Argentina. Vet. Parasitol. 1999, 86, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Dubey, J.P.; Lindsay, D.S.; Rezende, P.C.; Costa, A.J. Characterization of an Unidentified Sarcocystis falcatula-like Parasite from the South American Opossum, Didelphis albiventris from Brazil. J. Eukaryot. Microbiol. 2000, 47, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Gallo, S.S.M.; Lindsay, D.S.; Ederli, N.B.; Matteoli, F.P.; Venancio, T.M.; de Oliveira, F.C.R. Identification of Opossums Didelphis aurita (Wied-Neuweid, 1826) as a Definitive Host of Sarcocystis falcatula-like Sporocysts. Parasitol. Res. 2018, 117, 213–223. [Google Scholar] [CrossRef]
- Suedmeyer, W.K.; Bermudez, A.J.; Barr, B.C.; Marsh, A.E. Acute Pulmonary Sarcocystis falcatula-like Infection in three Victoria Crowned Pigeons (Goura victoria) Housed Indoors. J. Zoo Wildl. Med. 2001, 32, 252–256. [Google Scholar] [CrossRef]
- Dubey, J.P.; Garner, M.M.; Stetter, M.D.; Marsh, A.E.; Barr, B.C. Acute Sarcocystis falcatula-like Infection in a Carmine Bee-Eater (Merops nubicus) and Immunohistochemical Cross Reactivity between Sarcocystis falcatula and Sarcocystis neurona. J. Parasitol. 2001, 87, 824–832. [Google Scholar] [CrossRef]
- Luznar, S.L.; Avery, M.L.; Dame, J.B.; MacKay, R.J.; Greiner, E.C. Development of Sarcocystis falcatula in its Intermediate Host, the Brown-headed Cowbird (Molothrus ater). Vet. Parasitol. 2001, 95, 327–334. [Google Scholar] [CrossRef]
- Smith, J.H.; Meier, J.L.; Neill, P.J.; Box, E.D. Pathogenesis of Sarcocystis falcatula in the Budgerigar. I. Early Pulmonary Schizogony. Lab. Investig. 1987, 56, 60–71. [Google Scholar]
- Smith, J.H.; Meier, J.L.; Neill, P.J.; Box, E.D. Pathogenesis of Sarcocystis falcatula in the Budgerigar. II. Pulmonary Pathology. Lab. Investig. 1987, 56, 72–84. [Google Scholar]
- Smith, J.H.; Neill, P.J.; Box, E.D. Pathogenesis of Sarcocystis falcatula (Apicomplexa: Sarcocystidae) in the Budgerigar (Melopsittacus undulatus). III. Pathologic and Quantitative Parasitologic Analysis of Extrapulmonary Disease. J. Parasitol. 1989, 75, 270–287. [Google Scholar] [CrossRef] [PubMed]
- Bolon, B.; Greiner, E.C.; Mays, M.B. Microscopic Features of Sarcocystis falcatula in Skeletal Muscle from a Patagonian Conure. Vet. Pathol. 1989, 26, 282–284. [Google Scholar] [CrossRef] [PubMed]
- Dame, J.B.; MacKay, R.J.; Yowell, C.A.; Cutler, T.J.; Marsh, A.; Greiner, E.C. Sarcocystis falcatula from Passerine and Psittacine Birds: Synonymy with Sarcocystis neurona, Agent of Equine Protozoal Myeloencephalitis. J. Parasitol. 1995, 81, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Speer, C.A.; Dubey, J.P. Ultrastructure of Shizonts and Merozoites of Sarcocystis falcatula in the Lungs of Budgerigars (Melopsittacus undulatus). J. Parasitol. 1999, 85, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Godoy, S.N.; De Paula, C.D.; Cubas, Z.S.; Matushima, E.R.; Catão-Dias, J.L. Occurrence of Sarcocystis falcatula in Captive Psittacine Birds in Brazil. J. Avian Med. Surg. 2009, 23, 18–23. [Google Scholar] [CrossRef]
- Verma, S.K.; Trupkiewicz, J.G.; Georoff, T.; Dubey, J.P. Molecularly Confirmed Acute, Fatal Sarcocystis falcatula Infection in the Rainbow Lorikeets (Trichoglossus moluccanus) at the Philadelphia Zoo. J. Parasitol. 2018, 104, 710–712. [Google Scholar] [CrossRef]
- Origlia, J.; Unzaga, F.; Piscopo, M.; Moré, G. Fatal Sarcocystosis in Psittacine Birds from Argentina. Parasitol. Res. 2022, 121, 491–497. [Google Scholar] [CrossRef]
- Kirejczyk, S.G.M.; Burns, R.E.; Hyatt, M.W.; Yabsley, M.J.; Ter Beest, J.M.; Gyimesi, Z.S.; Ossiboff, R.J.; Waltman, A.; Seimon, T.A.; McManamon, R. Fatal Sarcocystis falcatula Infection in Three Penguins. Front. Vet. Sci. 2019, 6, 340. [Google Scholar] [CrossRef]
- Wünschmann, A.; Rejmanek, D.; Cruz-Martinez, L.; Barr, B.C. Sarcocystis falcatula-Associated Encephalitis in a Bree-Ranging Great Horned Owl (Bubo virginianus). J. Vet. Diagn. Investig. 2009, 21, 283–287. [Google Scholar] [CrossRef]
- Olias, P.; Gruber, A.D.; Heydorn, A.O.; Kohls, A.; Mehlhorn, H.; Hafez, H.M.; Lierz, M. A Novel Sarcocystis-Associated Encephalitis and Myositis in Racing Pigeons. Avian Pathol. 2009, 38, 121–2128. [Google Scholar] [CrossRef]
- Olias, P.; Gruber, A.D.; Heydorn, A.O.; Kohls, A.; Hafez, H.M.; Lierz, M. Unusual Biphasic Disease in Domestic Pigeons (Columba livia f. domestica) Following Experimental Infection with Sarcocystis calchasi. Avian Dis. 2010, 54, 1032–1037. [Google Scholar] [CrossRef] [PubMed]
- Olias, P.; Gruber, A.D.; Kohls, A.; Hafez, H.M.; Heydorn, A.O.; Mehlhorn, H.; Lierz, M. Sarcocystis Species Lethal for Domestic Pigeons. Emerg. Infect. Dis. 2010, 16, 497–499. [Google Scholar] [CrossRef] [PubMed]
- Olias, P.; Maier, K.; Wünschmann, A.; Reed, L.; Armién, A.G.; Shaw, D.P.; Gruber, A.D.; Lierz, M. Sarcocystis calchasi has an Expanded Host Range and Induces Neurological Disease in Cockatiels (Nymphicus hollandicus) and North American Rock Pigeons (Columbia livia f. dom.). Vet. Parasitol. 2014, 200, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Maier, K.; Olias, P.; Enderlein, D.; Klopfleisch, R.; Mayr, S.L.; Gruber, A.D.; Lierz, M. Parasite Distribution and Early-Stage Encephalitis in Sarcocystis calchasi Infections in Domestic Pigeons (Columba livia f. domestica). Avian Pathol. 2015, 44, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Parmentier, S.L.; Maier-Sam, K.; Failing, K.; Enderlein, D.; Gruber, A.D.; Lierz, M. Prevalence of Sarcocystis calchasi in Free-Ranging Host Species: Accipiter Hawks and Common Woodpigeon in Germany. Sci. Rep. 2018, 8, 17610. [Google Scholar] [CrossRef] [PubMed]
- Ushio, N.; Watanabe, K.; Chambers, J.K.; Shibato, T.; Nakayama, H.; Uchida, K. Sarcocystis calchasi Encephalitis in a Rock Pigeon. J. Vet. Med. Sci. 2015, 77, 1523–1526. [Google Scholar] [CrossRef] [PubMed]
- Trupkiewicz, J.G.; Calero-Bernal, R.; Verma, S.K.; Mowery, J.; Davison, S.; Habecker, P.; Georoff, T.A.; Ialeggio, D.M.; Dubey, J.P. Acute, fatal Sarcocystis calchasi-associated hepatitis in Roller pigeons (Columba livia f. dom.) at Philadelphia Zoo. Vet. Parasitol. 2016, 216, 52–58. [Google Scholar] [CrossRef]
- Maier-Sam, K.; Kaiponen, T.; Schmitz, A.; Schulze, C.; Bock, S.; Hlinak, A.; Olias, P. Encephalitis Associated with Sarcocystis halieti Infection in a Free-Ranging Little Owl (Athene noctua). J. Wildl. Dis. 2021, 57, 712–714. [Google Scholar] [CrossRef]
- Mayr, S.L.; Maier, K.; Müller, J.; Enderlein, D.; Gruber, A.D.; Lierz, M. Accipiter hawks (Accipitridae) confirmed as definitive hosts of Sarcocystis turdusi, Sarcocystis cornixi and Sarcocystis sp. ex Phalacrocorax carbo. Parasitol. Res. 2016, 115, 3041–3047. [Google Scholar] [CrossRef]
- Gjerde, B.; Vikøren, T.; Hamnes, I.S. Molecular Identification of Sarcocystis halieti n. sp., Sarcocystis lari and Sarcocystis truncata in the Intestine of a White-tailed Sea Eagle (Haliaeetus albicilla) in Norway. Int. J. Parasitol. Parasites Wildl. 2018, 7, 1–11. [Google Scholar] [CrossRef]
- Máca, O.; González-Solís, D. Role of three Bird Species in the Life Cycle of two Sarcocystis spp. (Apicomplexa, Sarcocystidae) in the Czech Republic. Int. J. Parasitol. Parasites Wildl. 2022, 17, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Prakas, P.; Butkauskas, D.; Švažas, S.; Stanevičius, V. Morphological and Genetic Characterisation of Sarcocystis halieti from the Great Cormorant (Phalacrocorax carbo). Parasitol. Res. 2018, 117, 3663–3667. [Google Scholar] [CrossRef] [PubMed]
- Prakas, P.; Butkauskas, D.; Juozaitytė-Ngugu, E. Molecular Identification of four Sarcocystis Species in the Herring Gull, Larus argentatus, from Lithuania. Parasit. Vectors 2020, 13, 2. [Google Scholar] [CrossRef] [PubMed]
- Juozaitytė-Ngugu, E.; Butkauskas, D.; Švažas, S.; Prakas, P. Investigations on Sarcocystis Species in the Leg Muscles of the Bird Family Corvidae in Lithuania. Parasitol. Res. 2022, 121, 703–711. [Google Scholar] [CrossRef]
- Logminas, V.; Nedzinskas, V.; Drobelis, E.; Petraitis, A.; Patapavičius, R.; Žalakevičius, M.; Valius, M.; Šablevičius, B.; Gražulevičius, G.; Raudonikis, L.; et al. Lithuanian Fauna. In Birds; Mokslas Press: Vilnius, Lithuania, 1990; Volume 2. [Google Scholar]
- Kurlavičius, P.; Preikša, Ž.; Skuja, S.; Kirstukas, M.; Brazaitis, G.; Stanevičius, V.; Mačiulis, M.; Jusys, V.; Butleris, A.; Raudonikis, L.; et al. Lithuanian Breeding Bird Atlas; Lututė Press: Kaunas, Lithuania, 2006. [Google Scholar]
- Duszynski, D.W.; Box, E.D. The Opossum (Didelphis virginiana) as a Host for Sarcocystis debonei from Cowbirds (Molothrus ater) and Grackles (Cassidix mexicanus, Quiscalus quiscula). J. Parasitol. 1978, 64, 326–329. [Google Scholar] [CrossRef]
- Odening, K. The Present State of Species-Systematics in Sarcocystis Lankester, 1882 (Protista, Sporozoa, Coccidia). Syst. Parasitol. 1998, 41, 209–233. [Google Scholar] [CrossRef]
- Singh, L.A.L.; Raisinghani, P.M.; Kumar, D.; Swarankar, C.P. Clinical and Haematobiochemical Changes in Dogs Experimentally Infected with Sarcocystis capracanis. Indian J. Anim. Sci. 1993, 63, 1055–1057. [Google Scholar]
- Porter, R.A.; Ginn, P.E.; Dame, J.B.; Greiner, E.C. Evaluation of the Shedding of Sarcocystis falcatula Sporocysts in Experimentally Infected Virginia Opossums (Didelphis virginiana). Vet. Parasitol. 2001, 95, 313–319. [Google Scholar] [CrossRef]
- Prakas, P.; Butkauskas, D.; Juozaitytė-Ngugu, E. Molecular and Morphological Description of Sarcocystis kutkienae sp. nov. from the Common Raven (Corvus corax). Parasitol. Res. 2020, 119, 4205–4210. [Google Scholar] [CrossRef]
- Máca, O.; González-Solís, D. White-tailed Eagle (Haliaeetus albicilla) as the Definitive Host of Sarcocystis lutrae in the Czech Republic. Front. Vet. Sci. 2022, 9, 981829. [Google Scholar] [CrossRef]
- Prakas, P.; Balčiauskas, L.; Juozaitytė-Ngugu, E.; Butkauskas, D. The Role of Mustelids in the Transmission of Sarcocystis spp. Using Cattle as Intermediate Hosts. Animals 2021, 11, 822. [Google Scholar] [CrossRef] [PubMed]
- Juozaitytė-Ngugu, E.; Švažas, S.; Šneideris, D.; Rudaitytė-Lukošienė, E.; Butkauskas, D.; Prakas, P. The Role of Birds of the Family Corvidae in Transmitting Sarcocystis Protozoan Parasites. Animals 2021, 11, 3258. [Google Scholar] [CrossRef] [PubMed]
- Prakas, P.; Rudaitytė-Lukošienė, E.; Šneideris, D.; Butkauskas, D. Invasive American mink (Neovison vison) as Potential Definitive Host of Sarcocystis elongata, S. entzerothi, S. japonica, S. truncata and S. silva using Different Cervid Species as Intermediate Hosts. Parasitol. Res. 2021, 120, 2243–2250. [Google Scholar] [CrossRef] [PubMed]
- Prakas, P.; Moskaliova, D.; Šneideris, D.; Juozaitytė-Ngugu, E.; Maziliauskaitė, E.; Švažas, S.; Butkauskas, D. Molecular Identification of Sarcocystis rileyi and Sarcocystis sp. (Closely Related to Sarcocystis wenzeli) in Intestines of Mustelids from Lithuania. Animals 2023, 13, 467. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.K.; Lindsay, D.S.; Grigg, M.E.; Dubey, J.P. Isolation, Culture and Cryopreservation of Sarcocystis species. Curr. Protoc. Microbiol. 2017, 45, 11–127. [Google Scholar] [CrossRef]
- Kutkienė, L.; Prakas, P.; Sruoga, A.; Butkauskas, D. Sarcocystis in the birds family Corvidae with description of Sarcocystis cornixi sp. nov. from the hooded crow (Corvus cornix). Parasitol. Res. 2009, 104, 329–336. [Google Scholar] [CrossRef]
- Kutkienė, L.; Prakas, P.; Sruoga, A.; Butkauskas, D. The mallard duck (Anas platyrhynchos) as intermediate host for Sarcocystis wobeseri sp. nov. from the barnacle goose (Branta leucopsis). Parasitol. Res. 2010, 107, 879–888. [Google Scholar] [CrossRef]
- Kutkienė, L.; Prakas, P.; Butkauskas, D.; Sruoga, A. Description of Sarcocystis turdusi sp. nov. from the Common Blackbird (Turdus merula). Parasitology 2012, 139, 1438–1443. [Google Scholar] [CrossRef]
- Prakas, P.; Kutkienė, L.; Butkauskas, D.; Sruoga, A.; Zalakevičius, M. Molecular and Morphological Investigations of Sarcocystis corvusi sp. nov. from the Jackdaw (Corvus monedula). Parasitol. Res. 2013, 112, 1163–1167. [Google Scholar] [CrossRef]
- Gjerde, B. Molecular Characterisation of Sarcocystis rileyi from a Common eider (Somateria mollissima) in Norway. Parasitol. Res. 2014, 113, 3501–3509. [Google Scholar] [CrossRef]
- Rozen, S.; Skaletsky, H.J. Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics Methods and Protocols. Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2000. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K. Estimation of the Number of Nucleotide Substitutions when there are Strong Transition-Transversion and G+ C-Content Biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar] [PubMed]
- Reiczigel, J. Confidence Intervals for the Binomial Parameter: Some New Considerations. Stat. Med. 2003, 22, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Reiczigel, J.; Abonyi-Tóth, Z.; Singer, J. An Exact Confidence Set for two Binomial Proportions and Exact Unconditional Confidence Intervals for the Difference and Ratio of Proportions. Comput. Stat. Data Anal. 2008, 52, 5046–5053. [Google Scholar] [CrossRef]
- Rózsa, L.; Reiczigel, J.; Majoros, G. Quantifying Parasites in Samples of Hosts. J. Parasitol. 2000, 86, 228–232. [Google Scholar] [CrossRef]
- Solonen, T.; Heikki, L.; Sulkava, S. Diet and Brood Size in Rural and Urban Northern Goshawks Accipiter gentilis in Southern Finland. Avian Biol. Res. 2019, 12, 3–9. [Google Scholar] [CrossRef]
- Kenward, R. The Goshawk; Poyser Press: London, UK, 2006. [Google Scholar]
- Prakas, P.; Butkauskas, D.; Sruoga, A.; Švažas, S.; Kutkienė, L. Identification of Sarcocystis columbae in Wood Pigeons (Columba palumbus) in Lithuania. Vet. Zootech. 2011, 55, 33–39. [Google Scholar]
- Shadbolt, T.; Pocknell, A.; Sainsbury, A.W.; Egerton-Read, S.; Blake, D.P. Molecular Identification of Sarcocystis wobeseri-like Parasites in a New Intermediate Host Species, the White-tailed Sea Eagle (Haliaeetus albicilla). Parasitol. Res. 2021, 120, 1845–1850. [Google Scholar] [CrossRef]
- Cramp, S.; Simmons, K. (Eds.) Handbook of the Birds of Europe, the Middle East and North Africa: The Birds of the Western Palearctic; Oxford University Press: Oxford, UK, 1980; Volume 2. [Google Scholar]
- Newton, I. The Sparrowhawk; Poyser Press: London, UK, 1986. [Google Scholar]
- Panter, C.; Amar, A. Sex and Age Differences in the Diet of the Eurasian Sparrowhawk (Accipiter nisus) using Web-Sourced Photographs: Exploring the Feasibility of a New Citizen Science Approach. Ibis 2021, 163, 928–947. [Google Scholar] [CrossRef]
- Prakas, P.; Bea, A.; Juozaitytė-Ngugu, E.; Olano, I.; Villanúa, D.; Švažas, S.; Butkauskas, D. Molecular Identification of Sarcocystis halieti in the Muscles of two Species of Birds of Prey from Spain. Parasit. Vectors 2021, 14, 414. [Google Scholar] [CrossRef]
- Zawadzka, D.; Zawadzki, J. Breeding Populations and Diets of the Sparrowhawk Accipiter nisus and the Hobby Falco subbuteo in Wigry National Park (NE Poland). Acta Ornithol. 2001, 36, 25–31. [Google Scholar] [CrossRef]
- Rimoldi, G.; Speer, B.; Wellehan, J.F., Jr.; Bradway, D.S.; Wright, L.; Reavill, D.; Barr, B.C.; Childress, A.; Shivaprasad, H.L.; Chin, R.P. An Outbreak of Sarcocystis calchasi Encephalitis in Multiple Psittacine Species within an Enclosed Zoological Aviary. J. Vet. Diagn. Investig. 2013, 25, 775–781. [Google Scholar] [CrossRef] [PubMed]



| Species | Sarcocystis spp. 23LTAcc | S. calchasi | S. wobeseri |
|---|---|---|---|
| Sarcocystis spp. 23LTAcc | 100% (0) | 0.044 | 0.054 |
| S. calchasi | 95.34–95.57% | 98.14–100% (0.005) | 0.071 |
| S. wobeseri | 93.26–93.95% | 91.86–92.79% | 99.07–100% (0.001) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Šukytė, T.; Butkauskas, D.; Juozaitytė-Ngugu, E.; Švažas, S.; Prakas, P. Molecular Confirmation of Accipiter Birds of Prey as Definitive Hosts of Numerous Sarcocystis Species, including Sarcocystis sp., Closely Related to Pathogenic S. calchasi. Pathogens 2023, 12, 752. https://doi.org/10.3390/pathogens12060752
Šukytė T, Butkauskas D, Juozaitytė-Ngugu E, Švažas S, Prakas P. Molecular Confirmation of Accipiter Birds of Prey as Definitive Hosts of Numerous Sarcocystis Species, including Sarcocystis sp., Closely Related to Pathogenic S. calchasi. Pathogens. 2023; 12(6):752. https://doi.org/10.3390/pathogens12060752
Chicago/Turabian StyleŠukytė, Tautvilė, Dalius Butkauskas, Evelina Juozaitytė-Ngugu, Saulius Švažas, and Petras Prakas. 2023. "Molecular Confirmation of Accipiter Birds of Prey as Definitive Hosts of Numerous Sarcocystis Species, including Sarcocystis sp., Closely Related to Pathogenic S. calchasi" Pathogens 12, no. 6: 752. https://doi.org/10.3390/pathogens12060752
APA StyleŠukytė, T., Butkauskas, D., Juozaitytė-Ngugu, E., Švažas, S., & Prakas, P. (2023). Molecular Confirmation of Accipiter Birds of Prey as Definitive Hosts of Numerous Sarcocystis Species, including Sarcocystis sp., Closely Related to Pathogenic S. calchasi. Pathogens, 12(6), 752. https://doi.org/10.3390/pathogens12060752








