Pathogenic Potential and Antibiotic Susceptibility: A Comprehensive Study of Enterococci from Different Ecological Settings
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Isolation of the Bacteria
2.2. DNA Preparation
2.3. Species Identification
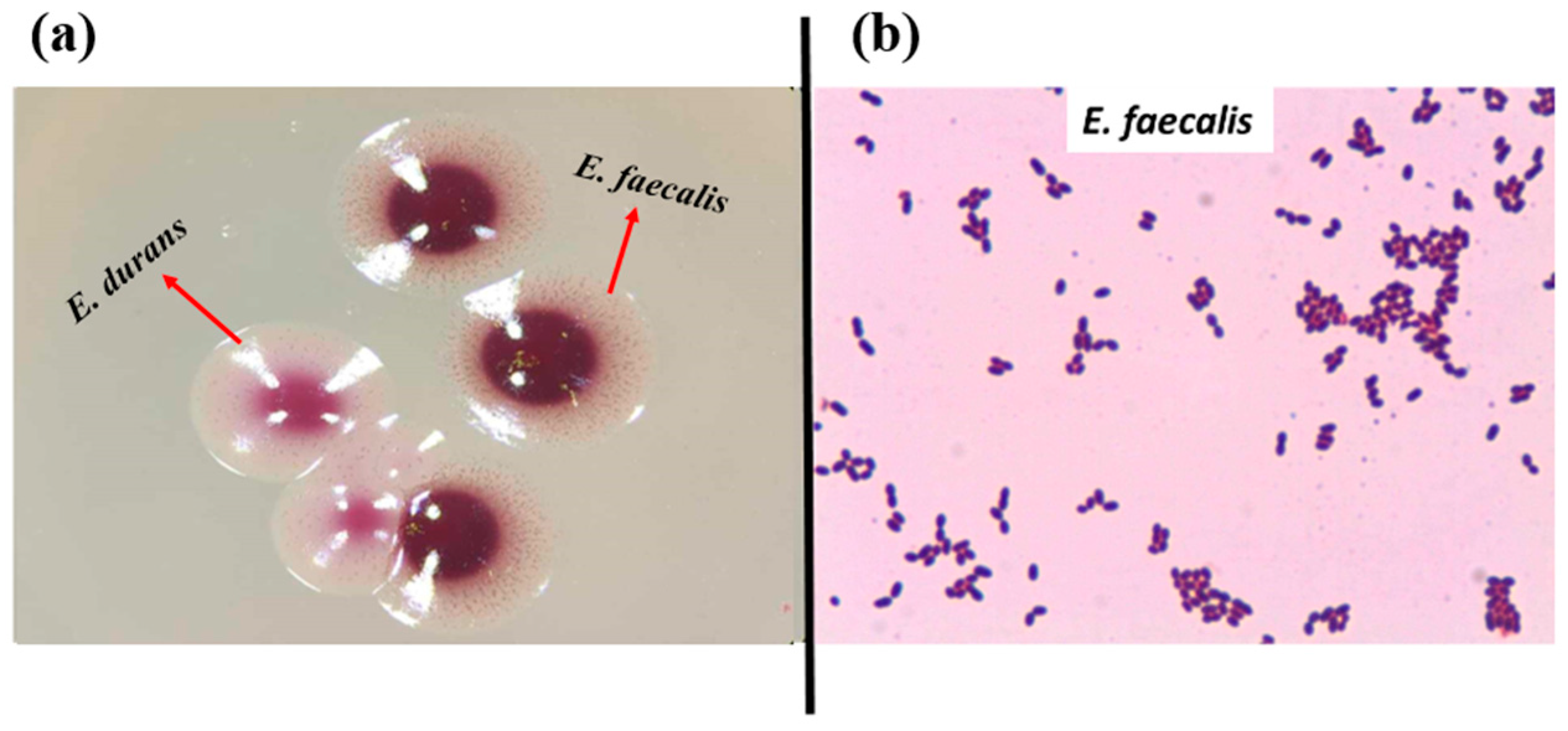
2.4. Phenotypic Hemolytic Activity Assay
2.5. Phenotypic Gelatinase Activity Assay
2.6. Antibiotic Susceptibility Testing
2.7. PCR Amplification of Virulence and Antibiotic Resistance Genes
2.8. Data Analysis
3. Results
3.1. Bacterial Isolation and Identification
3.2. Occurrence of cylB Gene and Production of Hemolysin
3.3. Occurrence of gelE and Production of Gelatinase
3.4. Phenotypic Antibiotic Resistance
3.5. Screening for Antibiotic Resistance Genes
3.6. Screening for Virulence-Associated Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jha, A.K.; Bais, H.P.; Vivanco, J.M. Enterococcus faecalis mammalian virulence-related factors exhibit potent pathogenicity in the Arabidopsis thaliana plant model. Infect Immun. 2005, 73, 464–475. [Google Scholar] [CrossRef] [PubMed]
- Micallef, S.A.; Rosenberg Goldstein, R.E.; George, A.; Ewing, L.; Tall, B.D.; Boyer, M.S.; Joseph, S.W.; Sapkota, A.R. Diversity, distribution and antibiotic resistance of Enterococcus spp. recovered from tomatoes, leaves, water and soil on U.S. Mid-Atlantic farms. Food Microbiol. 2013, 36, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Sadowy, E.; Luczkiewicz, A. Drug-resistant and hospital-associated Enterococcus faecium from wastewater, riverine estuary and anthropogenically impacted marine catchment basin. BMC Microbiol. 2014, 14, 66. [Google Scholar] [CrossRef] [PubMed]
- Nowakiewicz, A.; Ziółkowska, G.; Trościańczyk, A.; Zieba, P.; Gnat, S. Determination of resistance and virulence genes in Enterococcus faecalis and E. faecium strains isolated from poultry and their genotypic characterization by ADSRRS-fingerprinting. Poult. Sci. 2017, 96, 986–996. [Google Scholar] [CrossRef] [PubMed]
- Abriouel, H.; Omar, N.B.; Molinos, A.C.; López, R.L.; Grande, M.J.; Martínez-Viedma, P.; Ortega, E.; Cañamero, M.M.; Galvez, A. Comparative analysis of genetic diversity and incidence of virulence factors and antibiotic resistance among enterococcal populations from raw fruit and vegetable foods, water and soil, and clinical samples. Int. J. Food Microbiol. 2008, 123, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Dubin, K.; Pamer, E.G. Enterococci and Their Interactions with the Intestinal Microbiome. Microbiol Spectr. 2014, 5, 5–6. [Google Scholar] [CrossRef]
- Franz, C.M.A.P.; Schillinger, U.; Holzapfel, W.H. Production and characterization of enterocin 900, a bacteriocin produced by Enterococcus faecium BFE 900 from black olives. Int. J. Food Microbiol. 1996, 29, 255–270. [Google Scholar] [CrossRef]
- Kizheva, Y.; Georgiev, G.; Donchev, D.; Dimitrova, M.; Pandova, M.; Rasheva, I.; Hristova, P. Cross-Over Pathogenic Bacteria Detected in Infected Tomatoes (Solanum lycopersicum L.) and Peppers (Capsicum annuum L.) in Bulgaria. Pathogens 2022, 11, 1507. [Google Scholar] [CrossRef]
- Johnston, L.M.; Jaykus, L.A. Antimicrobial resistance of Enterococcus species isolated from produce. Appl. Environ. Microbiol. 2004, 70, 3133–3137. [Google Scholar] [CrossRef]
- Mundt, J.O. Occurrence of Enterococci on Plants in aWild Environment. Appl. Microbiol. 1963, 11, 141–144. [Google Scholar] [CrossRef]
- Müller, T.; Ulrich, A.; Ott, E.M.; Müller, M. Identification of plant-associated enterococci. J. Appl. Microbiol. 2001, 91, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.R.; Murray, B.E.; Rice, L.B.; Arias, C.A. Resistance in Vancomycin-Resistant Enterococci. Infect. Dis. Clin. N. Am. 2020, 34, 751–771. [Google Scholar] [CrossRef] [PubMed]
- Godfree, A.F.; Kay, D.; Wyer, M.D. Faecal streptococci as indicators of faecal contamination in water. Soc. Appl. Bacteriol. Symp. Ser. 1997, 26, 110S–119S. [Google Scholar] [CrossRef] [PubMed]
- Ott, E.M.; Müller, T.; Müller, M.; Franz, C.M.A.P.; Ulrich, A.; Gabel, M.; Seyfarth, W. Population dynamics and antagonistic potential of enterococci colonizing the phyllosphere of grasses. J. Appl. Microbiol. 2001, 91, 54–66. [Google Scholar] [CrossRef] [PubMed]
- Byappanahalli, M.N.; Nevers, M.B.; Korajkic, A.; Staley, Z.R.; Harwood, V.J. Enterococci in the Environment. Microbiol. Mol. Biol. 2012, 76, 685. [Google Scholar] [CrossRef] [PubMed]
- Švec, P.; Vandamme, P.; Bryndová, H.; Holochová, P.; Kosina, M.; Mašlaňová, I.; Sedláček, I. Enterococcus plantarum sp. nov., isolated from plants. Int. J. Syst. Evol. Microbiol. 2012, 62 Pt 7, 1499–1505. [Google Scholar] [CrossRef] [PubMed]
- Giraffa, G. Enterococci from foods. FEMS Microbiol. Rev. 2002, 26, 163–171. [Google Scholar] [CrossRef]
- Hammerum, A.M. Enterococci of animal origin and their significance for public health. Clin. Microbiol. Infect. 2012, 18, 619–625. [Google Scholar] [CrossRef]
- Leff, J.W.; Fierer, N. Bacterial communities associated with the surfaces of fresh fruits and vegetables. PLoS ONE 2013, 8, e59310. [Google Scholar] [CrossRef]
- Mogren, L.; Windstam, S.; Boqvist, S.; Vågsholm, I.; Söderqvist, K.; Rosberg, A.K.; Lindén, J.; Mulaosmanovic, E.; Karlsson, M.; Uhlig, E.; et al. The hurdle approach-A holistic concept for controlling food safety risks associated with pathogenic bacterial contamination of leafy green vegetables. A review. Front. Microbiol. 2018, 9, 1965. [Google Scholar] [CrossRef]
- Chajęcka-Wierzchowska, W.; Zarzecka, U.; Zadernowska, A. Enterococci isolated from plant-derived food—Analysis of antibiotic resistance and the occurrence of resistance genes. Food Sci. Technol. 2021, 139, 110549. [Google Scholar] [CrossRef]
- Jahan, M.; Zhanel, G.G.; Sparling, R.; Holley, R.A. Horizontal transfer of antibiotic resistance from Enterococcus faecium of fermented meat origin to clinical isolates of E. faecium and Enterococcus faecalis. Int. J. Food Microbiol. 2015, 199, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Saillant, V.; Lipuma, D.; Ostyn, E.; Joubert, L.; Boussac, A.; Guerin, H.; Brandelet, G.; Arnoux, P.; Lechardeur, D. A novel enterococcus faecalis heme transport regulator (Fhtr) senses host heme to control its intracellular homeostasis. MBio 2021, 12, e03392-e20. [Google Scholar] [CrossRef] [PubMed]
- Laissue, J.A.; Chappuis, B.B.; Müller, C.; Reubi, J.C.; Gebbers, J.O. The intestinal immune system and its relation to disease. Dig. Dis. 1993, 11, 298–312. [Google Scholar] [CrossRef] [PubMed]
- García-Díez, J.; Saraiva, C. Use of starter cultures in foods from animal origin to improve their safety. Int. J. Environ. Res. Public Health 2021, 18, 2544. [Google Scholar] [CrossRef] [PubMed]
- Gelsomino, R.; Vancanneyt, M.; Condon, S.; Swings, J.; Cogan, T.M. Enterococcal diversity in the environment of an Irish Cheddar-type cheesemaking factory. Int. J. Food Microbiol. 2001, 71, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Franz, C.M.A.P.; Huch, M.; Abriouel, H.; Holzapfel, W.; Gálvez, A. Enterococci as probiotics and their implications in food safety. Int. J. Food Microbiol. 2011, 151, 125–140. [Google Scholar] [CrossRef]
- Giraffa, G. Functionality of enterococci in dairy products. Int. J. Food Microbiol. 2003, 88, 215–222. [Google Scholar] [CrossRef]
- Lengfelder, I.; Sava, I.G.; Hansen, J.J.; Kleigrewe, K.; Herzog, J.; Neuhaus, K.; Ho_man, T.; Sartor, R.B.; Haller, D. Complex bacterial consortia reprogram the colitogenic activity of Enterococcus faecalis in a gnotobiotic mouse model of chronic, immune-mediated colitis. Front. Immunol. 2019, 10, 1420. [Google Scholar] [CrossRef]
- Staley, C.; Dunny, G.M.; Sadowsky, M.J. Environmental and Animal-Associated Enterococci. Adv. Appl. Microbiol. 2014, 87, 147–186. [Google Scholar] [CrossRef]
- Shankar, P.; Chung, R.; Frank, D.A. Association of food insecurity with children’s behavioral, emotional, and academic outcomes: A systematic review. J. Dev. Pediatr. 2017, 38, 135–150. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.A.; Murray, B.E. The rise of the Enterococcus: Beyond vancomycin resistance. Nat. Rev. Microbiol. 2012, 10, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Klein, G.; Pack, A.; Reuter, G. Antibiotic resistance patterns of enterococci and occurrence of vancomycin-resistant enterococci in raw minced beef and pork in Germany. Appl. Environ. Microbiol. 1998, 64, 1825–1830. [Google Scholar] [CrossRef] [PubMed]
- Tendolkar, P.M.; Baghdayan, A.S.; Shankar, N. Pathogenic enterococci: New developments in the 21st century. Cell. Mol. Life Sci. 2003, 60, 2622–2636. [Google Scholar] [CrossRef] [PubMed]
- Giacobbe, D.R.; Battaglini, D.; Ball, L.; Brunetti, I.; Bruzzone, B.; Codda, G.; Crea, F.; De Maria, A.; Dentone, C.; Di Biagio, A.; et al. Bloodstream infections in critically ill patients with COVID-19. Eur. J. Clin. Investig. 2020, 50, e13319. [Google Scholar] [CrossRef] [PubMed]
- Kokkoris, S.; Papachatzakis, I.; Gavrielatou, E.; Ntaidou, T.; Ischaki, E.; Malachias, S.; Vrettou, C.; Nichlos, C.; Kanavou, A.; Zervakis, D.; et al. ICU-acquired bloodstream infections in critically ill patients with COVID-19. J. Hosp. Infect. 2021, 107, 95–97. [Google Scholar] [CrossRef] [PubMed]
- Patrier, J.; Villageois-Tran, K.; Szychowiak, P.; Ruckly, S.; Gschwind, R.; Wicky, P.H.; Gueye, S.; Armand-Lefevre, L.; Marzouk, M.; Sonneville, R.; et al. Oropharyngeal and intestinal concentrations of opportunistic pathogens are independently associated with death of SARS-CoV-2 critically ill adults. J. Crit. Care. 2022, 26, 300. [Google Scholar] [CrossRef]
- Protonotariou, E.; Mantzana, P.; Meletis, G.; Tychala, A.; Kassomenaki, A.; Vasilaki, O.; Kagkalou, G.; Gkeka, I.; Archonti, M.; Kati, S.; et al. Microbiological characteristics of bacteremias among COVID-19 hospitalized patients in a tertiary referral hospital in Northern Greece during the second epidemic wave. FEMS Microbes 2022, 2, xtab021. [Google Scholar] [CrossRef]
- Tao, S.; Chen, H.; Li, N.; Fang, Y.; Xu, Y.; Liang, W. Association of CRISPR-Cas System with the Antibiotic Resistance and Virulence Genes in Nosocomial Isolates of Enterococcus. Infect. Drug Resist. 2022, 15, 6939–6949. [Google Scholar] [CrossRef]
- Toc, D.A.; Butiuc-Keul, A.L.; Iordache, D.; Botan, A.; Mihaila, R.M.; Costache, C.A.; Colosi, I.A.; Chiorean, C.; Neagoe, D.S.; Gheorghiu, L.; et al. Descriptive Analysis of Circulating Antimicrobial Resistance Genes in Vancomycin-Resistant Enterococcus (VRE) during the COVID-19 Pandemic. Biomedicines 2022, 10, 1122. [Google Scholar] [CrossRef]
- Lopes, M.; Simões, A.; Tenreiro, R.; Joaquim, J.; Marques, F.; Crespo, M. Activity and expression of a virulence factor, gelatinase, in dairy enterococci. Int. J. Food Microbiol. 2006, 112, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Leavis, H.; Top, J.; Shankar, N.; Borgen, K.; Bonten, M.; Van Embden, J.; Willems, R.J.L. A novel putative enterococcal pathogenicity island linked to the esp virulence gene of Enterococcus faecium and associated with epidemicity. J. Bacteriol. 2004, 186, 672–682. [Google Scholar] [CrossRef] [PubMed]
- Kiruthiga, A.; Padmavathy, K.; Shabana, P.; Naveenkumar, V.; Gnanadesikan, S.; Malaiyan, J. Improved detection of esp, hyl, asa1, gelE, cylA virulence genes among clinical isolates of Enterococci. BMC Res. Notes. 2020, 13, 170. [Google Scholar] [CrossRef] [PubMed]
- Frazzon, A.P.; Gama, B.A.; Hermes, V.; Bierhals, C.G.; Pereira, R.I.; Guedes, A.G.; d’Azevedo, P.A.; Frazzon, J. Prevalence of antimicrobial resistance and molecular characterization of tetracycline resistance mediated by tet(M) and tet(L) genes in Enterococcus spp. isolated from food in Southern Brazil. World J. Microbiol. Biotechnol. 2010, 26, 365–370. [Google Scholar] [CrossRef]
- Cattoir, V. The multifaceted lifestyle of enterococci: Genetic diversity, ecology and risks for public health. Curr. Opin. Microbiol. 2022, 65, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Koleva, Z.; Dedov, I.; Kizheva, J.; Lipovanska, R.; Moncheva, P.; Hristova, P. Lactic acid microflora of the gut of snail Cornu aspersum. Biotechnol. Biotechnol. Equip. 2014, 28, 627–634. [Google Scholar] [CrossRef]
- Jackson, C.R.; Fedorka-Cray, P.J.; Barrett, J.B. Use of a genus- and species-specific multiplex PCR for identification of enterococci. J. Clin. Microbiol. 2004, 42, 3558–3565. [Google Scholar] [CrossRef]
- Yoon, J.H.; Lee, S.T.; Park, Y.H. Inter- and intraspecific phylogenetic analysis of the genus Nocardioides and related taxa based on 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 1998, 48, 187–194. [Google Scholar] [CrossRef]
- Carrillo, P.G.; Mardaraz, C.; Pitta-Alvarez, S.I.; Giulietti, A.M. Isolation and selection of biosurfactant-producing bacteria. World J. Microbiol. Biotechnol. 1996, 12, 82–84. [Google Scholar] [CrossRef]
- Mangia, N.P.; Saliba, L.; Deiana, P. Functional and safety characterization of autochthonous Lactobacillus paracasei FS103 isolated from sheep cheese and its survival in sheep and cow fermented milks during cold storage. Ann. Microbiol. 2019, 69, 161–170. [Google Scholar] [CrossRef]
- Bauer, A.W.; Kirby, W.M.M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standard single disk method. Am. J. Clin. Pathol. 1966, 36, 493–496. [Google Scholar] [CrossRef]
- EUCAST Version 13.1, 2023. (n.d.). Available online: http://www.eucast.org/expert_rules_and_intrinsic_resistance (accessed on 10 August 2023).
- McBride, S.M.; Fischetti, V.A.; LeBlanc, D.J.; Moellering, R.C.; Gilmore, M.S. Genetic diversity among Enterococcus faecalis. PLoS ONE 2007, 2, e582. [Google Scholar] [CrossRef] [PubMed]
- Eaton, T.J.; Gasson, M.J. Molecular Screening of Enterococcus virulence Determinants and Potential for Genetic Exchange between Food and Medical Isolates. Appl. Environ. Microbiol. 2001, 67, 1628–1635. [Google Scholar] [CrossRef] [PubMed]
- Mannu, L.; Paba, A.; Daga, E.; Comunian, R.; Zanetti, S.; Duprè, I.; Sechi, L.A. Comparison of the incidence of virulence determinants and antibiotic resistance between Enterococcus faecium strains of dairy, animal and clinical origin. Int. J. Food Microbiol. 2003, 88, 291–304. [Google Scholar] [CrossRef] [PubMed]
- van Asselt, G.J.; Vliegenthart, J.S.; Petit, P.L.C.; van de Klundert, J.A.M.; Mouton, R.P. High-level aminoglycoside resistance among enterococci and group A streptococci. J. Antimicrob. Chemother. 1992, 30, 651–659. [Google Scholar] [CrossRef]
- Garofalo, C.; Vignaroli, C.; Zandri, G.; Aquilanti, L.; Bordoni, D.; Osimani, A.; Clementi, F.; Biavasco, F. Direct detection of antibiotic resistance genes in specimens of chicken and pork meat. Int. J. Food Microbiol. 2007, 113, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Aarestrup, F.M.; Agerso, Y.; Gerner–Smidt, P.; Madsen, M.; Jensen, L.B. Comparison of antimicrobial resistance phenotypes and resistance genes in Enterococcus faecalis and Enterococcus faecium from humans in the community, broilers, and pigs in Denmark. Diagn. Microbiol. Infect. Dis. 2000, 37, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.B.; Frimodt-Møller, N.; Aarestrup, F.M. Presence of erm gene classes in gram-positive bacteria of animal and human origin in Denmark. FEMS Microbiol. Lett. 1999, 170, 151–158. [Google Scholar] [CrossRef]
- Jia, W.; Li, G.; Wang, W. Prevalence and Antimicrobial Resistance of Enterococcus Species: A Hospital-Based Study in China. Int. J. Environ. 2014, 11, 3424–3442. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, Z.Y.; Dong, K.; Yuan, J.P.; Guo, X.K. Antibiotic resistance of probiotic strains of lactic acid bacteria isolated from marketed foods and drugs. Biomed. Environ. Sci. 2009, 22, 401–412. [Google Scholar] [CrossRef]
- Ferchichi, M.; Sebei, K.; Boukerb, A.M.; Karray-Bouraoui, N.; Chevalier, S.; Feuilloley, M.G.J.; Connil, N.; Zommiti, M. Enterococcus spp.: Is It a Bad Choice for a Good Use—A Conundrum to Solve? Microorganisms 2021, 9, 2222. [Google Scholar] [CrossRef] [PubMed]
- Gunnar Kahlmeter and the EUCAST Steering Committee 2019, Redefining Susceptibility Testing Categories S, I and R. Available online: https://www.eucast.org/newsiandr (accessed on 10 August 2023).
- Hiltunen, T.; Virta, M.; Laine, A.L. Antibiotic resistance in the wild: An eco-evolutionary perspective. Philos. Trans. R Soc. Lond B Biol. Sci. 2017, 372, 20160039. [Google Scholar] [CrossRef] [PubMed]
- de Kraker, M.E.; Jarlier, V.; Monen, J.C.; Heuer, O.E.; van de Sande, N.; Grundmann, H. The changing epidemiology of bacteraemias in Europe: Trends from the European Antimicrobial Resistance Surveillance System. Clin. Microbiol. Infect. 2013, 19, 860–868. [Google Scholar] [CrossRef] [PubMed]
- McAuley, C.M.; Britz, M.L.; Gobius, K.S.; Craven, H.M. Prevalence, seasonality, and growth of enterococci in raw and pasteurized milk in Victoria, Australia. J. Dairy Sci. 2015, 98, 8348–8358. [Google Scholar] [CrossRef]
- Gołaś-Prądzyńska, M.; Łuszczyńska, M.; Rola, J.G. Dairy Products: A Potential Source of Multidrug-Resistant Enterococcus faecalis and Enterococcus faecium Strains. Foods 2022, 11, 4116. [Google Scholar] [CrossRef]
- Martín, R.; Langa, S.; Reviriego, C.; Jiménez, E.; Marín, M.L.; Xaus, J.; Fernández, L.; Rodríguez, J.M. Human milk is a source of lactic acid bacteria for the infant gut. J. Pediatr. 2003, 143, 754–758. [Google Scholar] [CrossRef]
- Laursen, M.F.; Larsson, M.W.; Lind, M.V.; Larnkjær, A.; Mølgaard, C.; Michaelsen, K.F.; Bahl, M.I.; Licht, T.R. Intestinal Enterococcus abundance correlates inversely with excessive weight gain and increased plasma leptin in breastfed infants. FEMS Microbiol. Ecol. 2021, 96, fiaa066. [Google Scholar] [CrossRef]
- Wajda, Ł.; Ostrowski, A.; Błasiak, E.; Godowska, P. Enterococcus faecium Isolates Present in Human Breast Milk Might Be Carriers of Multi-Antibiotic Resistance Genes. Bacteria 2022, 1, 66–87. [Google Scholar] [CrossRef]
- Huang, M.S.; Cheng, C.C.; Tseng, S.Y.; Lin, Y.L.; Lo, H.M.; Chen, P.W. Most commensally bacterial strains in human milk of healthy mothers display multiple antibiotic resistance. MicrobiologyOpen 2019, 8, e00618. [Google Scholar] [CrossRef]
- Franz, C.M.; Muscholl-Silberhorn, A.B.; Yousif, N.M.; Vancanneyt, M.; Swings, J.; Holzapfel, W.H. Incidence of virulence factors and antibiotic resistance among Enterococci isolated from food. Appl. Environ. Microbiol. 2001, 67, 4385–4389. [Google Scholar] [CrossRef]
- Santana, L.A.M.; Andrade, N.N.N.; da Silva, L.S.C.; Oliveira, C.N.T.; de Brito, B.B.; de Melo, F.F.; Souza, C.L.; Marques, L.M.; Oliveira, M.V. Identification and characterization of resistance and pathogenicity of Enterococcus spp. in samples of donor breast milk. World J. Clin. Pediatr. 2020, 9, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Huycke, M.M.; Spiegel, C.A.; Gilmore, M.S. Bacteremia caused by hemolytic, high-level gentamicin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 1991, 35, 1626–1634. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, M.S.; Segarra, R.A.; Booth, M.C.; Bogie, C.P.; Hall, L.R.; Clewell, D.B. Genetic structure of the Enterococcus faecalis plasmid pAD1-encoded cytolytic toxin system and its relationship to lantibiotic determinants. J. Bacteriol. 1994, 176, 7335–7344. [Google Scholar] [CrossRef] [PubMed]
- Haas, W.; Shepard, B.D.; Gilmore, M.S. Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction. Nature 2002, 415, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Haas, W.; Gilmore, M.S. Molecular nature of a novel bacterial toxin: The cytolysin of Enterococcus faecalis. Med. Microbiol. Immunol. 1999, 187, 183–190. [Google Scholar] [CrossRef]
- Ike, Y.; Clewell, D.B.; Segarra, R.A.; Gilmore, M.S. Genetic analysis of the pAD1 hemolysin/bacteriocin determinant in Enterococcus faecalis: Tn917 insertional mutagenesis and cloning. J. Bacteriol. 1990, 172, 155–163. [Google Scholar] [CrossRef]
- Vergis, E.N.; Shankar, N.; Chow, J.W.; Hayden, M.K.; Snydman, D.R.; Zervos, M.J.; Linden, P.K.; Wagener, M.M.; Muder, R.R. Association between the presence of enterococcal virulence factors gelatinase, hemolysin, and enterococcal surface protein and mortality among patients with bacteremia due to Enterococcus faecalis. Clin. Infect. Dis. 2002, 35, 570–575. [Google Scholar] [CrossRef]
- Semedo, T.; Santos, M.A.; Lopes, M.F.; Figueiredo Marques, J.J.; Barreto Crespo, M.T.; Tenreiro, R. Virulence factors in food, clinical and reference Enterococci: A common trait in the genus? Syst. Appl. Microbiol. 2003, 26, 13–22. [Google Scholar] [CrossRef]
- Creti, R.; Imperi, M.; Bertuccini, L.; Fabretti, F.; Orefici, G.; Di Rosa, R.; Baldassarri, L. Survey for virulence determinants among Enterococcus faecalis isolated from different sources. J. Med. Microbiol. 2004, 53 Pt 1, 13–20. [Google Scholar] [CrossRef]






| Primer | Sequence (5′ to 3′) | Tm (°C) | Product Size (bp) | Reference |
|---|---|---|---|---|
| Primers for virulence-related genes | ||||
| cylB-F | GGAGAATTAGTGTTTAGAGCG | 57 | 522 | [53] |
| cylB-R | GCTTCATAACCATTGTTACTATAGAAAC | |||
| esp-F | CGATAAAGAGAGAGCGGAG | 57 | 539 | [53] |
| esp-R | GCAAACTCTACATCCACGTC | |||
| gls24-F | GCATTAGATGAGATTGATGGTC | 54 | 446 | [53] |
| gls24-R | GCGAGGTTCAGTTTCTTC | |||
| psaA-F | CTATTTTGCAGCAAGTGATG | 54 | 540 | [53] |
| psaA-R | CGCATAGTAACTATCACCATCTTG | |||
| agg-F | AAGAAAAAGAAGTAGACCAAC | 54 | 1553 | [54] |
| agg-R | AAACGGCAAGACAAGTAAATA | |||
| ace-F | AAAGTAGAATTAGATCACAC | 51 | 320 | [55] |
| ace-R | TCTATCACATTCGGTTGCG | |||
| gelE-F | ACCCCGTATCATTGGTTT | 51 | 419 | [54] |
| gelE-R | ACGCATTGCTTTTCCATC | |||
| nucl-F | GTGTAAAAGAAGTTACTGAAAATGTTACTC | 62 | 332 | [53] |
| nucl-R | GCGTTTTTTGTAGTAATGTTCCATCTACG | |||
| Primers for antibiotic resistance-related genes | ||||
| aac6′-aph2″-F | CTGATGAGATAGTCTATGGTATGGATC | 65 | 375 | [53] |
| aac6′-aph2″-R | GCCACACTATCATAACCACTACCG | |||
| aphA-F | GCCGATGTGGATTGCGAAAA | 55 | 292 | [56] |
| aphA-R | GCTTGATCCCCAGTAAGTCA | |||
| blaZ-F | ACTTCAACACCTGCTGCTTTC | 60 | 240 | [57] |
| blaZ-R | TAGGTTCAGATTGGCCCTTAG | |||
| catpIP501-F | GGATATGAAATTTATCCCTC | 50 | 486 | [58] |
| catpIP501-R | CAATCATCTACCCTATGAAT | |||
| gyrA-F | ACTTGAAGATGTTTTAGGTGAT | 55 | 559 | [59] |
| gyrA-R | TTAGGAAATCTTGATGGCAA | |||
| erm-F | CATTTAACGACGAAACTGGC | 55 | 726 | [59] |
| erm-R | GGAACATCTGTGGTATGGCG | |||
| ermB-F | CATTTAACGACGAAACTGGC | 52 | 405 | [59] |
| ermB-R | GGAACATCTGTGGTATGGCG | |||
| mefA-F | ACTATCATTAATCACTAGTGC | 52 | 346 | [60] |
| mefA-R | TTCTTCTGGTACTAAAAGTGG | |||
| vanA36-F | TTGCTCAGAGGAGCATGACG | 65 | 957 | [61] |
| vanA992-R | TCGGGAAGTGCAATACCTGC | |||
| No | Isolate | Origin | Species | Method of Identification | No. | Isolate | Origin | Species | Method of Identification |
|---|---|---|---|---|---|---|---|---|---|
| 1 | CA1 | C. aspersum | E. mundtii | MALDI-TOF | 37 | BY8 | Bulgarian yogurt | Enterococcus sp. | Sequencing |
| 2 | CA2 | C. aspersum | E. casseliflavus | PCR, Sequencing | 38 | BY9 | Bulgarian yogurt | E. casseliflavus | PCR, Sequencing |
| 3 | CA3 | C. aspersum | E. gilvus | MALDI-TOF | 39 | BY10 | Bulgarian yogurt | E. faecalis | PCR |
| 4 | CA4 | C. aspersum | E. mundtii | MALDI-TOF | 40 | BY11 | Bulgarian yogurt | E. faecalis | PCR |
| 5 | CA5 | C. aspersum | E. casseliflavus | PCR, Sequencing | 41 | BY12 | Bulgarian yogurt | E. faecium | PCR |
| 6 | CA6 | C. aspersum | E. mundtii | Sequencing | 42 | BY13 | Bulgarian yogurt | E. faecium | PCR |
| 7 | CA7 | C. aspersum | E. mundtii | MALDI-TOF | 43 | BY14 | Bulgarian yogurt | E. faecium | PCR |
| 8 | CA8 | C. aspersum | E. pseudoavium | Sequencing | 44 | BY15 | Bulgarian yogurt | E. faecium | PCR |
| 9 | CA9 | C. aspersum | E. pseudoavium | Sequencing | 45 | BY16 | Bulgarian yogurt | E. faecium | PCR |
| 10 | CA10 | C. aspersum | E. pallens | Sequencing | 46 | BY17 | Bulgarian yogurt | E. gallinarum | MALDI-TOF |
| 11 | CA11 | C. aspersum | E. malodoratus | MALDI-TOF | 47 | BY18 | Bulgarian yogurt | E. casseliflavus | PCR, Sequencing |
| 12 | CA12 | C. aspersum | E. casseliflavus | PCR, Sequencing | 48 | BY19 | Bulgarian yogurt | E. casseliflavus | PCR, Sequencing |
| 13 | CA13 | C. aspersum | E. devriesei | Sequencing | 49 | BY20 | Bulgarian yogurt | E. casseliflavus | PCR, Sequencing |
| 14 | CA14 | C. aspersum | E. gallinarum | Sequencing | 50 | BY21 | Bulgarian yogurt | E. casseliflavus | PCR, Sequencing |
| 15 | CA15 | C. aspersum | E. gallinarum | Sequencing | 51 | BY22 | Bulgarian yogurt | E. faecalis | PCR |
| 16 | CA16 | C. aspersum | E. devriesei | MALDI-TOF | 52 | BY23 | Bulgarian yogurt | E. faecalis | PCR |
| 17 | CA17 | C. aspersum | E. mundtii | MALDI-TOF | 53 | BY24 | Bulgarian yogurt | E. faecalis | PCR |
| 18 | CM1 | Cow milk | E. faecium | PCR | 54 | BY25 | Bulgarian yogurt | E. faecalis | PCR |
| 19 | CM2 | Cow milk | E. durans | MALDI-TOF | 55 | BY26 | Bulgarian yogurt | E. faecalis | PCR |
| 20 | CM3 | Cow milk | E. durans | MALDI-TOF | 56 | BY27 | Bulgarian yogurt | E. faecalis | PCR |
| 21 | CM4 | Cow milk | E. faecalis | MALDI-TOF | 57 | BM1 | Breast milk | E. faecalis | PCR |
| 22 | YFC1 | Young feta cheese | E. faecalis | PCR | 58 | BM2 | Breast milk | E. faecalis | PCR |
| 23 | YFC2 | Young feta cheese | E. durans | PCR | 59 | BM3 | Breast milk | E. faecalis | PCR |
| 24 | YFC3 | Young feta cheese | E. faecalis | PCR | 60 | BM4 | Breast milk | E. faecalis | PCR |
| 25 | YFC4 | Young feta cheese | E. durans | PCR | 61 | BM5 | Breast milk | E. faecalis | PCR |
| 26 | YFC5 | Young feta cheese | E. durans | PCR | 62 | BM6 | Breast milk | E. faecalis | PCR |
| 27 | MFC1 | Matured feta cheese | E. faecium | PCR | 63 | BM7 | Breast milk | E. faecalis | PCR |
| 28 | MFC2 | Matured feta cheese | E. faecium | PCR | 64 | BM8 | Breast milk | E. faecalis | PCR |
| 29 | DK1 | Doner kebab | E. faecium | PCR | 65 | BM9 | Breast milk | E. faecalis | PCR |
| 30 | BY1 | Bulgarian yogurt | E. faecium | MALDI-TOF | 66 | BM10 | Breast milk | E. faecalis | PCR |
| 31 | BY2 | Bulgarian yogurt | E. faecalis | PCR | 67 | BM11 | Breast milk | E. faecalis | PCR |
| 32 | BY3 | Bulgarian yogurt | E. faecalis | PCR | 68 | BM12 | Breast milk | E. faecalis | PCR |
| 33 | BY4 | Bulgarian yogurt | E. faecalis | PCR | 69 | BM13 | Breast milk | E. faecalis | PCR |
| 34 | BY5 | Bulgarian yogurt | E. faecalis | PCR | 70 | BM14 | Breast milk | E. faecalis | PCR |
| 35 | BY6 | Bulgarian yogurt | E. faecalis | PCR | 71 | BM15 | Breast milk | E. faecalis | PCR |
| 36 | BY7 | Bulgarian yogurt | Enterococcus sp. | Sequencing | 72 | BM16 | Breast milk | E. faecalis | PCR |
| ABR Phenotype | Number of Isolates | Species Identification | Origin of Isolation |
|---|---|---|---|
| One Antibiotic | |||
| AMP | 12 | E. faecium DK1 | Doner kebab |
| E. gallinarum BY17 | Bulgarian yogurt | ||
| E. mundtii CA1 | C. aspersum | ||
| E. malodoratus CA11 | |||
| E. devriesei CA13 | |||
| E. faecalis BM3 | Human breast milk | ||
| E. faecalis BM4 | |||
| E. faecalis BM5 | |||
| E. faecalis BM6 | |||
| E. faecalis BM9 | |||
| E. faecalis BM12 | |||
| E. faecalis BM14 | |||
| Two antibiotics | |||
| AMP + ERV | 1 | E. faecium CM1 | Cow milk |
| AMP + TG | 1 | E. faecalis YFC1 | Young feta cheese |
| GEN + RP | 1 | E. faecalis BM7 | Human breast milk |
| Three antibiotics | |||
| AMP + HLS + RP | 1 | E. faecalis BM15 | Human breast milk |
| Strains | Virulence Genes | Strains | Virulence Genes | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cyl B | esp | gls 24 | nucl | psa | agg | gel E | ace | cyl B | esp | gls 24 | nucl | psa | agg | gel E | ace | ||
| E. faecium CM1 | E. faecalis BY25 | ||||||||||||||||
| E. durans CM2 | E. faecalis BY26 | ||||||||||||||||
| E. durans CM3 | E. faecalis BY27 | ||||||||||||||||
| E. faecalis CM4 | E. mundtii CA1 | ||||||||||||||||
| E. faecalis YFC1 | E. casseliflavus CA2 | ||||||||||||||||
| E. durans YFC2 | E. gilvus CA3 | ||||||||||||||||
| E. faecalis YFC3 | E. mundtii CA4 | ||||||||||||||||
| E. durans YFC4 | E. casseliflavus CA5 | ||||||||||||||||
| E. durans YFC5 | E. mundtii CA6 | ||||||||||||||||
| E. faecium MFC1 | E. mundtii CA7 | ||||||||||||||||
| E. faecium MFC2 | E. pseudoavium CA8 | ||||||||||||||||
| E. faecium DK1 | E. pseudoavium CA9 | ||||||||||||||||
| E. faecium BY1 | E. pallens CA10 | ||||||||||||||||
| E. faecalis BY2 | E. maloduratus CA11 | ||||||||||||||||
| E. faecalis BY3 | E. casseliflavus CA12 | ||||||||||||||||
| E. faecalis BY4 | E. devriesei CA13 | ||||||||||||||||
| E.faecalis BY5 | E. gallinarum CA14 | ||||||||||||||||
| E.faecalis BY6 | E. gallinarum CA15 | ||||||||||||||||
| E. species BY7 | E. devriesei CA16 | ||||||||||||||||
| E. species BY8 | E. mundtii CA17 | ||||||||||||||||
| E. casseliflavus BY9 | E. faecalis BM1 | ||||||||||||||||
| E. faecalis BY10 | E. faecalis BM2 | ||||||||||||||||
| E. faecalis BY11 | E. faecalis BM3 | ||||||||||||||||
| E. faecium BY12 | E. faecalis BM4 | ||||||||||||||||
| E. faecium BY13 | E. faecalis BM5 | ||||||||||||||||
| E. faecium BY14 | E. faecalis BM6 | ||||||||||||||||
| E. faecium BY15 | E. faecalis BM7 | ||||||||||||||||
| E. faecium BY16 | E. faecalis BM8 | ||||||||||||||||
| E. gallinarum BY17 | E. faecalis BM9 | ||||||||||||||||
| E. casseliflavus BY18 | E. faecalis BM10 | ||||||||||||||||
| E. casseliflavus BY19 | E. faecalis BM11 | ||||||||||||||||
| E. casseliflavus BY20 | E. faecalis BM12 | ||||||||||||||||
| E. casseliflavus BY21 | E. faecalis BM13 | ||||||||||||||||
| E. faecalis BY22 | E. faecalis BM14 | ||||||||||||||||
| E. faecalis BY23 | E. faecalis BM15 | ||||||||||||||||
| E. faecalis BY24 | E. faecalis BM16 | ||||||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandova, M.; Kizheva, Y.; Tsenova, M.; Rusinova, M.; Borisova, T.; Hristova, P. Pathogenic Potential and Antibiotic Susceptibility: A Comprehensive Study of Enterococci from Different Ecological Settings. Pathogens 2024, 13, 36. https://doi.org/10.3390/pathogens13010036
Pandova M, Kizheva Y, Tsenova M, Rusinova M, Borisova T, Hristova P. Pathogenic Potential and Antibiotic Susceptibility: A Comprehensive Study of Enterococci from Different Ecological Settings. Pathogens. 2024; 13(1):36. https://doi.org/10.3390/pathogens13010036
Chicago/Turabian StylePandova, Maria, Yoana Kizheva, Margarita Tsenova, Mariya Rusinova, Tsvetomira Borisova, and Petya Hristova. 2024. "Pathogenic Potential and Antibiotic Susceptibility: A Comprehensive Study of Enterococci from Different Ecological Settings" Pathogens 13, no. 1: 36. https://doi.org/10.3390/pathogens13010036
APA StylePandova, M., Kizheva, Y., Tsenova, M., Rusinova, M., Borisova, T., & Hristova, P. (2024). Pathogenic Potential and Antibiotic Susceptibility: A Comprehensive Study of Enterococci from Different Ecological Settings. Pathogens, 13(1), 36. https://doi.org/10.3390/pathogens13010036






