Abstract
Previously, we described the RNA recombinants accumulating in tissues infected with the bromoviruses BMV (Brome mosaic virus) and CCMV (Cowpea chlorotic mottle virus). In this work, we characterize the recombinants encapsidated inside the purified virion particles of BMV and CCMV. By using a tool called the Viral Recombination Mapper (ViReMa) that detects recombination junctions, we analyzed a high number of high-throughput sequencing (HTS) short RNA sequence reads. Over 28% of BMV or CCMV RNA reads did not perfectly map to the viral genomes. ViReMa identified 1.40% and 1.83% of these unmapped reads as the RNA recombinants, respectively, in BMV and CCMV. Intra-segmental crosses were more frequent than the inter-segmental ones. Most intra-segmental junctions carried short insertions/deletions (indels) and caused frameshift mutations. The mutation hotspots clustered mainly within the open reading frames. Substitutions of various lengths were also identified, whereas a small fraction of crosses occurred between viral and their host RNAs. Our data reveal that the virions can package detectable amounts of multivariate recombinant RNAs, contributing to the flexible nature of the viral genomes.
Keywords:
RNA viruses; RNA-seq; mutation profiling; RNA recombination; substitutions; deletions; SNPs; Bowtie; ViReMa 1. Introduction
Plus-sense RNA viruses rapidly adapt to new environments, and the processes of RNA recombination diversify viral populations [1,2]. Homologous and non-homologous RNA recombination can lead to the accumulation of beneficial mutations but can also eliminate the harmful ones. These recombinations can reshuffle larger portions of the viral genome, generating defective RNAs, or cause crossovers among different viral or viral and host RNAs [3,4]. RNA recombinants have been described in various animal viruses [5,6,7,8,9,10], plant viruses [11,12], bacteriophages [13], negative-sense RNA viruses [14], and retroviruses [15]. Defective (D) or defective interfering (DI) RNAs have been identified for both animal and plant RNA viruses, either in natural infections or in cell culture.
Bromoviridae are tripartite plus-stranded RNA viruses of plants. The Brome mosaic virus (BMV) is one of the best-characterized bromoviruses. Its virions separately encapsidate RNA1, RNA2, or RNA3, plus subgenomic [sg] RNA4 components [16]. RNA recombination has been studied extensively for BMV. Both homologous and nonhomologous crossovers were detected among RNA segments or within the same RNA segment [4]. Mapping the cross sites revealed recombination hot spots near the secondary structures [17], while mutations in 1a and 2a proteins changed the crossover hotspots [18,19]. RNA recombination was also demonstrated in the Cowpea chlorotic mottle virus (CCMV) using transgenically-expressed viral RNA fragments [20]. These and other observations strongly suggested replicase-, but also the coat protein (CP)-, mediated template switching processes [16,21,22,23].
The various kinds of BMV RNA recombinants described to date were analyzed in total RNA extracts from the infected tissue. We previously described that BMV and broad bean mottle virus (BBMV) virions could package the host cellular RNA sequences [24,25]. To find out if virions could package the recombinant variants, in this work we analyzed BMV or CCMV RNAs extracted from extensively purified viral preparations. By using the techniques of high-throughput sequencing (HTS) and a recently developed algorithm named Viral Recombination Mapper (ViReMa), described in [26], we characterized groups of encapsidated recombinant RNAs. These included intra- and inter-segmental crossings, virus-to-host recombinants, as well as larger insertions and substitutions. Overall, our results reveal populations of RNA variants (RNA cloud) that can be carried in viral particles, in addition to their canonical genomic RNA components.
2. Materials and Methods
2.1. Virus Propagation, Purification, and RNA Extraction
BMV (Russian strain) was propagated in barley seedlings and purified by ultracentrifugation in a sucrose gradient, essentially as described in [27]. Similar protocols were utilized for purification of CCMV (Bawden strain) that was grown in cowpea seedlings (cv. California blackeye #5). After collecting the distinct opalescent virus bands from sucrose gradients, the virions were pelleted down and the glassy pellet re-suspended in the virus buffer (VB; 0.05 M NaOAc, 0.008 M Mg(OAc)2, pH 4.5), by soaking overnight. Then, the virus preparations were treated with RNase and DNase to eliminate co-purifying host nucleic acids, and the nucleases were removed by centrifugal concentration in Amicon Ultra 4–5 mL concentrators (UFC810024), as described in [24]. The viruses were finally washed and re-suspended in storage buffer (10× diluted VB). The purified virions were lysed and the encapsidated viral RNA extracted and purified, as described in [24]. The final RNA preparations were dissolved in RNase-free water, and the integrity of the RNA was confirmed by electrophoresis in a denatured (formamide/formaldehyde) agarose gel (not shown).
2.2. High-Throughput Sequencing (HTS)
To prepare the RNA sequencing library, viral RNAs were treated with the PrepX RNA-Seq Library Kit by Wafergen Biosystems (Fremont, CA, USA). Briefly, the protocol involved limited digestion with the RNaseIII enzyme, followed by the cDNA synthesis with the superscript III reverse transcriptase, PCR amplification with LongAmp Taq polymerase and primer mix, and purification of the library. For details see at http://www.wafergen.com (accessed on 15 January 2024) and Takara.com (accessed on 15 January 2024). This non-stranded RNA-Seq library preparation allowed us to address global reads at both direct and complementary cDNA levels. The nucleotide sequences were then determined on an Illumina HiSeq2000 instrument (Illumina, San Diego, CA, USA), at the University of Illinois at Chicago, Center for Genomic Research (http://tinyurl.com/dnas-ilab) (accessed on 15 January 2024).
2.3. Sequence Analysis with Bioinformatics Tools
The HTS data were then analyzed with bioinformatics tools. First, to avoid incorrect assemblies, low-quality score reads (p < 0.05) as well as short ones (<50 nts) were eliminated by using DynamicTrim and LengthSort v. 2.5 in Solexa QA software [28]. Then the cleaned reads were mapped to both viral RNA genomes published in GenBank, shown in Table 1, and the plant host mRNAs and rRNAs: (i) BMV host Hordeum vulgare (barley), from JGI (https://phytozome.jgi.doe.gov/pz/portal.html#!infoalias=Org_Hvulgare_er) (accessed on 15 January 2024); (ii) CCMV host Vigna unguiculata (cowpea), from JGI (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Vunguiculata_er) (accessed on 15 January 2024). The cleaned BMV and CCMV reads were deposited into GenBank (PRJNA565451).

Table 1.
Reference genome statistics.
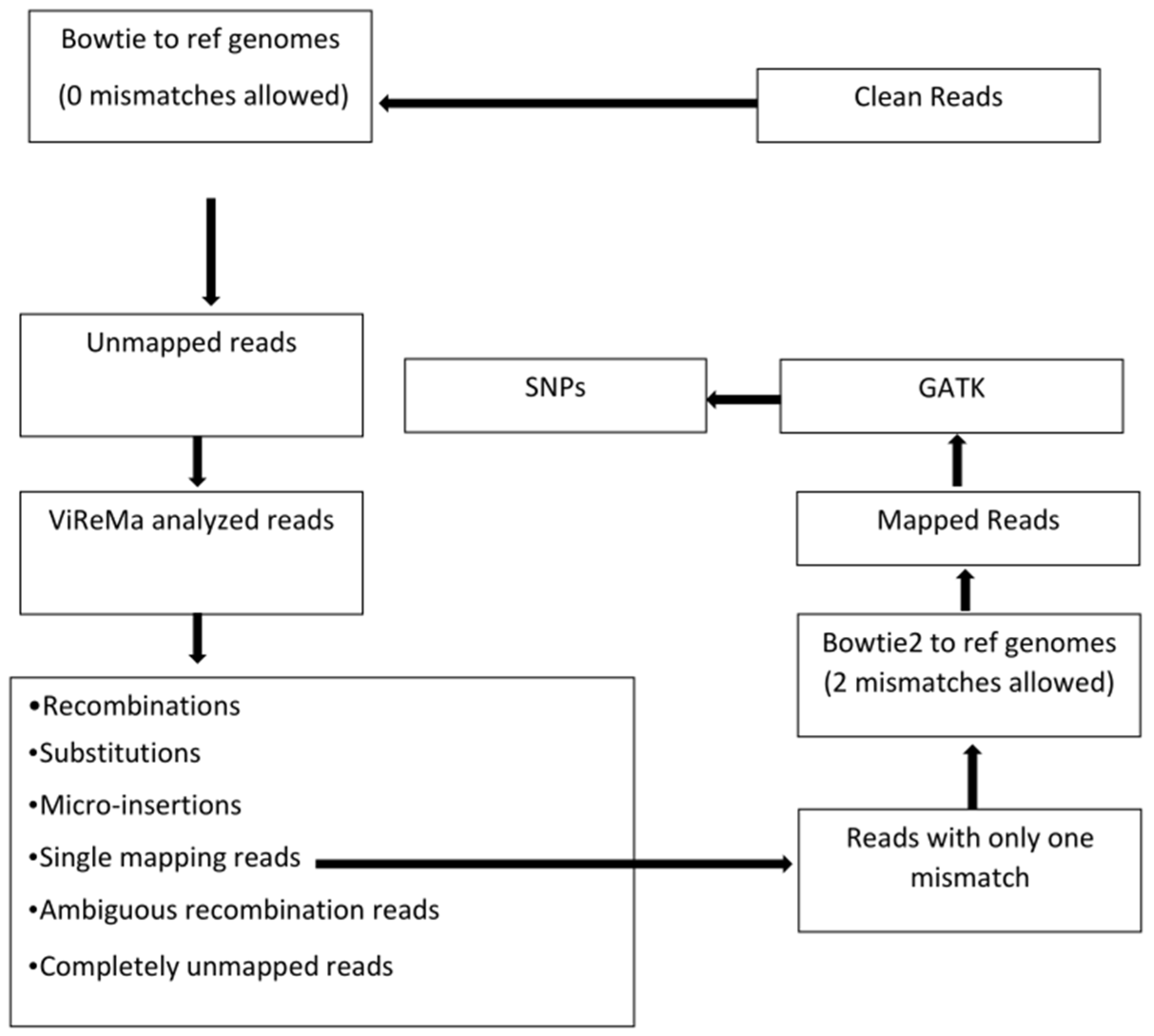
As outlined in Figure 1, the trimmed reads were mapped to the reference sequences (viral genomes and the host transcriptomes) using Bowtie version 1 [29] with the following command: bowtie –v 0 –S reference_index fastq_file –un unmapped_read_output_file mapped_read_output_file. Mismatches allowed were set to zero. The results from Bowtie mapping are shown in Table 2.
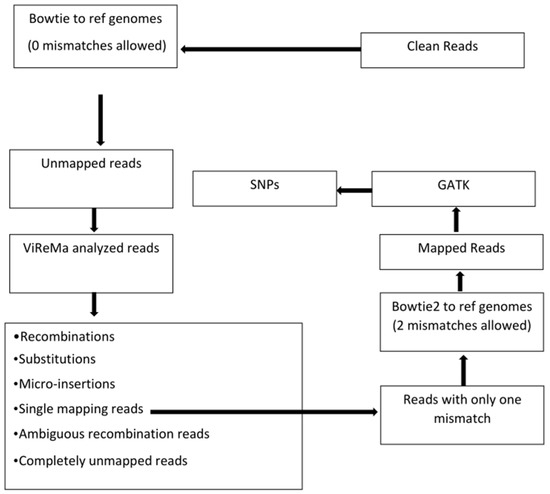
Figure 1.
Flow chart of bioinformatics data analyses. The reference genomes for Bowtie mapping and ViReMa analysis include not only the viral genomes but also plant hosts’ transcriptomes (mRNAs and rRNAs). Bowtie version 1 was used before ViReMa analysis because Bowtie1 allows a perfect match without mismatches, and we aimed to collect unmapped reads with all kinds of variations for ViReMa analysis. Only reads indicated to have one mismatch from single alignment reads were fed into Bowtie2, which allows mismatches for SNP identification. The different categories of read outputs from ViReMa are explained in Section 3.

Table 2.
Bowtie mapping results using all reads as input.
The Bowtie unmapped reads were processed by ViReMa [26]. Instead of mapping the full-length reads, the ViReMa algorithm attempts to split the reads and iteratively maps the 5′ and 3′ segments of the reads to the references using Bowtie. Hence, ViReMa identifies reads that are derived from (i) recombination crosses as shown in Figure 2; (ii) small insertions; (iii) small deletions (≤5 nts); (iv) multi-base substitutions (alignment with pads—a short stretch of nucleotides (<25 nts) that did not align to reference), and (v) completely unmapped reads. ViReMa was run with default parameters by using the viral genome and the host transcriptome (mRNAs + rRNAs) as the reference index. The statistics of ViReMa result can be found in Table 3.
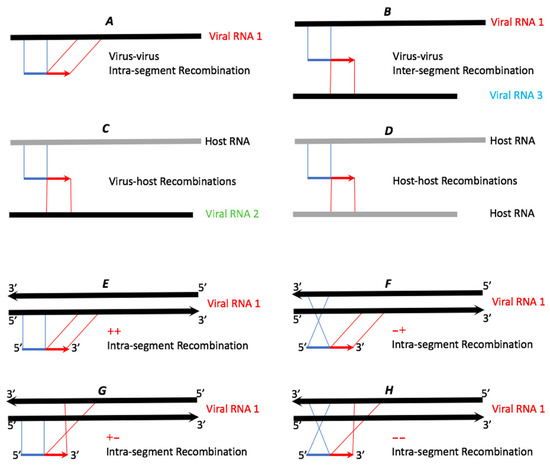
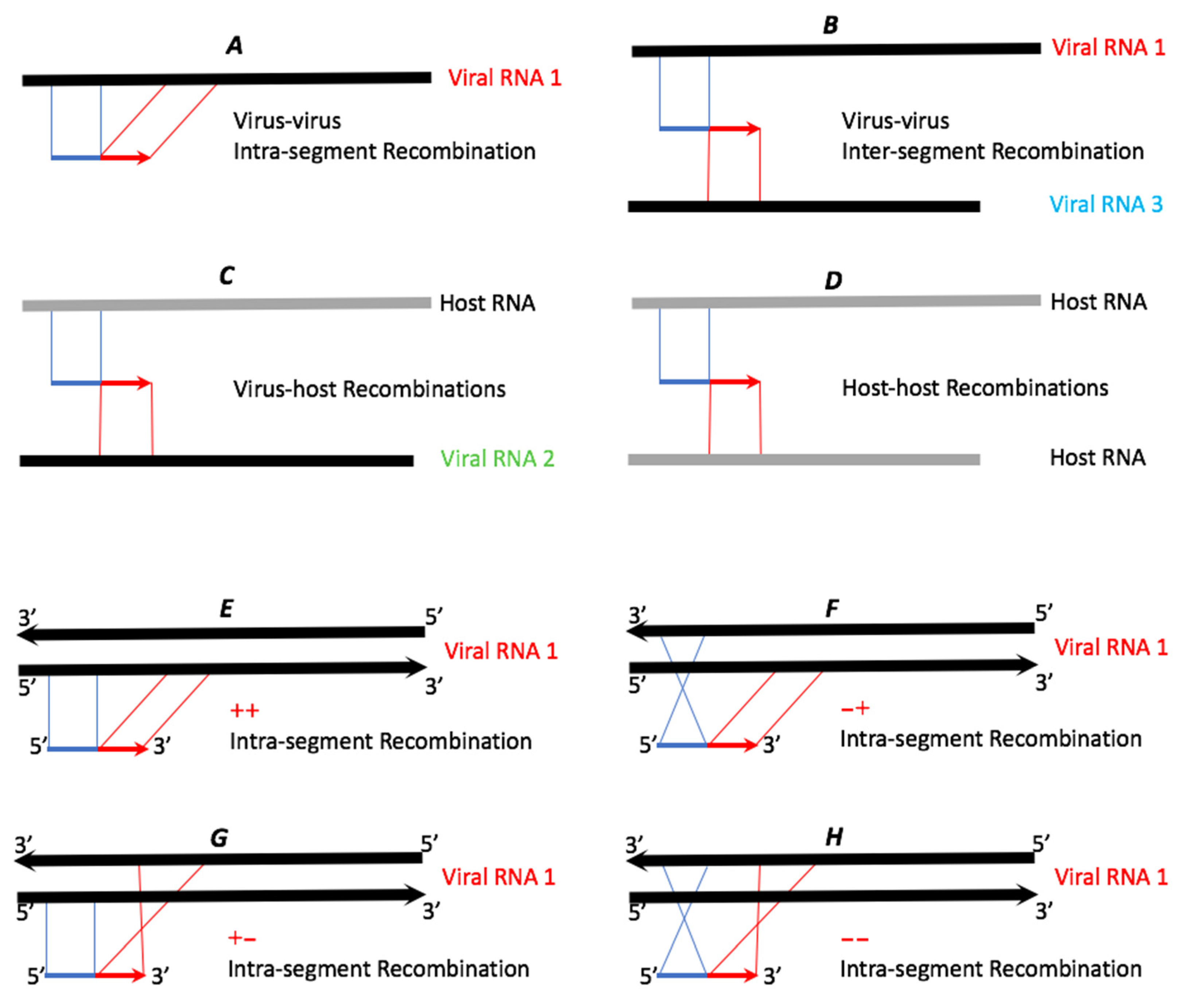
Figure 2.
Schematic examples of potential types of recombinant reads. The thicker lines symbolize reference RNAs (black for viral RNAs and grey for host RNAs). The thinner blue and red lines represent recombinant reads, with blue (5′) and red (3′) representing two recombined portions of the reference RNAs. Please note that since the non-stranded sequencing library was prepared, cross types E and H cannot be assigned to the replicating + or − viral RNA strands.

Table 3.
Bowtie-unmapped reads analyzed and classified by ViReMa.
In order to identify SNPs, the output file from ViReMa was parsed to extract reads with only one mismatch (1×) and no other unmapped regions. These reads were then extracted and subjected to another round of Bowtie mapping with two allowed mismatches. Bowtie-mapped reads were then analyzed using the Genome Analysis Toolkit [30] in order to identify single nucleotide polymorphisms (SNPs).
Unix Bash and Perl scripts were developed to parse the output files from ViReMa to generate data for graphs. For all figures, only the events with greater than 10 supporting reads were used. We also tried plotting events with greater than 5 supporting reads, and the conclusions did not change. R version 3.2.5, specifically the ‘ggplot2’ package (https://ggplot2.tidyverse.org/) (accessed on 15 January 2024), was used to make figures.
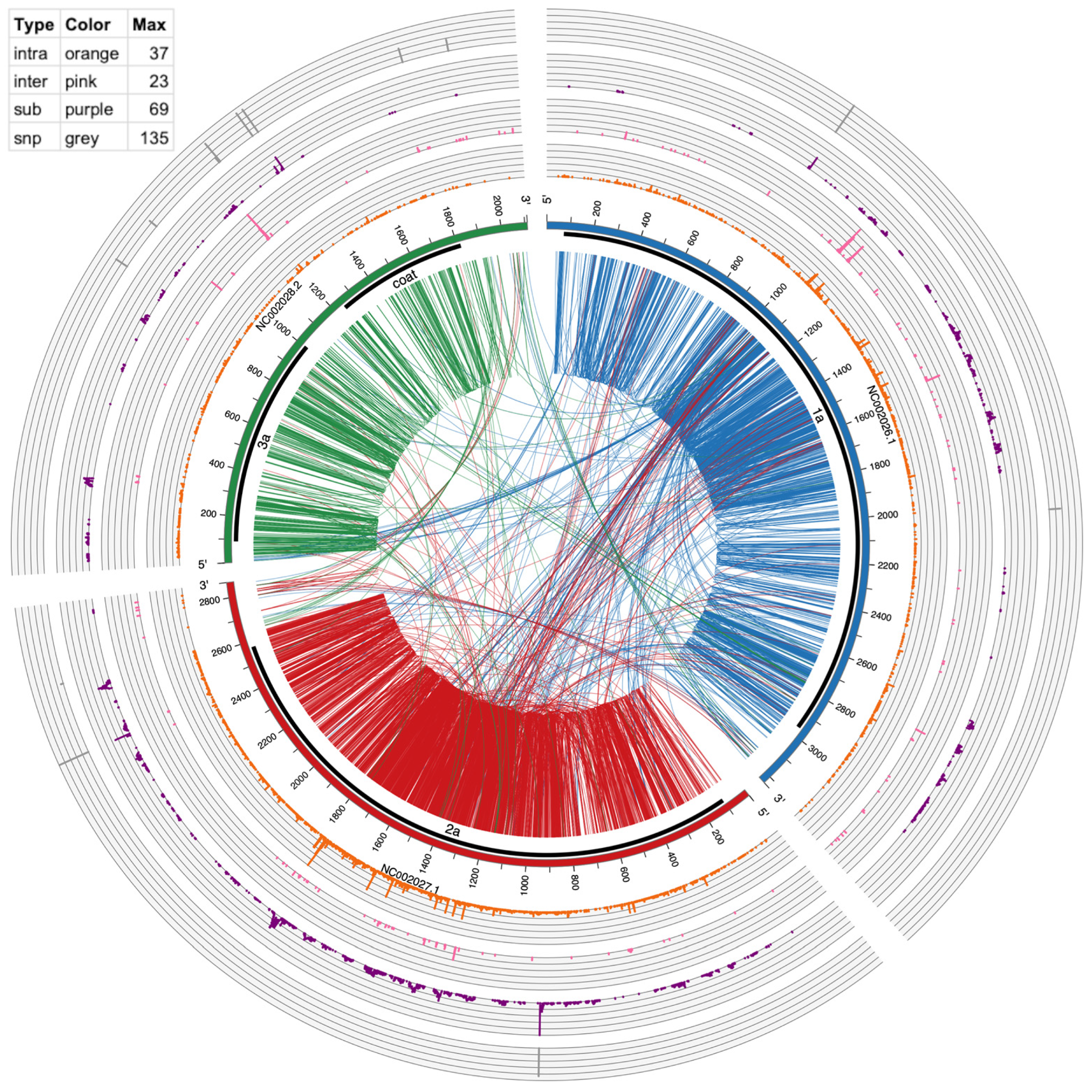
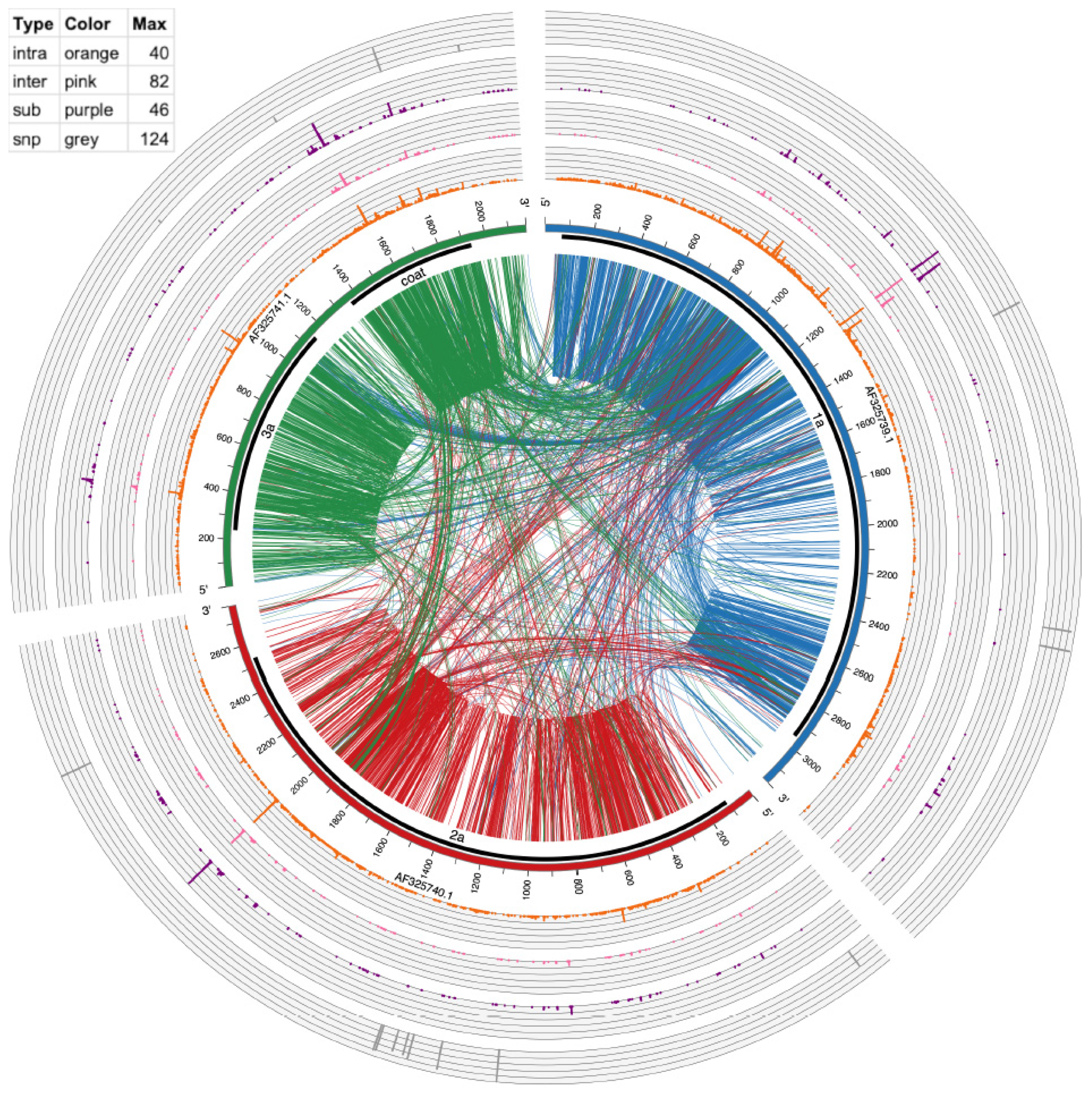
Circos version 0.69–5 [31] was used to make Figure 3 and Figure 4. The links in the inside of the plot visualize actual recombination events. The circular graphs around the outside of the plot represent frequency of substitutions, inter-recombination, and intra-recombination crosses. Intra-recombination events were defined as those occurring within the same viral segment (e.g., RNA1); inter-recombination events were defined as events between two different viral segments (e.g., RNA1 and RNA2).
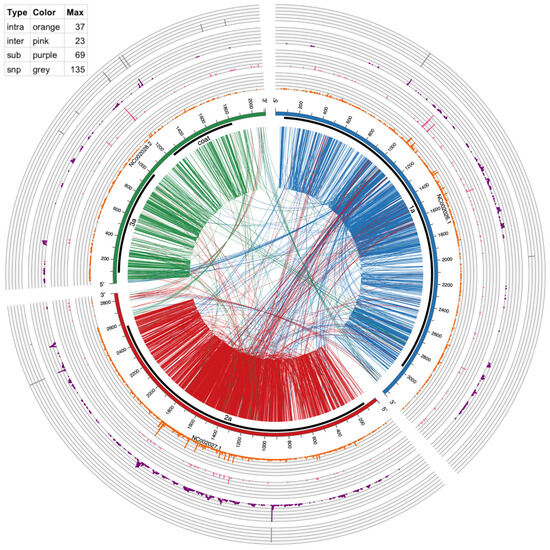
Figure 3.
Circos plot of mutation events in BMV RNAs. From outside to inside: SNPs (gray bars), multi-base substitution events (≥10 supporting reads, purple histogram), inter-genome recombination (≥10 supporting reads, pink histogram), intra-genome recombination (≥10 supporting reads, orange histogram), genome ideogram (with ticks marking the nucleotide position), protein coding region shown as thick black lines, and encoded protein product labels, lines connecting two positions within an RNA (intra-genome crosses) and between two RNAs (inter-genome crosses). These recombination lines are directional, being color-coded according to which RNA the line starts from. For example, lines starting from NC_002026 are coded in dark blue, representing that the recombination has its 5′ end in this RNA. The table in the top left shows the height and spacing of the four histograms, representing the number of events found in each position. Higher means more events or hot mutation spots.
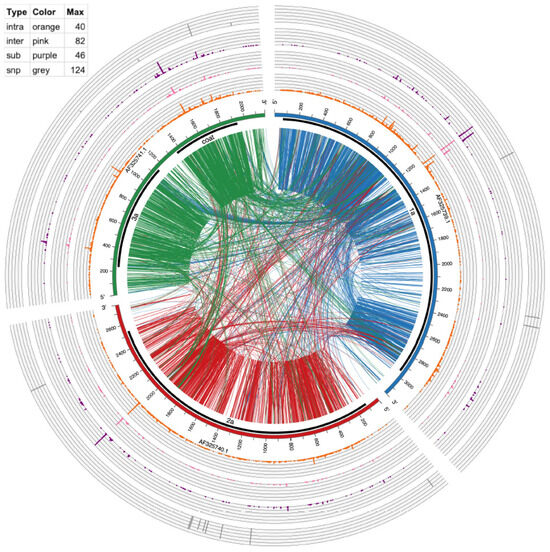
Figure 4.
Circos plot of mutation events in CCMV RNAs. From outside to inside: SNPs (gray bars), multi-base substitution events (≥10 supporting reads, purple histogram), inter-genome recombination (≥10 supporting reads, pink histogram), intra-genome recombination (≥10 supporting reads, orange histogram), genome ideogram (with ticks marking the nucleotide position), protein coding region shown as thick black lines and encoded protein product labels, lines connecting two positions within an RNA (intra-genome crosses) and between two RNAs (inter-genome crosses). These recombination lines are directional, being color-coded according to which RNA the line starts from. For example, lines starting from AF325739 are coded in dark blue, representing that the recombination has its 5′ end in this RNA. The table in the top left shows the height and spacing of the four histograms, representing the number of events found in each position. Higher means more events or hot mutation spots.
3. Results
3.1. High Numbers of HTS Reads Do Not Map to the Canonical Viral Genomes
Two bromoviruses, BMV and CCMV, were propagated in barley and cowpea plants, respectively, and purified, and the encapsidated viral RNAs were extracted, as described in Section 2. The RNAs were subjected to high throughput sequencing, generating over 100 million 76 nt single-end reads as shown in Table 2 (NCBI BioProject ID: PRJNA565451). Considering the total RNA genome size of 8210 nts for BMV and 8122 nts for CCMV, the sequence depth was over 106, enough to detect very rare mutations (rate 10−6).
The reads were mapped to the published references by applying a developed analysis pipeline (Figure 1). The use of the Bowtie program revealed nearly 70% reads mapping to the viral RNAs, but at different proportions per RNA segment, reaching respectively 22.63%, 40.17%, and 8.28% for BMV, and 34.69%, 16.08%, and 16.78% for CCMV RNAs as shown in the Table 2. The different ratios of encapsidated viral RNAs could be due to different molecular requirements, e.g., during RNA replication, translation or encapsidation. Aside from the reads matching the viral RNA sequences, a significant percentage of reads remained to be unmapped for both BMV (28.85%) and CCMV (32.31%), which are the focus of this study.
3.2. ViReMa Identified Six Classes of RNA Variants among the Bowtie-Unmapped Reads
The mutations on the viral RNAs generate variant reads that do not map to the reference. Those include point mutations, insertions or deletions, and recombination events. To identify these variants, the Bowtie unmapped reads were further analyzed by ViReMa [26] (see Section 2). As shown in Table 3, ViReMa cataloged the reads into the following six categories
- Recombination crosses that include reads split into at least two fragments in the reference RNAs (illustrated in Figure 2). The crossovers that occur in the same RNA segment will appear as insertions or deletions (see below). Out of 31,429,207 BMV reads and 35,192,299 CCMV reads, 1.40% and 1.83% were reported as such recombinant reads.
- Ambiguous recombinants included reads with pads longer than 25 nts at either end, or reads with pads of any size in the middle (12.73% and 9.45% of reads, respectively).
- Nucleotide substitutions in reads carrying larger than two consecutive nucleotide mismatches. We found that 1.25% and 1.21% reads of BMV and CCMV fell into this group. The single nucleotide mismatches (or SNPs) were not reported by ViReMa (see the additional analysis of SNPs below).
- Micro-insertions included reads mapped to a single reference after exclusion of a small number (≤4 nts) of nucleotides in the middle, involving 0.01% and 0.23% reads in the two viruses. Reads with longer than 5 nts insertions were reported as ambiguous recombinants by ViReMa.
- Single mapping reads carried the pads of unmapped regions shorter than 25 nts, as well as reads with single-base mismatches in the middle. Those involved, 80.40% and 82.08% reads for BMV and CCMV, respectively.
- Unmapped reads. Only 4.21% and 5.20% reads fell into this category.
3.3. Inter-Segmental vs. Intra-Segmental Recombinants
Different groups of recombination events illustrated in Figure 2 were parsed from the ViReMa recombination output file of 440,975 and 644,596 reads (Table 3) of BMV and CCMV. Theoretically, the recombinant reads can be derived from intra-segmental or inter-segmental viral crossovers. Additionally, reads can be mapped to RNAs between the host and the virus, or between two RNAs of the host. As summarized in Table 4, the number of recombination events varied, depending on the RNA segment. Among the total events for CCMV (4983) and BMV (4009), the latter supported more events in RNA2, followed by RNA1 and RNA3, whereas in CCMV the order was RNA1, RNA3, and RNA2 (consistent with the Bowtie data of Table 2). In both viruses, there were higher numbers of intra-segmental than the inter-segmental events: 91% (3642 out of 4009) events in BMV, compared to 78% (3888 out of 4983) in CCMV (see also Section 4).

Table 4.
Counts of recombination events among different RNA segments of BMV and CCMV.
A characteristic of intra-segmental recombinants was that most of them carried short insertions/deletions (indels). Table 5 shows that out of the 3642 intra-segmental crosses in BMV, 1493 led to insertions and the rest to deletions. The 91.8% deletions had a length ≤5 nts. Among 3888 intra-segmental crosses in CCMV, 1365 caused insertions and 2523 caused deletions; 2155 (85.4%) deletions had a length ≤5 nts. The length of indels was either a multitude of 3, 3 plus 1, or 3 plus 2, leading to six types of indels (Table 5). Del1 was the most abundant (42.83% in BMV and 45.65% in CCMV), together with ins2 accounting for over 50% of events and leading to 1->2 frame shift. Overall, most events caused the frameshifts within the coding region. Distribution of length of indels are shown in Supplementary Figure S1.

Table 5.
Indel types and frame shifts caused by intra-recombination events.
3.4. Mapping of Substitutions to the Reference Genomes
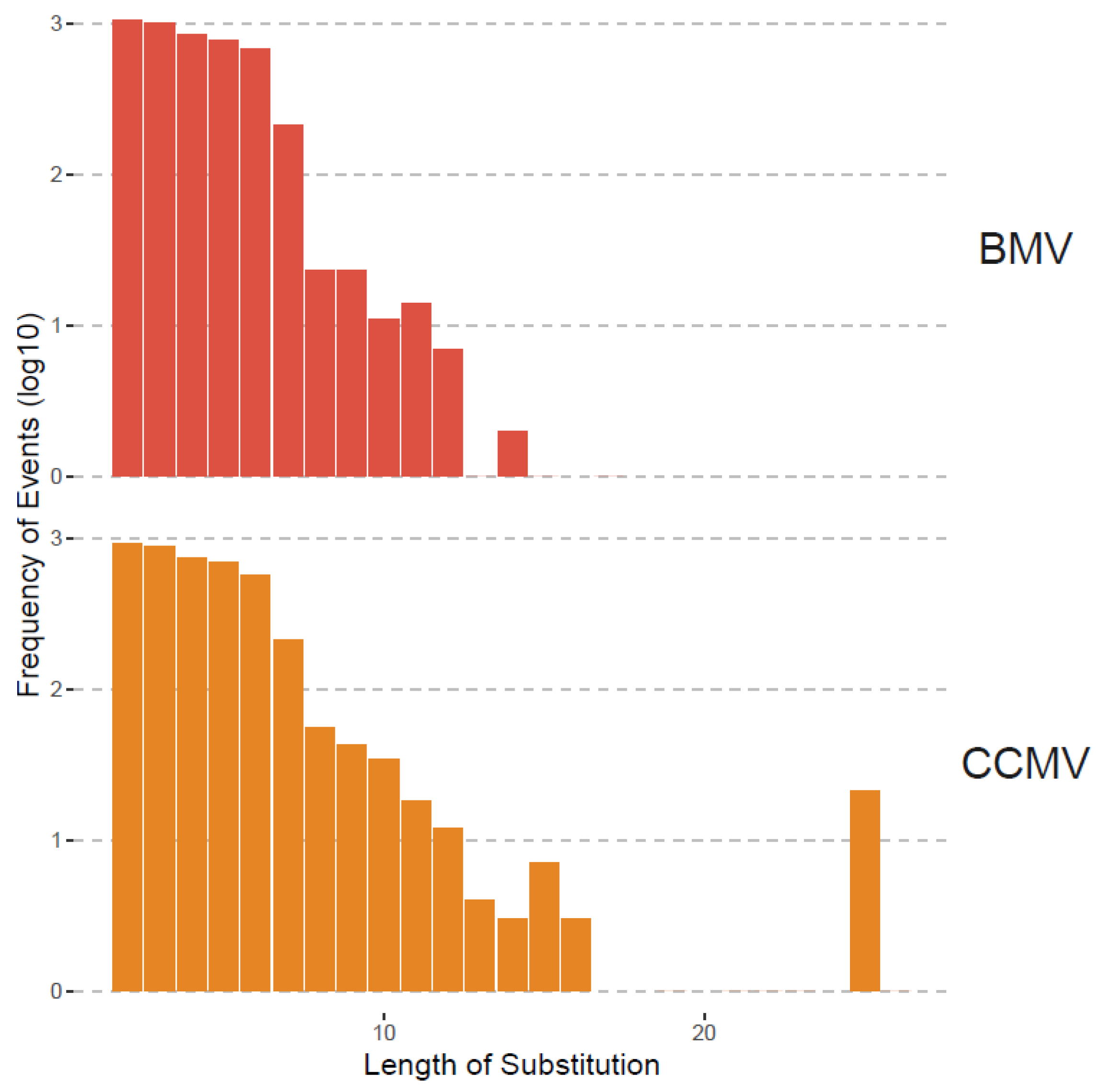
Although recombination was the focus of this study, we also analyzed substitutions as shown in Table 3 and Figure 3 and Figure 4. Substitutions were mapped by ViReMa Version 0.6 as mismatches longer than two consecutive nucleotides in the reference genome. Table 3 provides the total numbers of reads derived from such multi-base substitutions, whereas Figure 5 shows the distribution of their length. Apparently, for BMV, the lengths of substitutions are evenly distributed for two to six nts, but gradually diminished to zero at 14 nts and longer. For CCMV, the number of events diminishes at a slower rate to an outlying category at 25 nts.
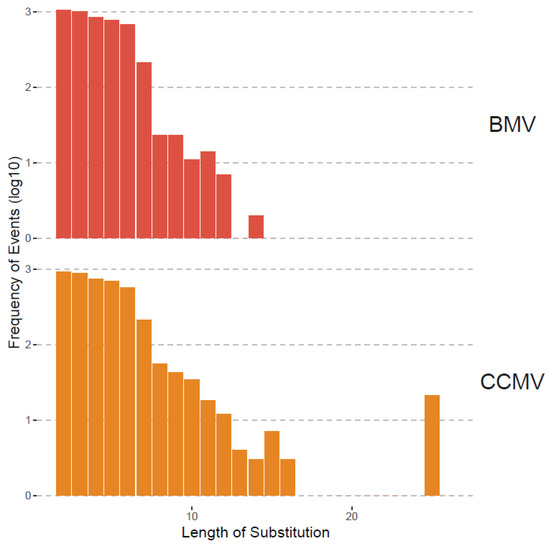
Figure 5.
Length distribution of substitutions in BMV and CCMV. The x-axis is the length starting at 2 nts. The y-axis is the frequency (i.e., the number of events having that length of substitution) shown in log10 scale.
The largest category of ViReMa-mapped reads were single mapping mismatches, Single nucleotide polymorphisms (SNPs) from these reads were identified by using a separate approach, as described in Section 2. As illustrated in the outermost ring in Figure 3 and Figure 4, after filtering for ≥10 supporting reads, a total of 25 SNPs was found in BMV (RNA1: 4, RNA2: 5, RNA3: 16), and 30 in CCMV (RNA1: 6, RNA2: 18, RNA3: 6). This shows the RNA variants can also arise from the SNPs along with the other recombinations described above.
3.5. Visualizing of Recombination Hotspots with Circos Plots
To provide a global mapping of RNA crosses, as shown in Table 4 and Table 6, all inter- and intra-recombination events that were supported by ≥10 reads were plotted as a circular diagram illustrated by Figure 3 for BMV and Figure 4 for CCMV. The connecting lines represent recombination events within and between different RNA segments, whereas the concentric outer ring histograms summarize the frequency of intra- and inter-recombination events, multi-base substitutions, and SNPs, respectively. CCMV reveals more hotspots than BMV, particularly in RNA3 at several locations. Examples in Figure 3, Figure 4 and Figure S2 illustrate the distribution of hotspots on BMV-RNA1 (position 1097) versus CCMV-RNA1 (1103) (Table 6). The hotspots for intra- and inter-crossovers were scattered in CCMV RNA1 at positions 1231, 1260 and 1261; CCMV-RNA2 at 2016 has hot spots for both inter- and intra-crosses (Table 6). Interestingly, in CCMV, some of the highest hotspot peaks are also found on the multi-base substitution ring (purple ring in Figure 4), suggesting that these regions are highly mutable. Further studies are required to explain the observed differences between BMV and CCMV.

Table 6.
Recombination hotspots in BMV and CCMV.
3.6. Recombination Events between Virus and Host RNAs
Table 2 and Table 3 show that a small percentage of BMV and CCMV recombinant reads (respectively 0.07% and 0.04%) mapped to their respective host mRNAs or rRNAs. In addition, there were host-derived reads carrying mutations parsed from the ViReMa output files. There were 170 (BMV) and 138 (CCMV) host–host or virus–host cross events (see detailed breakdown of numbers in Table 7 (count of events) and Table S2 (count of reads)), Generally, BMV carried more virus–host than host–host recombinants than did CCMV. Interestingly, micro-insertions were not found in the host sequences. Surprisingly, 2.21% host–host cross events in CCMV (as shown in Table 7) were covered by 28.31% of reads (Table S2), suggesting some enhancing processes or possible PCR chimera artifacts. To resolve the two possibilities, additional work will be needed in the future to include control experiments with mixed-host RNAs and encapsidated viral RNAs for sequencing without the PCR amplification. Also, among the BMV virus-mRNA cross events, 76 occurred at the intercistronic region (positions 1185–1200) (NC_002028.1 pink peak, Figure 3). In addition to recombinants, there was a higher percentage (0.1%) reads mapped just to host RNAs, extending previous results about the ability of packaging of host RNAs in virus particles (6).

Table 7.
Breakdown counts of mutation events supported by ≥10 reads*.
4. Discussion
In this work, we characterize a population of the encapsidated RNAs in two bromoviruses, BMV and CCMV. Among several software packages available, we focused on the program ViReMa that initially finds a seed-based alignment, and then identifies a new read segment at the 3′ side vis-á-vis the reference genomes. This then detects multiple recombinants within a read, including insertions and substitutions at the junctions [26]. Various types of RNA recombinants were detected, from unambiguous recombinants to the unidentifiable (ambiguous) variants. For both viruses, a vast majority of reads was mapped to the viral genomes but some to the host cellular RNAs, as shown in Table 2.
Our previous studies on homologous recombination using two BMV strains [17] revealed recombination hot spots between marker mutations mostly at the coding regions in RNAs 1 and 2. In RNA3, the crosses occurred within both the inter-cistronic region and the 3a open reading frame (ORF) (the CP-binding region) but were much lower within the CP ORF. Since the marker mutations designed were positioned far apart, mapping of the crossover sites to a narrow region was not possible. Here, we narrowed the cross sites by using HTS of the encapsidated RNAs. The major clusters of crosses were mapped at multiple spots (Table 6) within the ORFs but less within the untranslated regions (UTRs.) Lower number of crosses at the UTRs were at the conserved sequences, which is likely associated with the functional relevance of the regions (see below).
Viral and host factors can impact recombination activity at hot spots [32,33,34,35,36]. We demonstrated previously that binding of CP to cis-acting RNA motifs debilitated recombination in BMV RNA3 [22]. Also, the structure of viral RNAs per se can affect cross sites, as shown for HIV-1 [37] or BMV [38]. Yet selection pressure for functional sequences is also important. For instance, portions of the 3′ UTR and around the middle of 3a ORF in BMV RNA3 serve as the encapsidation and RNA replication signals [39], and indeed they support lower crossover frequency, shown in Figure 3. Also, reduced crosses occurred at the mapped RNA3 replication enhancer site [40], positions 1043–1167. Other modifications such as SNPs, insertions/substitutions, or inter-segmental crosses as shown in Figure 3 and Figure 4 are also reduced within these regions, which suggests the functionality of the preserved sequences.
Two major categories of recombinants were the intra-segmental and the inter-segmental variants. The majority of BMV recombinants, as shown in Table 4 and Table S1, were intra-segmental. Circos plots represented by Figure 3 and Figure 4 illustrate the crosses in a global form. Intersegmental crosses were less frequent but detectable, with the most apparent ones within the RNA1 and RNA2 ORFs (Table 4), but less frequent between the 3′ UTRs of RNA2 and RNA3 (Figure 3). The latter might reflect a negative selection for recombinants during encapsidation because the 3′ UTRs contain both the encapsidation (at least for BMV RNA3) and the replication initiation signals. Also, the BMV RNA3 ORFs were less active than for CCMV, likely reflecting some differences in requirements for host factors between two viruses [41]. In addition, in this work we observed disparities between BMV and CCMV regarding viral–host recombinants (Table 7, Table S2, and Figure S2), perhaps due to different host contributions. Interestingly, the crosses between virus and host rRNAs were very limited despite known abundant concentration of rRNAs (Table 7 and Table S2), possibly reflecting the separation of recombination and translation functions. Overall, there are limited reports about viral–host RNA recombination events [4,42,43].
Reads that carried micro-insertions (less than 4 nts long) were identified at about 50 times higher in CCMV compared to BMV, as shown in Table 3. However, the number of events encompassed within these reads were similar for the two viruses (28 insertions in BMV and 33 in CCMV: Table 7). These non-templated insertions might imply that either double (multiple) crosses with host RNAs or some form of reiterative RNA synthesis occurred more in CCMV, possibly because of less precise CCMV replicase and/or RNA editing in comparison to BMV. Apparently, CCMV seems more prone to supporting insertions in its genomic RNAs. In contrast to insertions, for both viruses, there were similar numbers of reads carrying two or more nucleotide substitutions as shown in Table 3 and Table 7. The mechanism of such replacements is probably different from that for micro-insertions and might involve a precise process, e.g., RNA re-ligation with non-templated copying.
Single nucleotide substitutions (SNPs) were mapped in all of the RNA segments of both viruses (Figure 3 and Figure 4), with the highest number for BMV RNA3 (in CP ORF) and for CCMV RNA2 (in 2a ORF).
A number of reads (4.21% of total reads in BMV and 5.20% in CCMV) remained unmapped to the viral genome and to host references (Table 3), and hence their origin is unclear. Possibilities include (i) reads from parts of the host’s genome other than the transcriptome (we used only mRNAs and tRNAs); (ii) low-quality reads; or (iii) contaminated reads.
In summary, these studies reveal the dynamic character of the viral RNA genome, generating the encapsidated RNA variants. A question remains whether the HTS methodology detects false-positive reads, as described for other cases [28,44]. In this work, we attempted to reduce potential noise by requiring that a junction be represented by at least ten overlapping reads (Table 4 and Table 7). Future analyses with other controls must be used to eliminate more false positives. One baseline control would involve mixing separate RNA samples preceding the reverse transcription reaction. Nevertheless, our findings encourage the use of these novel approaches by inspiring a closer look at where the viral RNAs recombine and what types of events predominate. In addition, this methodology can be used to reveal potentially functional domains in viral RNAs, helpful for novel anti-viral therapies via targeting recombination hotspots or functional motifs.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pathogens13010096/s1, Figure S1: Length distribution of indels caused by intra-segmental recombination in BMV and CCMV.; Figure S2: An example of recombination hotspots in BMV and CCMV RNA1. Table S1: Counts of recombination events among different RNA segments of BMV and CCMV; Table S2: Breakdown counts of reads of mutation events supported by ≥10 reads.
Author Contributions
Conceptualization, Y.Y. and J.J.B.; Methodology, N.S. (Wet Lab and RNA Sequencing), S.D. (Bioinformatics) and B.S.C. (Bioinformatics); Software, S.D. and B.S.C.; Formal analysis, S.D., N.S., Y.Y. and J.J.B.; Resources, Y.Y. and J.J.B.; Data curation, S.D.; Writing—original draft, Y.Y.; Writing—review & editing, N.S., Y.Y. and J.J.B.; Visualization, S.D.; Supervision, Y.Y. and J.J.B.; Funding acquisition, Y.Y. and J.J.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded mainly by the National Institutes of Health (NIH) AREA award (1R15GM114706) and the University of Nebraska—Lincoln, and partially by the National Science Foundation (NSF) CAREER award (DBI-1652164), the NIH R21 award (R21AI171952), the NIH R01 award (R01GM140370), the United States Department of Agriculture (USDA) award (58-8042-7-089) to Y.Y.; J.J.B. was supported by a grant from National Science Foundation (MCB-0920617) and through the Plant Molecular Biology Center and the Graduate School, both at Northern Illinois University. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The cleaned BMV and CCMV reads were deposited into GenBank (PRJNA565451): https://www.ncbi.nlm.nih.gov/sra/SRX6849035 for CCMV and https://www.ncbi.nlm.nih.gov/sra/SRX6849034 for BMV.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Koonin, E.V.; Dolja, V.V.; Krupovic, M. Origins and evolution of viruses of eukaryotes: The ultimate modularity. Virology 2015, 479, 2–25. [Google Scholar] [CrossRef]
- Simon-Loriere, E.; Holmes, E.C. Why do RNA viruses recombine? Nat. Rev. Microbiol. 2011, 9, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Sztuba-Solińska, J.; Urbanowicz, A.; Figlerowicz, M.; Bujarski, J.J. RNA-RNA recombination in plant virus replication and evolution. Annu. Rev. Phytopathol. 2011, 49, 415–443. [Google Scholar] [CrossRef]
- Bujarski, J.J. Genetic recombination in plant-infecting messenger-sense RNA viruses: Overview and research perspectives. Front. Plant Sci. 2013, 4, 68. [Google Scholar] [CrossRef] [PubMed]
- Graham, R.L.; Baric, R.S. Recombination, reservoirs, and the modular spike: Mechanisms of coronavirus cross-species transmission. J. Virol. 2010, 84, 3134–3146. [Google Scholar] [CrossRef] [PubMed]
- Lam, T.T.-Y.; Wang, J.; Shen, Y.; Zhou, B.; Duan, L.; Cheung, C.-L.; Ma, C.; Lycett, S.J.; Leung, C.Y.-H.; Chen, X. The genesis and source of the H7N9 influenza viruses causing human infections in China. Nature 2013, 502, 241–244. [Google Scholar] [CrossRef] [PubMed]
- Lukashev, A. Recombination among picornaviruses. Rev. Med. Virol. 2010, 20, 327–337. [Google Scholar] [CrossRef]
- Martin-Valls, G.; Kvisgaard, L.K.; Tello, M.; Darwich, L.; Cortey, M.; Burgara-Estrella, A.; Hernández, J.; Larsen, L.E.; Mateu, E. Analysis of ORF5 and full-length genome sequences of porcine reproductive and respiratory syndrome virus isolates of genotypes 1 and 2 retrieved worldwide provides evidence that recombination is a common phenomenon and may produce mosaic isolates. J. Virol. 2014, 88, 3170–3181. [Google Scholar] [CrossRef]
- Villabona-Arenas, C.J.; Zanotto, P.M.d.A. Worldwide spread of dengue virus type 1. PLoS ONE 2013, 8, e62649. [Google Scholar] [CrossRef]
- Weiss, B.G.; Schlesinger, S. Recombination between sindbis virus RNAs. J. Virol. 1991, 65, 4017–4025. [Google Scholar] [CrossRef]
- Pagán, I.; Holmes, E.C. Long-term evolution of the Luteoviridae: Time scale and mode of virus speciation. J. Virol. 2010, 84, 6177–6187. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ahlquist, P. Filling a GAP(DH) in asymmetric viral RNA synthesis. Cell Host Microbe 2008, 3, 124–125. [Google Scholar] [CrossRef]
- Palasingam, K.; Shaklee, P.N. Reversion of Q beta RNA phage mutants by homologous RNA recombination. J. Virol. 1992, 66, 2435–2442. [Google Scholar] [CrossRef] [PubMed]
- Han, G.-Z.; Worobey, M. Homologous recombination in negative sense RNA viruses. Viruses 2011, 3, 1358–1373. [Google Scholar] [CrossRef] [PubMed]
- Delviks-Frankenberry, K.; Galli, A.; Nikolaitchik, O.; Mens, H.; Pathak, V.K.; Hu, W.S. Mechanisms and factors that influence high frequency retroviral recombination. Viruses 2011, 3, 1650–1680. [Google Scholar] [CrossRef] [PubMed]
- Rao, A.L. Genome packaging by spherical plant RNA viruses. Annu. Rev. Phytopathol. 2006, 44, 61–87. [Google Scholar] [CrossRef]
- Kolondam, B.; Rao, P.; Sztuba-Solinska, J.; Weber, P.H.; Dzianott, A.; Johns, M.A.; Bujarski, J.J. Co-infection with two strains of Brome mosaic bromovirus reveals common RNA recombination sites in different hosts. Virus Evol. 2015, 1, vev021. [Google Scholar] [CrossRef]
- Figlerowicz, M.; Nagy, P.D.; Bujarski, J.J. A mutation in the putative RNA polymerase gene inhibits nonhomologous, but not homologous, genetic recombination in an RNA virus. Proc. Natl. Acad. Sci. USA 1997, 94, 2073–2078. [Google Scholar] [CrossRef]
- Figlerowicz, M.; Nagy, P.D.; Tang, N.; Kao, C.C.; Bujarski, J.J. Mutations in the N terminus of the brome mosaic virus polymerase affect genetic RNA-RNA recombination. J. Virol. 1998, 72, 9192–9200. [Google Scholar] [CrossRef]
- Allison, R.; Thompson, C.; Ahlquist, P. Regeneration of a functional RNA virus genome by recombination between deletion mutants and requirement for cowpea chlorotic mottle virus 3a and coat genes for systemic infection. Proc. Natl. Acad. Sci. USA 1990, 87, 1820–1824. [Google Scholar] [CrossRef]
- Ranjith-Kumar, C.T.; Kim, Y.C.; Gutshall, L.; Silverman, C.; Khandekar, S.; Sarisky, R.T.; Kao, C.C. Mechanism of de novo initiation by the hepatitis C virus RNA-dependent RNA polymerase: Role of divalent metals. J. Virol. 2002, 76, 12513–12525. [Google Scholar] [CrossRef] [PubMed]
- Sztuba-Solinska, J.; Fanning, S.W.; Horn, J.R.; Bujarski, J.J. Mutations in the coat protein-binding cis-acting RNA motifs debilitate RNA recombination of Brome mosaic virus. Virus Res. 2012, 170, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Weber, P.H.; Bujarski, J.J. Multiple functions of capsid proteins in (+) stranded RNA viruses during plant-virus interactions. Virus Res. 2015, 196, 140–149. [Google Scholar] [CrossRef]
- Shrestha, N.; Weber, P.H.; Burke, S.V.; Wysocki, W.P.; Duvall, M.R.; Bujarski, J.J. Next generation sequencing reveals packaging of host RNAs by brome mosaic virus. Virus Res. 2018, 252, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, N.; Duvall, M.R.; Bujarski, J.J. Variability among the Isolates of Broad Bean Mottle Virus and Encapsidation of Host RNAs. Pathogens 2022, 11, 817. [Google Scholar] [CrossRef] [PubMed]
- Routh, A.; Johnson, J.E. Discovery of functional genomic motifs in viruses with ViReMa—A Virus Recombination Mapper-for analysis of next-generation sequencing data. Nucleic Acids Res. 2014, 42, e11. [Google Scholar] [CrossRef] [PubMed]
- Ni, P.; Vaughan, R.C.; Tragesser, B.; Hoover, H.; Kao, C.C. The plant host can affect the encapsidation of brome mosaic virus (BMV) RNA: BMV virions are surprisingly heterogeneous. J. Mol. Biol. 2014, 426, 1061–1076. [Google Scholar] [CrossRef][Green Version]
- Cox, M.P.; Peterson, D.A.; Biggs, P.J. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinform. 2010, 11, 485. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, 1–10. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Chao, M.; Wang, T.C.; Lin, C.C.; Yung-Liang Wang, R.; Lin, W.B.; Lee, S.E.; Cheng, Y.Y.; Yeh, C.T.; Iang, S.B. Analyses of a whole-genome inter-clade recombination map of hepatitis delta virus suggest a host polymerase-driven and viral RNA structure-promoted template-switching mechanism for viral RNA recombination. Oncotarget 2017, 8, 60841–60859. [Google Scholar] [CrossRef] [PubMed]
- Haasnoot, P.C.; Olsthoorn, R.C.; Bol, J.F. The Brome mosaic virus subgenomic promoter hairpin is structurally similar to the iron-responsive element and functionally equivalent to the minus-strand core promoter stem-loop C. RNA 2002, 8, 110–122. [Google Scholar] [CrossRef][Green Version]
- Knies, J.L.; Dang, K.K.; Vision, T.J.; Hoffman, N.G.; Swanstrom, R.; Burch, C.L. Compensatory evolution in RNA secondary structures increases substitution rate variation among sites. Mol. Biol. Evol. 2008, 25, 1778–1787. [Google Scholar] [CrossRef] [PubMed]
- Shapka, N.; Nagy, P.D. The AU-rich RNA recombination hot spot sequence of Brome mosaic virus is functional in tombusviruses: Implications for the mechanism of RNA recombination. J. Virol. 2004, 78, 2288–2300. [Google Scholar] [CrossRef] [PubMed]
- Nagy, P.D.; Pogany, J. The dependence of viral RNA replication on co-opted host factors. Nat. Rev. Microbiol. 2012, 10, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Simon-Loriere, E.; Martin, D.P.; Weeks, K.M.; Negroni, M. RNA structures facilitate recombination-mediated gene swapping in HIV-1. J. Virol. 2010, 84, 12675–12682. [Google Scholar] [CrossRef]
- Nagy, P.D.; Bujarski, J.J. Efficient system of homologous RNA recombination in brome mosaic virus: Sequence and structure requirements and accuracy of crossovers. J. Virol. 1995, 69, 131–140. [Google Scholar] [CrossRef]
- Annamalai, P.; Rao, A.L. In vivo packaging of brome mosaic virus RNA3, but not RNAs 1 and 2, is dependent on a cis-acting 3’ tRNA-like structure. J. Virol. 2007, 81, 173–181. [Google Scholar] [CrossRef]
- Baumstark, T.; Ahlquist, P. The brome mosaic virus RNA3 intergenic replication enhancer folds to mimic a tRNA TpsiC-stem loop and is modified in vivo. RNA 2001, 7, 1652–1670. [Google Scholar]
- Sibert, B.S.; Navine, A.K.; Pennington, J.; Wang, X.; Ahlquist, P. Cowpea chlorotic mottle bromovirus replication proteins support template-selective RNA replication in Saccharomyces cerevisiae. PLoS ONE 2018, 13, e0208743. [Google Scholar] [CrossRef] [PubMed]
- Becher, P.; Tautz, N. RNA recombination in pestiviruses: Cellular RNA sequences in viral genomes highlight the role of host factors for viral persistence and lethal disease. RNA Biol. 2011, 8, 216–224. [Google Scholar] [CrossRef] [PubMed]
- Weiss, R.A. Exchange of Genetic Sequences Between Viruses and Hosts. Curr. Top. Microbiol. Immunol. 2017, 407, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Gorzer, I.; Guelly, C.; Trajanoski, S.; Puchhammer-Stockl, E. The impact of PCR-generated recombination on diversity estimation of mixed viral populations by deep sequencing. J. Virol. Methods 2010, 169, 248–252. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).