Multivalent Epigraph Hemagglutinin Vaccine Protects against Influenza B Virus in Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Viruses
2.3. Immunogen Gene Design
2.4. Phylogenetic and Sequence Analysis
2.5. Recombinant Adenovirus Type 5 Plasmid Construction and Confirmation
2.6. Epigraph Immunogen Rosette Analysis
2.7. Western Blotting
2.8. Tissues for Humoral and Cellular Assays
2.9. Hemagglutination Inhibition Titers
2.10. Microneutralization Titers
2.11. ELISpot Assay
2.12. Vaccination and Lethal IBV Challenges in Mice
2.13. Statistical Analysis
3. Results
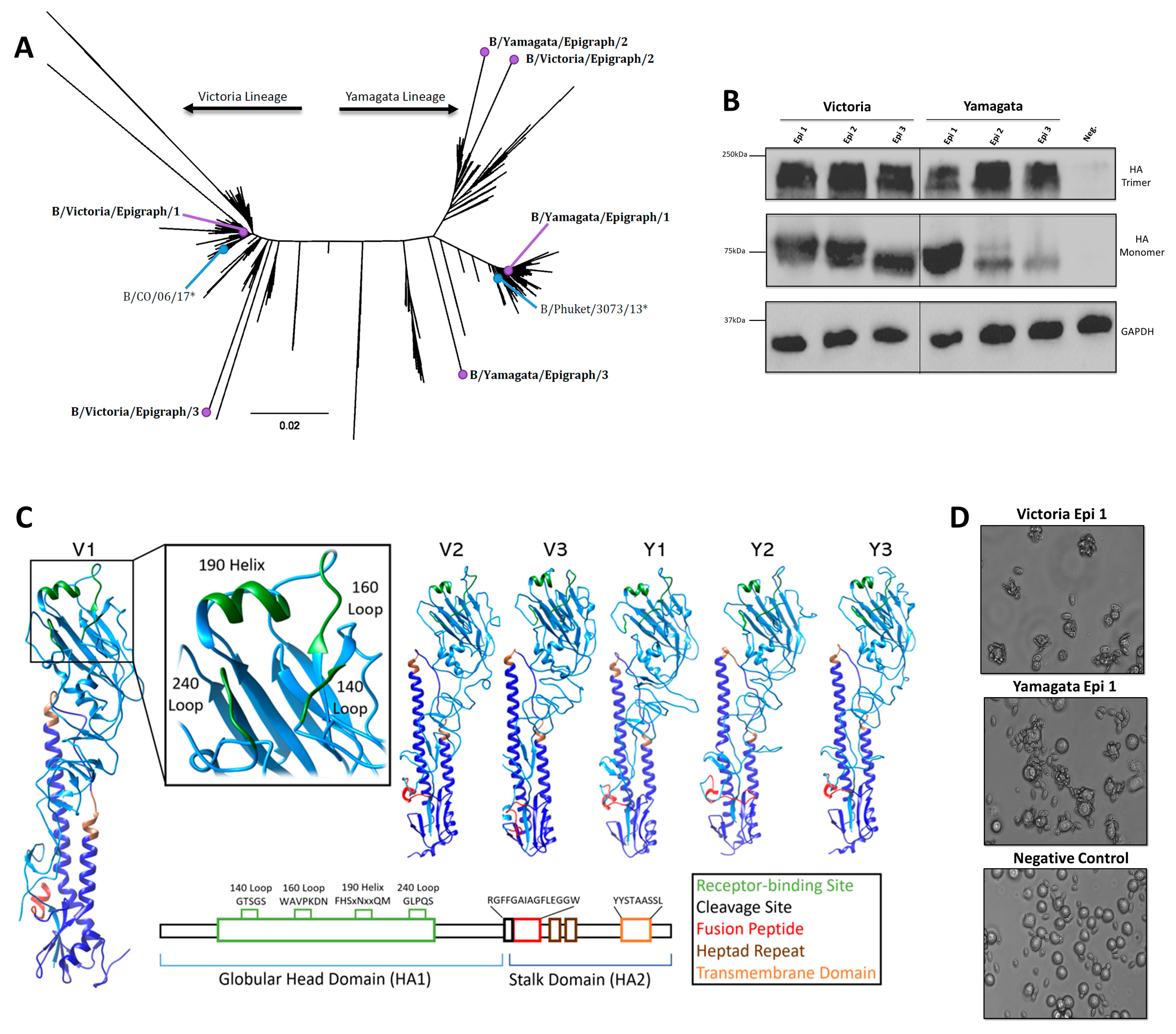
3.1. Design and Characterization of the IBV Epigraph Immunogens
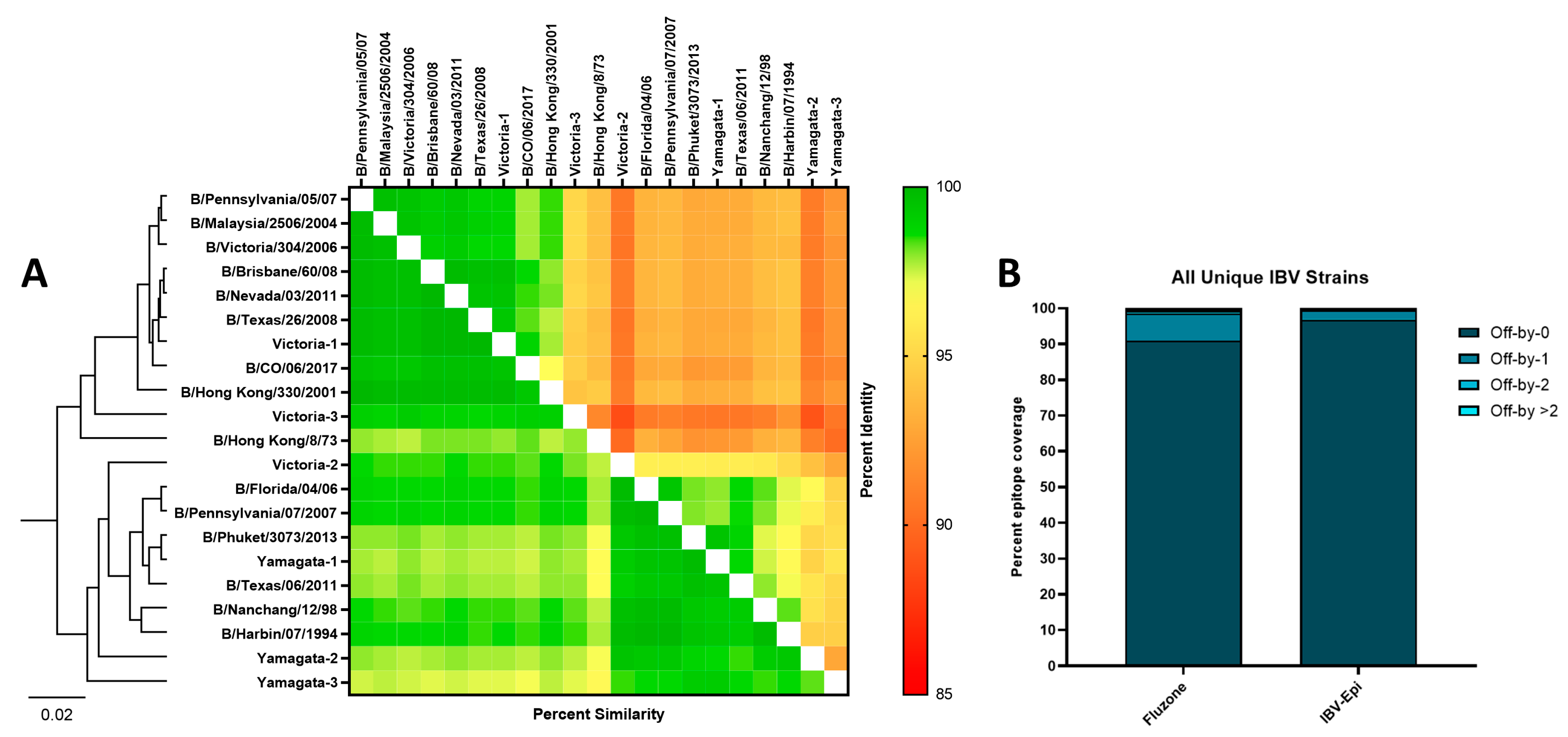
3.2. Bioinformatic Predictions of IBV Epigraph Immunogen Cross-Reactivity
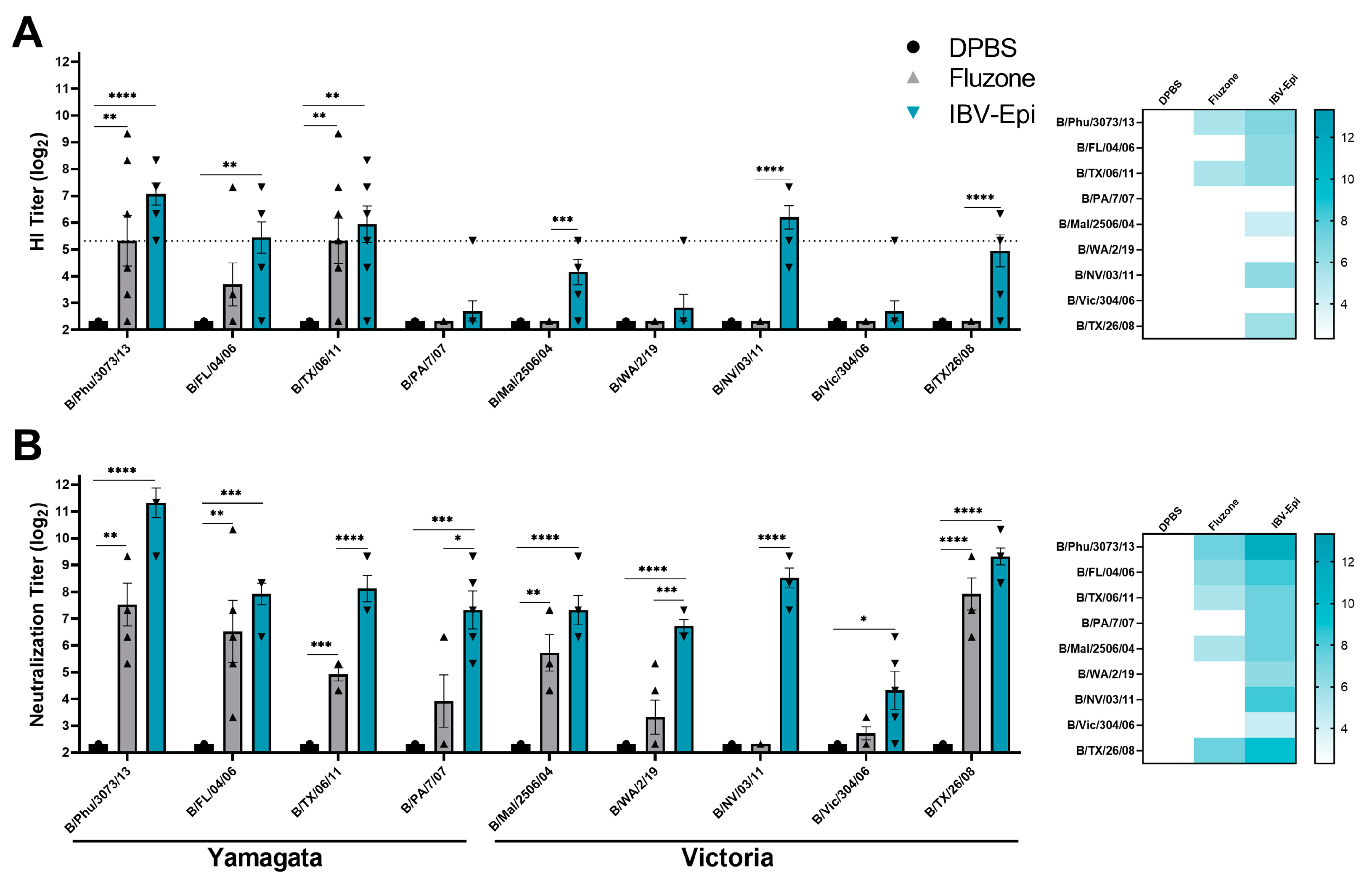
3.3. IBV Epigraph Vaccine Elicits Broad Humoral Immune Responses
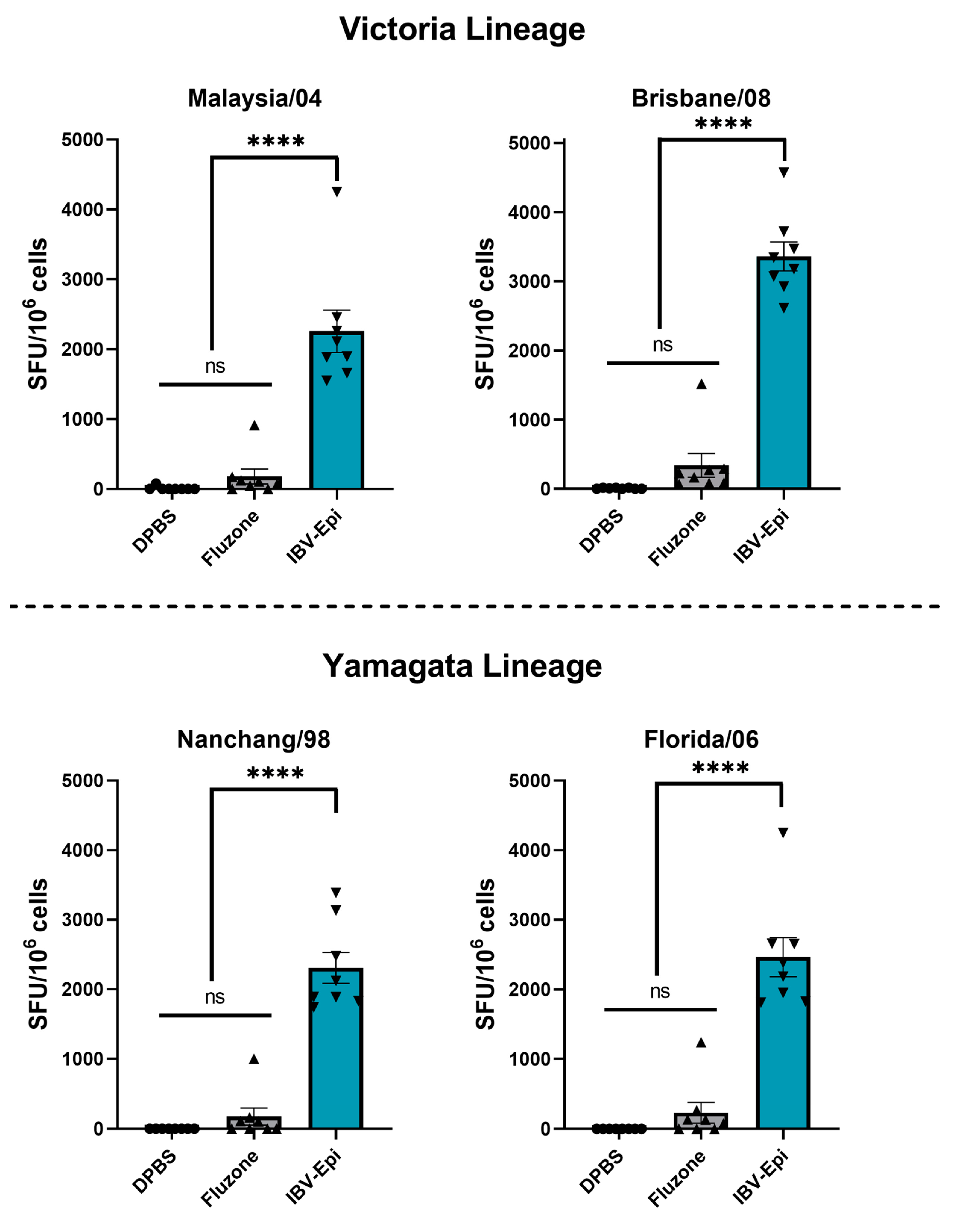
3.4. IBV Epigraph Vaccine Provides Broadly Cross-Reactive Cellular-Mediated Immune Responses
3.5. IBV Epigraph Completely Protects against Lethal IBV Challenge
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Howley, P.M.; Knipe, D.M.; Whelan, S. Fields Virology, 7th ed.; Orthomyxoviridae; Wolters Kluwer: Philadelphia, PA, USA, 2021; Volume 1, p. 826. [Google Scholar]
- Caini, S.; Kusznierz, G.; Garate, V.V.; Wangchuk, S.; Thapa, B.; de Paula Junior, F.J.; Ferreira de Almeida, W.A.; Njouom, R.; Fasce, R.A.; Bustos, P.; et al. The epidemiological signature of influenza B virus and its B/Victoria and B/Yamagata lineages in the 21st century. PLoS ONE 2019, 14, e0222381. [Google Scholar] [CrossRef] [PubMed]
- Brottet, E.; Vandroux, D.; Gauzere, B.A.; Antok, E.; Jaffar-Bandjee, M.C.; Michault, A.; Filleul, L. Influenza season in Reunion dominated by influenza B virus circulation associated with numerous cases of severe disease, France, 2014. Eurosurveillance 2014, 19, 20916. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kosasih, H.; Roselinda; Nurhayati; Klimov, A.; Xiyan, X.; Lindstrom, S.; Mahoney, F.; Beckett, C.; Burgess, T.H.; Blair, P.J. Surveillance of influenza in Indonesia, 2003–2007. Influenza Other Respir. Viruses 2012, 7, 312–320. [Google Scholar] [CrossRef][Green Version]
- Garten, R.; Blanton, L.; Elal, A.I.A.; Alabi, N.; Barnes, J.; Biggerstaff, M.; Brammer, L.; Budd, A.P.; Burns, E.; Cummings, C.N.; et al. Update: Influenza Activity in the United States During the 2017–18 Season and Composition of the 2018–19 Influenza Vaccine. MMWR Morb. Mortal. Wkly. Rep. 2018, 67, 634–642. [Google Scholar] [CrossRef]
- Epperson, S.; Blanton, L.; Kniss, K.; Mustaquim, D.; Steffens, C.; Wallis, T.; Dhara, R.; Leon, M.; Perez, A.; Chaves, S.S.; et al. Influenza activity—United States, 2013–2014 season and composition of the 2014–15 influenza vaccines. MMWR Morb. Mortal. Wkly. Rep. 2014, 63, 483–490. [Google Scholar]
- Su, S.; Chaves, S.S.; Perez, A.; D’Mello, T.; Kirley, P.D.; Yousey-Hindes, K.; Farley, M.M.; Harris, M.; Sharangpani, R.; Lynfield, R.; et al. Comparing clinical characteristics between hospitalized adults with laboratory-confirmed influenza A and B virus infection. Clin. Infect. Dis. 2014, 59, 252–255. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Muscatello, D.J.; Wang, Q.; Yang, P.; Pan, Y.; Huo, D.; Liu, Z.; Zhao, X.; Tang, Y.; Li, C.; et al. Hospitalizations for Influenza-Associated Severe Acute Respiratory Infection, Beijing, China, 2014–2016. Emerg. Infect. Dis. 2018, 24, 2098–2102. [Google Scholar] [CrossRef]
- Paul Glezen, W.; Schmier, J.K.; Kuehn, C.M.; Ryan, K.J.; Oxford, J. The burden of influenza B: A structured literature review. Am. J. Public Health 2013, 103, e43–e51. [Google Scholar] [CrossRef]
- Shang, M.; Blanton, L.; Brammer, L.; Olsen, S.J.; Fry, A.M. Influenza-Associated Pediatric Deaths in the United States, 2010–2016. Pediatrics 2018, 141, e20172918. [Google Scholar] [CrossRef]
- Read, J.M.; Zimmer, S.; Vukotich, C., Jr.; Schweizer, M.L.; Galloway, D.; Lingle, C.; Yearwood, G.; Calderone, P.; Noble, E.; Quadelacy, T.; et al. Influenza and other respiratory viral infections associated with absence from school among schoolchildren in Pittsburgh, Pennsylvania, USA: A cohort study. BMC Infect. Dis. 2021, 21, 291. [Google Scholar] [CrossRef]
- Han, S.B.; Rhim, J.W.; Kang, J.H.; Lee, K.Y. Clinical features and outcomes of influenza by virus type/subtype/lineage in pediatric patients. Transl. Pediatr. 2021, 10, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Yazici Ozkaya, P.; Turanli, E.E.; Metin, H.; Aydin Uysal, A.; Cicek, C.; Karapinar, B. Severe influenza virus infection in children admitted to the PICU: Comparison of influenza A and influenza B virus infection. J. Med. Virol. 2022, 94, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Fan, K.; Zhang, L.; Yang, M.; Yu, Q.; Liu, L.; Leung, L. Risk factors for influenza B virus-associated pneumonia in adults. Am. J. Infect. Control 2020, 48, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Avni, T.; Babich, T.; Nir, A.; Yahav, D.; Shaked, H.; Sorek, N.; Zvi, H.B.; Bishara, J.; Atamna, A. Comparison of clinical outcomes of influenza A and B at the 2017–2018 influenza season: A cohort study. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1109–1114. [Google Scholar] [CrossRef] [PubMed]
- Rota, P.A.; Hemphill, M.L.; Whistler, T.; Regnery, H.L.; Kendal, A.P. Antigenic and genetic characterization of the haemagglutinins of recent cocirculating strains of influenza B virus. J. Gen. Virol. 1992, 73, 2737–2742. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Prevention and control of seasonal influenza with vaccines. Recommendations of the Advisory Committee on Immunization Practices—United States, 2013–2014. MMWR Recomm. Rep. 2013, 62, 1–43. [Google Scholar]
- Tricco, A.C.; Chit, A.; Soobiah, C.; Hallett, D.; Meier, G.; Chen, M.H.; Tashkandi, M.; Bauch, C.T.; Loeb, M. Comparing influenza vaccine efficacy against mismatched and matched strains: A systematic review and meta-analysis. BMC Med. 2013, 11, 153. [Google Scholar] [CrossRef]
- Morimoto, N.; Takeishi, K. Change in the efficacy of influenza vaccination after repeated inoculation under antigenic mismatch: A systematic review and meta-analysis. Vaccine 2018, 36, 949–957. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Past Seasons’ Vaccine Effectiveness Estimates. 2023. Available online: https://www.cdc.gov/flu/vaccines-work/past-seasons-estimates.html (accessed on 29 November 2023).
- Bianchi, E.; Liang, X.; Ingallinella, P.; Finotto, M.; Chastain, M.A.; Fan, J.; Fu, T.M.; Song, H.C.; Horton, M.S.; Freed, D.C.; et al. Universal influenza B vaccine based on the maturational cleavage site of the hemagglutinin precursor. J. Virol. 2005, 79, 7380–7388. [Google Scholar] [CrossRef]
- Ermler, M.E.; Kirkpatrick, E.; Sun, W.; Hai, R.; Amanat, F.; Chromikova, V.; Palese, P.; Krammer, F. Chimeric Hemagglutinin Constructs Induce Broad Protection against Influenza B Virus Challenge in the Mouse Model. J. Virol. 2017, 91, 10–1128. [Google Scholar] [CrossRef]
- Sun, W.; Kirkpatrick, E.; Ermler, M.; Nachbagauer, R.; Broecker, F.; Krammer, F.; Palese, P. Development of Influenza B Universal Vaccine Candidates Using the “Mosaic” Hemagglutinin Approach. J. Virol. 2019, 93, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Savic, M.; Dembinski, J.L.; Kim, Y.; Tunheim, G.; Cox, R.J.; Oftung, F.; Peters, B.; Mjaaland, S. Epitope specific T-cell responses against influenza A in a healthy population. Immunology 2016, 147, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Hillaire, M.L.B.; van Trierum, S.E.; Kreijtz, J.; Bodewes, R.; Geelhoed-Mieras, M.M.; Nieuwkoop, N.J.; Fouchier, R.A.M.; Kuiken, T.; Osterhaus, A.; Rimmelzwaan, G.F. Cross-protective immunity against influenza pH1N1 2009 viruses induced by seasonal influenza A (H3N2) virus is mediated by virus-specific T-cells. J. Gen. Virol. 2011, 92, 2339–2349. [Google Scholar] [CrossRef] [PubMed]
- Grant, E.J.; Chen, L.; Quinones-Parra, S.; Pang, K.; Kedzierska, K.; Chen, W. T-cell immunity to influenza A viruses. Crit. Rev. Immunol. 2014, 34, 15–39. [Google Scholar] [CrossRef] [PubMed]
- Hemann, E.A.; Kang, S.M.; Legge, K.L. Legge, Protective CD8 T cell-mediated immunity against influenza A virus infection following influenza virus-like particle vaccination. J. Immunol. 2013, 191, 2486–2494. [Google Scholar] [CrossRef] [PubMed]
- Bullard, B.L.; Corder, B.N.; DeBeauchamp, J.; Rubrum, A.; Korber, B.; Webby, R.J.; Weaver, E.A. Epigraph hemagglutinin vaccine induces broad cross-reactive immunity against swine H3 influenza virus. Nat. Commun. 2021, 12, 1203. [Google Scholar] [CrossRef] [PubMed]
- Bullard, B.L.; DeBeauchamp, J.; Pekarek, M.J.; Petro-Turnquist, E.; Vogel, P.; Webby, R.J.; Weaver, E.A. An epitope-optimized human H3N2 influenza vaccine induces broadly protective immunity in mice and ferrets. NPJ Vaccines 2022, 7, 65. [Google Scholar] [CrossRef]
- Pekarek, M.J.; Petro-Turnquist, E.M.; Rubrum, A.; Webby, R.J.; Weaver, E.A. Expanding Mouse-Adapted Yamagata-like Influenza B Viruses in Eggs Enhances In Vivo Lethality in BALB/c Mice. Viruses 2022, 14, 1299. [Google Scholar] [CrossRef]
- He, T.C.; Zhou, S.; da Costa, L.T.; Yu, J.; Kinzler, K.W.; Vogelstein, B. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 1998, 95, 2509–2514. [Google Scholar] [CrossRef]
- Plunkett, M.L.; Springer, T.A. Purification and characterization of the lymphocyte function-associated-2 (LFA-2) molecule. J. Immunol. 1986, 136, 4181–4187. [Google Scholar] [CrossRef]
- Theiler, J.; Korber, B. Graph-based optimization of epitope coverage for vaccine antigen design. Stat. Med. 2018, 37, 181–194. [Google Scholar] [CrossRef]
- Theiler, J.; Yoon, H.; Yusim, K.; Picker, L.J.; Fruh, K.; Korber, B. Epigraph: A Vaccine Design Tool Applied to an HIV Therapeutic Vaccine and a Pan-Filovirus Vaccine. Sci. Rep. 2016, 6, 33987. [Google Scholar] [CrossRef] [PubMed]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Gallo Cassarino, T.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Velkov, T. The specificity of the influenza B virus hemagglutinin receptor binding pocket: What does it bind to? J. Mol. Recognit. 2013, 26, 439–449. [Google Scholar] [CrossRef]
- Thurmond, J.; Yoon, H.; Kuiken, C.; Yusim, K.; Perkins, S.; Theiler, J.; Bhattacharya, T.; Korber, B.; Fischer, W. Web-based design and evaluation of T-cell vaccine candidates. Bioinformatics 2008, 24, 1639–1640. [Google Scholar] [CrossRef]
- Hobson, D.; Curry, R.L.; Beare, A.S.; Ward-Gardner, A. The role of serum haemagglutination-inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. J. Hyg. 1972, 70, 767–777. [Google Scholar] [CrossRef]
- Coudeville, L.; Bailleux, F.; Riche, B.; Megas, F.; Andre, P.; Ecochard, R. Relationship between haemagglutination-inhibiting antibody titres and clinical protection against influenza: Development and application of a bayesian random-effects model. BMC Med. Res. Methodol. 2010, 10, 18. [Google Scholar] [CrossRef]
- Heeringa, M.; Leav, B.; Smolenov, I.; Palladino, G.; Isakov, L.; Matassa, V. Comparability of Titers of Antibodies against Seasonal Influenza Virus Strains as Determined by Hemagglutination Inhibition and Microneutralization Assays. J. Clin. Microbiol. 2020, 58, 10–1128. [Google Scholar] [CrossRef]
- Verschoor, C.P.; Singh, P.; Russell, M.L.; Bowdish, D.M.; Brewer, A.; Cyr, L.; Ward, B.J.; Loeb, M. Microneutralization assay titres correlate with protection against seasonal influenza H1N1 and H3N2 in children. PLoS ONE 2015, 10, e0131531. [Google Scholar] [CrossRef]
- Koutsakos, M.; Nguyen, T.H.; Barclay, W.S.; Kedzierska, K. Knowns and unknowns of influenza B viruses. Future Microbiol. 2016, 11, 119–135. [Google Scholar] [CrossRef] [PubMed]
- Terajima, M.; Babon, J.A.; Co, M.D.; Ennis, F.A. Cross-reactive human B cell and T cell epitopes between influenza A and B viruses. Virol. J. 2013, 10, 244. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Liu, C.; Lu, X.; Ling, Z.; Yi, C.; Zhang, Z.; Li, Z.; Jin, M.; Wang, W.; Tang, S.; et al. Unique binding pattern for a lineage of human antibodies with broad reactivity against influenza A virus. Nat. Commun. 2022, 13, 2378. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F.; Margine, I.; Tan, G.S.; Pica, N.; Krause, J.C.; Palese, P. A carboxy-terminal trimerization domain stabilizes conformational epitopes on the stalk domain of soluble recombinant hemagglutinin substrates. PLoS ONE 2012, 7, e43603. [Google Scholar] [CrossRef] [PubMed]
- McCann, N.; O’Connor, D.; Lambe, T.; Pollard, A.J. Viral vector vaccines. Curr. Opin. Immunol. 2022, 77, 102210. [Google Scholar] [CrossRef] [PubMed]
- Ghattas, M.; Dwivedi, G.; Lavertu, M.; Alameh, M.G. Vaccine Technologies and Platforms for Infectious Diseases: Current Progress, Challenges, and Opportunities. Vaccines 2021, 9, 1490. [Google Scholar] [CrossRef] [PubMed]
- Gravel, C.; Muralidharan, A.; Duran, A.; Zetner, A.; Pfeifle, A.; Zhang, W.; Hashem, A.; Tamming, L.; Farnsworth, A.; Loemba, H.; et al. Synthetic vaccine affords full protection to mice against lethal challenge of influenza B virus of both genetic lineages. iScience 2021, 24, 103328. [Google Scholar] [CrossRef]
- Dhanasekaran, V.; Sullivan, S.; Edwards, K.M.; Xie, R.; Khvorov, A.; Valkenburg, S.A.; Cowling, B.J.; Barr, I.G. Human seasonal influenza under COVID-19 and the potential consequences of influenza lineage elimination. Nat. Commun. 2022, 13, 1721. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Recommended Composition of Influenza Virus Vaccines for Use in the 2024 Southern Hemisphere Influenza Season. 2023. Available online: https://cdn.who.int/media/docs/default-source/influenza/who-influenza-recommendations/vcm-southern-hemisphere-recommendation-2024/202309_recommendation.pdf?sfvrsn=2c2cbebd_8&download=true (accessed on 23 November 2023).
- Huang, Q.S.; Wood, T.; Jelley, L.; Jennings, T.; Jefferies, S.; Daniells, K.; Nesdale, A.; Dowell, T.; Turner, N.; Campbell-Stokes, P.; et al. Impact of the COVID-19 nonpharmaceutical interventions on influenza and other respiratory viral infections in New Zealand. Nat. Commun. 2021, 12, 1001. [Google Scholar] [CrossRef]
- Paget, J.; Caini, S.; Del Riccio, M.; van Waarden, W.; Meijer, A. Has influenza B/Yamagata become extinct and what implications might this have for quadrivalent influenza vaccines? Eurosurveillance 2022, 27, 2200753. [Google Scholar] [CrossRef]
- Chen, R.; Holmes, E.C. The evolutionary dynamics of human influenza B virus. J. Mol. Evol. 2008, 66, 655–663. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petro-Turnquist, E.; Corder Kampfe, B.; Gadeken, A.; Pekarek, M.J.; Weaver, E.A. Multivalent Epigraph Hemagglutinin Vaccine Protects against Influenza B Virus in Mice. Pathogens 2024, 13, 97. https://doi.org/10.3390/pathogens13020097
Petro-Turnquist E, Corder Kampfe B, Gadeken A, Pekarek MJ, Weaver EA. Multivalent Epigraph Hemagglutinin Vaccine Protects against Influenza B Virus in Mice. Pathogens. 2024; 13(2):97. https://doi.org/10.3390/pathogens13020097
Chicago/Turabian StylePetro-Turnquist, Erika, Brigette Corder Kampfe, Amber Gadeken, Matthew J. Pekarek, and Eric A. Weaver. 2024. "Multivalent Epigraph Hemagglutinin Vaccine Protects against Influenza B Virus in Mice" Pathogens 13, no. 2: 97. https://doi.org/10.3390/pathogens13020097
APA StylePetro-Turnquist, E., Corder Kampfe, B., Gadeken, A., Pekarek, M. J., & Weaver, E. A. (2024). Multivalent Epigraph Hemagglutinin Vaccine Protects against Influenza B Virus in Mice. Pathogens, 13(2), 97. https://doi.org/10.3390/pathogens13020097




