Role of Recognition MicroRNAs in Hemaphysalis longicornis and Theileria orientalis Interactions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Tick Collection and RNA Extraction
2.3. Construction of the Tick Infection Model
2.4. High-Throughput Sequencing of Small RNAs
2.5. Small RNA Analysis
2.6. miRNA Targets and Gene Ontology Analysis
2.7. Real-Time Quantitative PCR
2.8. Analysis of the Dual-Luciferase Reporter System
2.9. Statistical Analysis
3. Results
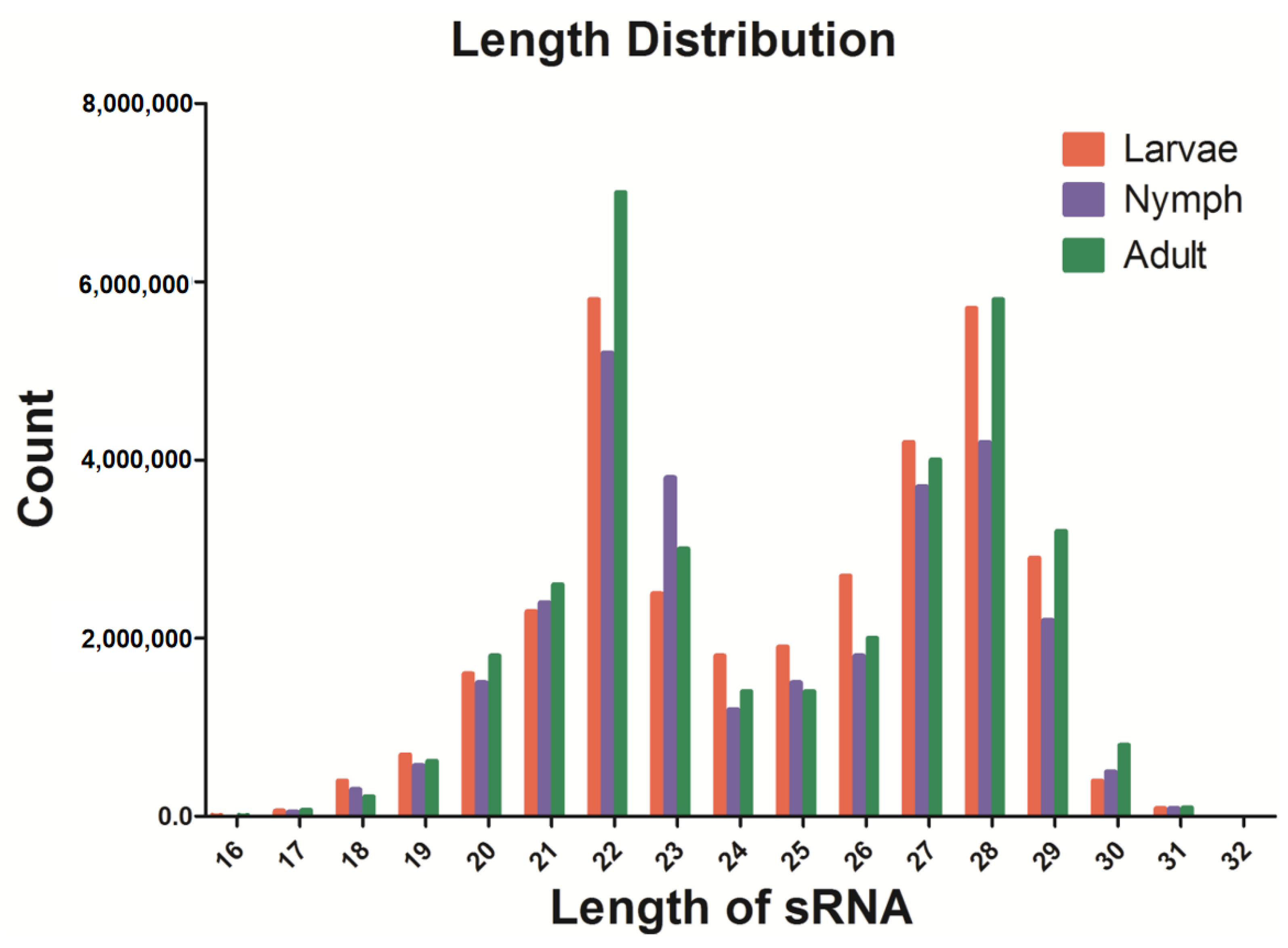
3.1. Small RNA Library Construction and Solexa Sequencing
3.2. Differential Expression Analysis
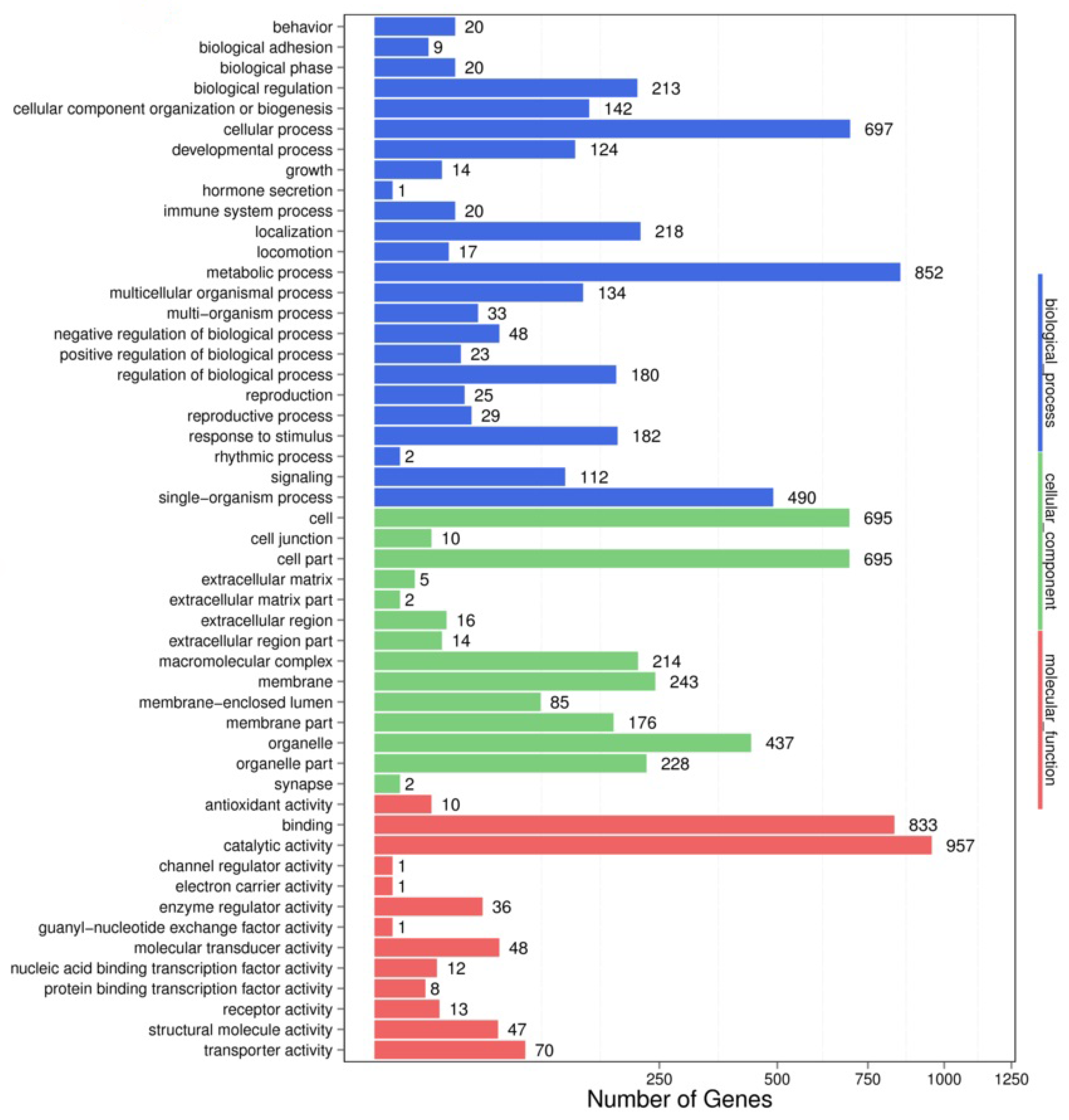
3.3. GO Enrichment Analysis of the miR-5309 Target Genes
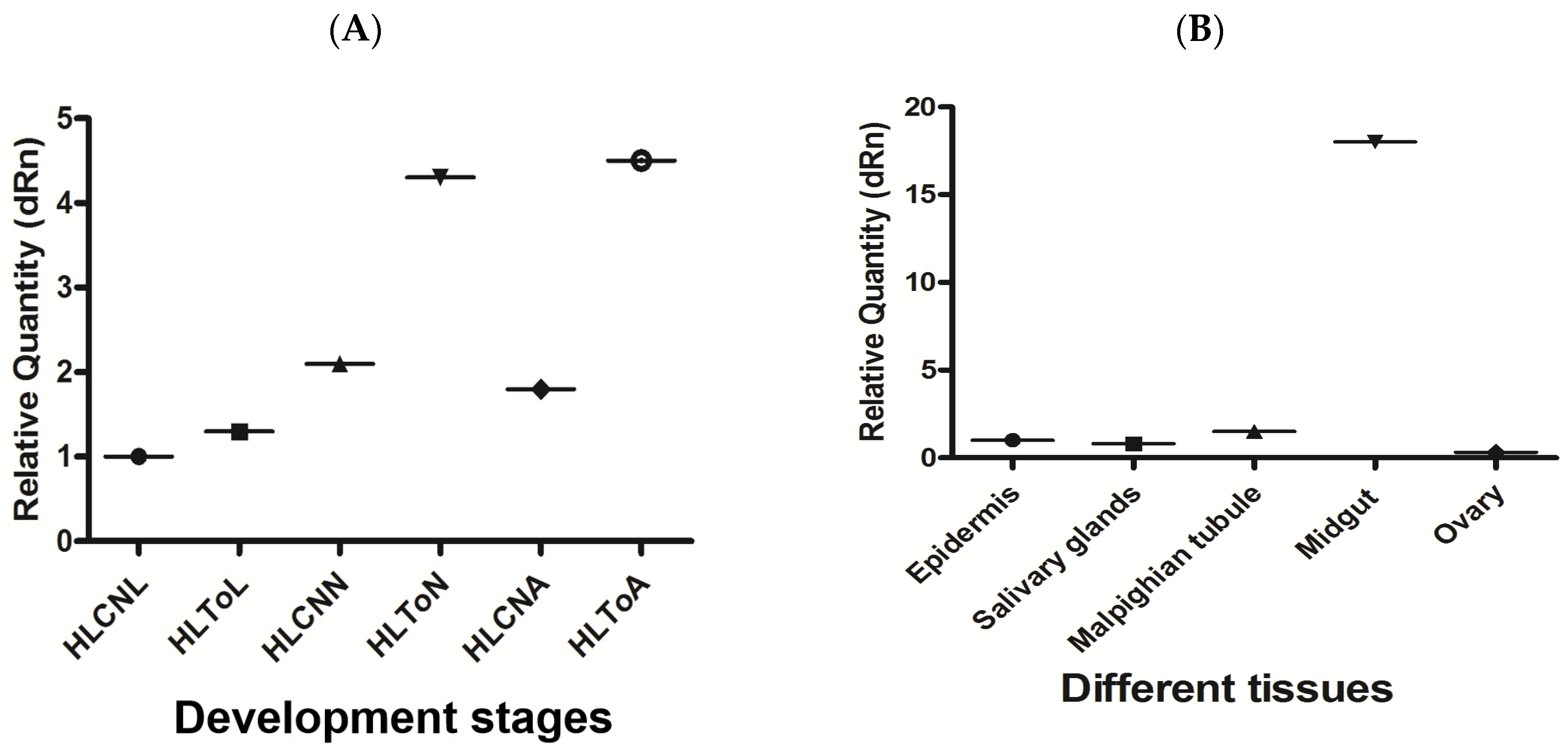
3.4. Analysis of the Expression of miR-5309 and Its Target Longipain in Infected Ticks
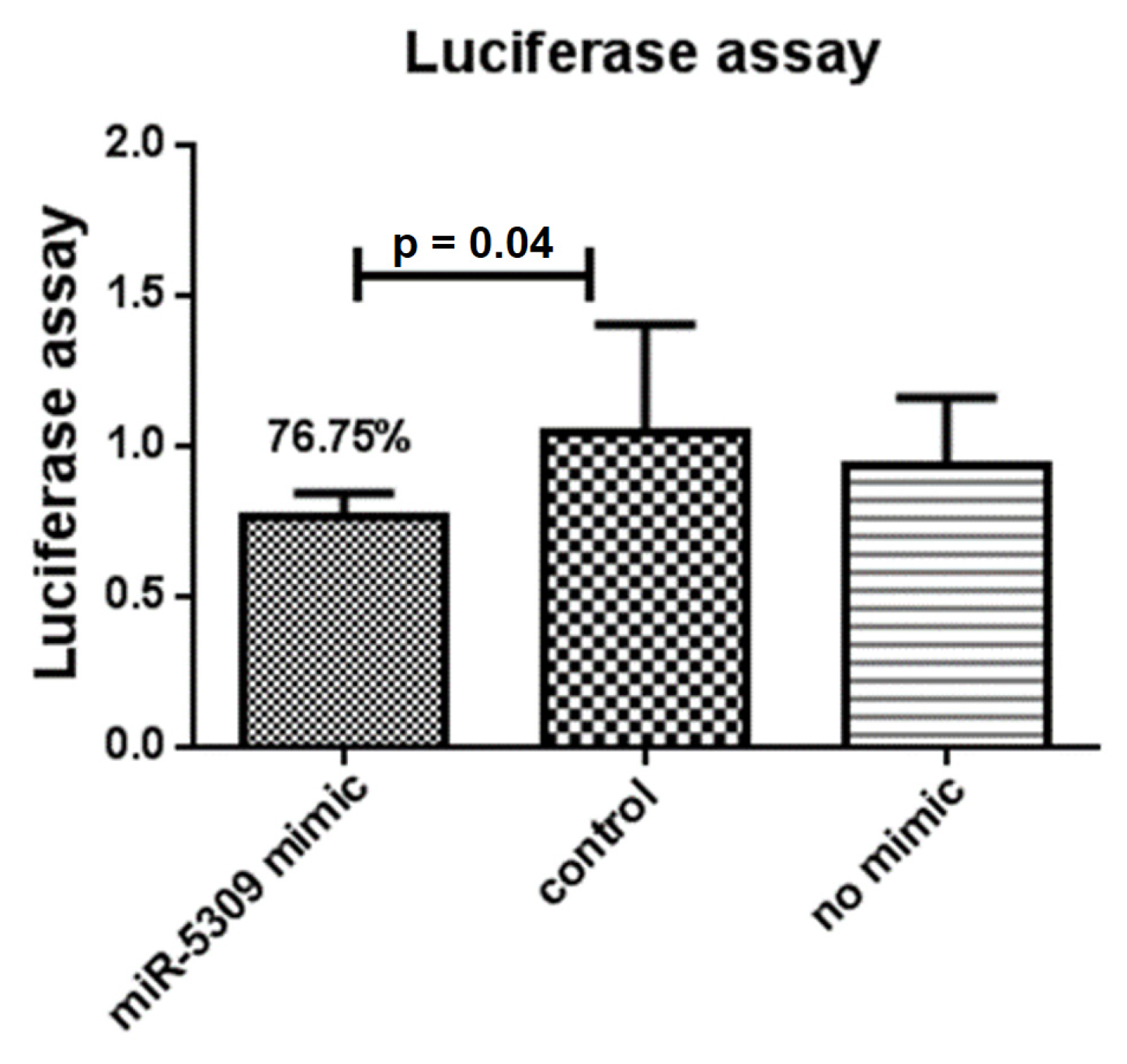
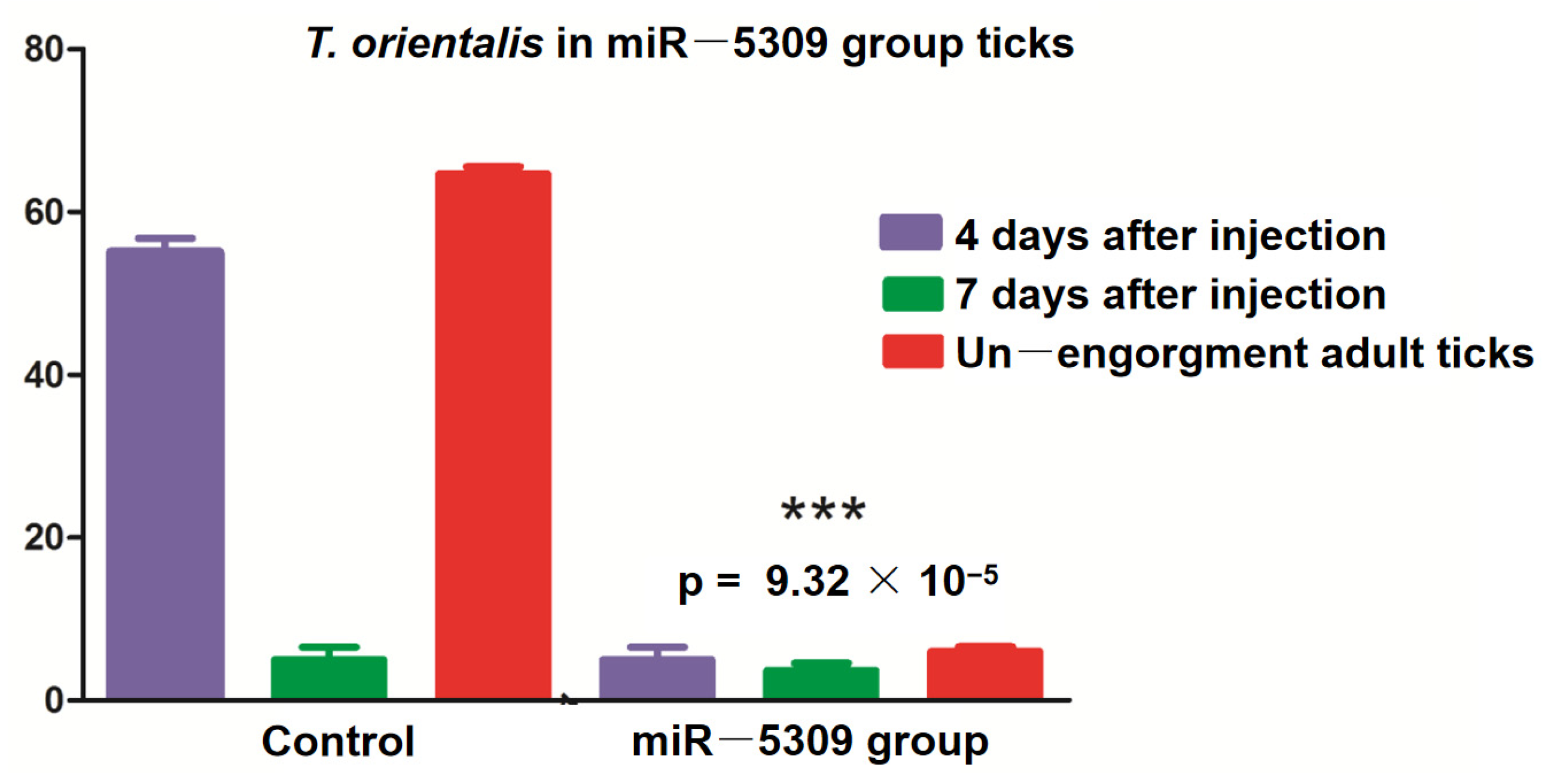
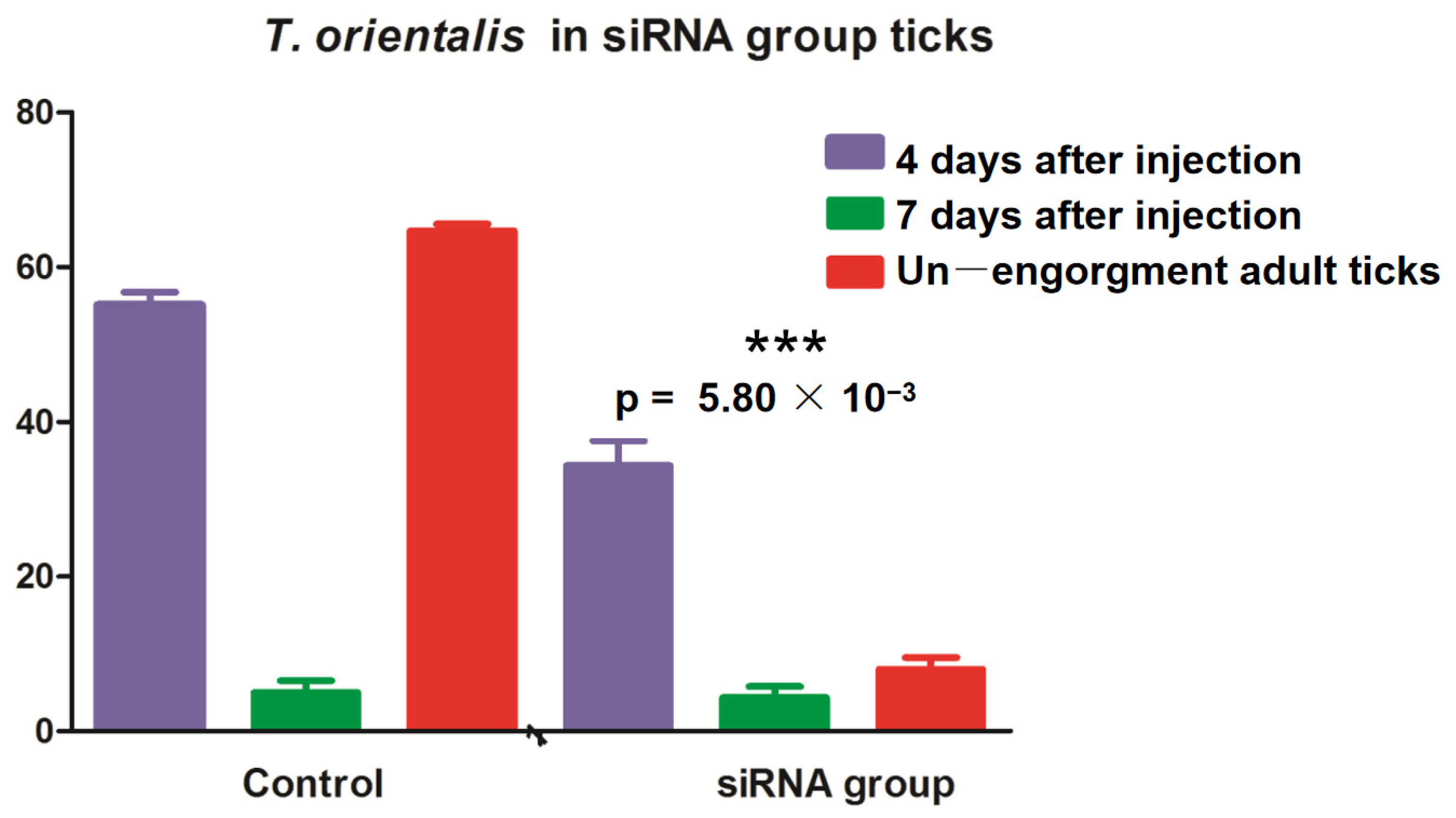
3.5. Effects of miRNA-Blocking Inhibition on Ticks
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berezikov, E.; Guryev, V.; Van de Belt, J.; Wienholds, E.; Plasterk, R.H.A.; Cuppen, E. Phylogenetic Shadowing and Computational Identification of Human microRNA Genes- ScienceDirect. Cell 2005, 120, 21–24. [Google Scholar] [CrossRef] [PubMed]
- Engels, B.M.; Hutvagner, G. Principles and effects of microRNA-mediated posttranscriptional gene regulation. Oncogene 2006, 25, 6163–6169. [Google Scholar] [CrossRef] [PubMed]
- Lakew, B.T.; Eastwood, S.; Walkden-Brown, S.W. Epidemiology and Transmission of Theileria orientalis in Australasia. Pathogens 2023, 12, 1187. [Google Scholar] [CrossRef] [PubMed]
- Dinkel, K.D.; Herndon, D.R.; Noh, S.M.; Lahmers, K.K.; Todd, S.M.; Ueti, M.W.; Scoles, G.A.; Mason, K.L.; Fry, L.M. A U.S. isolate of Theileria orientalis, Ikeda genotype, is transmitted to cattle by the invasive Asian longhorned tick, Hemaphysalis longicornis. Parasites Vectors 2021, 14, 157. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.G.; Playford, M.C.; Hickey, K.L. Theileria orientalis: A review. N. Z. Vet. J. 2016, 64, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Damian, D.; Damas, M.; Wensman, J.J.; Berg, M. Molecular Diversity of Hard Tick Species from Selected Areas of a Wildlife-Livestock Interface Ecosystem at Mikumi National Park, Morogoro Region, Tanzania. Vet. Sci. 2021, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Li, J.C.; Zhao, J.; Li, H.; Fang, L.Q.; Liu, W. Epidemiology, clinical characteristics, and treatment of severe fever with thrombocytopenia syndrome. Infect. Med. 2022, 1, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Couret, J.; Schofield, S.; Narasimhan, S. The environment, the tick, and the pathogen—It is an ensemble. Front. Cell Infect. Microbiol. 2022, 12, 1049646. [Google Scholar] [CrossRef]
- Fogaça, A.C.; Sousa, G.; Pavanelo, D.B.; Esteves, E.; Martins, L.A.; Urbanová, V.; Kopáček, P.; Daffre, S. Tick Immune System: What Is Known, the Interconnections, the Gaps, and the Challenges. Front. Immunol. 2021, 12, 628054. [Google Scholar] [CrossRef]
- Luo, J.; Wu, F.; Liu, W.; Ren, Q.; Diao, P.; Guan, G.; Luo, J.; Yin, H.; Liu, G. A Novel MicroRNA and the Target Gene TAB2 Can Regulate the Process of Sucking Blood in and the Spawn Rate of Hyalomma asiaticum (Acari: Ixodidae) Ticks. Front. Immunol. 2022, 13, 930532. [Google Scholar] [CrossRef]
- Shao, X.; Gong, W.; Wang, Q.; Wang, P.; Shi, T.; Mahmut, A.; Qin, J.; Yao, Y.; Yan, W.; Chen, D.; et al. Atrophic skeletal muscle fiber-derived small extracellular vesicle miR-690 inhibits satellite cell differentiation during aging. J. Cachexia Sarcopenia Muscle 2022, 13, 3163–3180. [Google Scholar] [CrossRef] [PubMed]
- Adil, M.S.; Khulood, D.; Somanath, P.R. Targeting Akt-associated microRNAs for cancer therapeutics. Biochem. Pharmacol. 2021, 189, 114384. [Google Scholar] [CrossRef] [PubMed]
- Selinger, M.; Věchtová, P.; Tykalová, H.; Ošlejšková, P.; Rumlová, M.; Štěrba, J.; Grubhoffer, L. Integrative RNA profiling of TBEV-infected neurons and astrocytes reveals potential pathogenic effectors. Comput. Struct. Biotechnol. J. 2022, 20, 2759–2777. [Google Scholar] [CrossRef]
- Liu, X.; Xiao, H.; Peng, X.; Chai, Y.; Wang, S.; Wen, G. Identification and comprehensive analysis of circRNA–miRNA–mRNA regulatory networks in osteoarthritis. Front. Immunol. 2023, 13, 1050743. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.M.; Xu, L.; Yau, M.Y. Alternative mRNA Splicing in the Pathogenesis of Obesity. Int. J. Mol. Sci. 2018, 19, 632. [Google Scholar] [CrossRef] [PubMed]
- Yagi, Y.; Ohnuma, A.; Shiono, H.; Chikayama, Y.; Ito, T. Cytokine and inducible nitric oxide synthase gene expressions in peripheral blood mononuclear cells and related clinical characteristics in Theileria orientalis sergenti-infected calves. J. Vet. Med. Sci. 2003, 65, 1355–1359. [Google Scholar] [CrossRef]
- Nandakumar, M.; Malathi, P.; Sundar, A.R.; Rajadurai, C.P.; Philip, M.; Viswanathan, R. Role of miRNAs in the host–interaction between sugarcane and Colletotrichum falcatum, the red rot pathogen. Plant Cell Rep. 2021, 40, 851–870. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Li, G.; Guo, Y.; Gao, Y.; Zhu, L.; Liu, Z.; Tian, R.; Gao, C.; Han, P.; Wang, N.; et al. A fungal microRNA-like RNA subverts host immunity and facilitates pathogen infection by silencing two host receptor-like kinase genes. New Phytol. 2022, 233, 2503–2519. [Google Scholar] [CrossRef]
- Raziyeva, K.; Kim, Y.; Zharkinbekov, Z.; Kassymbek, K.; Jimi, S.; Saparov, A. Immunology of Acute and Chronic Wound Healing. Biomolecules 2021, 11, 700. [Google Scholar] [CrossRef]
- Liu, W.; Rodgers, G.P. Olfactomedin 4 expression and functions in innate immunity, inflammation, and cancer. Cancer Metastasis Rev. 2016, 35, 201–212. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, L.; Hong, L.; Zheng, W.; Cui, J.; Liu, X.; Xu, T. MicroRNA-181b-2 and MicroRNA-21-1 Negatively Regulate NF-κB and IRF3-Mediated Innate Immune Responses via Targeting TRIF in Teleost. Front. Immunol. 2021, 12, 734520. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Yang, Y.; Suen, A.; Zhu, J.; Williams, B.; Hu, J.; Chen, F.; Kozar, R.; Shen, S.; Li, Z.; et al. Role of extracellular microRNA-146a-5p in host innate immunity and bacterial sepsis. IScience 2021, 24, 103441. [Google Scholar] [CrossRef] [PubMed]
- Holla, S.; Balaji, K.N. Epigenetics and miRNA during bacteria-induced host immune responses. Epigenomics 2015, 7, 1197–1212. [Google Scholar] [CrossRef]
- Duval, M.; Cossart, P.; Lebreton, A. Mammalian microRNAs and long noncoding RNAs in the host-bacterial pathogen crosstalk. Semin. Cell Dev. Biol. 2017, 65, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Francés, R.; Mata-Garrido, J.; de la Fuente, R.; Carcelén, M.; Lafarga, M.; Berciano, M.T.; García, R.; Hurlé, M.A.; Tramullas, M. Identification of Epigenetic Interactions between MicroRNA-30c-5p and DNA Methyltransferases in Neuropathic Pain. Int. J. Mol. Sci. 2022, 23, 13994. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Huang, J.; Wu, S.; Li, Y.; Pan, Y. Integrative analysis of miRNA and mRNA expression associated with the immune response in the intestine of rainbow trout (Oncorhynchus mykiss) infected with infectious hematopoietic necrosis virus. Fish. Shellfish. Immunol. 2022, 131, 54–66. [Google Scholar] [CrossRef] [PubMed]
- Haidar, M.; Tajeri, S.; Momeux, L.; Mourier, T.; Ben-Rached, F.; Mfarrej, S.; Rchiad, Z.; Pain, A.; Langsley, G. miR-34c-3p Regulates Protein Kinase A Activity Independent of cAMP by Dicing prkar2b Transcripts in Theileria annulata-Infected Leukocytes. mSphere 2023, 8, e0052622. [Google Scholar] [CrossRef] [PubMed]
- Haidar, M.; Rchiad, Z.; Ansari, H.R.; Ben-Rached, F.; Tajeri, S.; Latre De Late, P.; Langsley, G.; Pain, A. miR-126-5p by direct targeting of JNK-interacting protein-2 (JIP-2) plays a key role in Theileria-infected macrophage virulence. PLoS Pathog. 2018, 14, e1006942. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, N.; Miyoshi, T.; Battsetseg, B.; Matsuo, T.; Xuan, X.; Fujisaki, K. A cysteine protease is critical for Babesia spp. transmission in Hemaphysalis ticks. PLoS Pathog. 2008, 4, e1000062. [Google Scholar] [CrossRef]
- Luo, J.; Liu, M.X.; Ren, Q.Y.; Chen, Z.; Tian, Z.C.; Hao, J.W.; Wu, F.; Liu, X.C.; Luo, J.X.; Yin, H.; et al. Micropathogen Community Analysis in Hyalomma rufipes via High-Throughput Sequencing of Small RNAs. Front. Cell Infect. Microbiol. 2017, 7, 374. [Google Scholar] [CrossRef]
- Clop, A.; Marcq, F.; Takeda, H.; Pirottin, D.; Tordoir, X.; Bibé, B.; Bouix, J.; Caiment, F.; Elsen, J.M.; Eychenne, F.; et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat. Genet. 2006, 38, 813–818. [Google Scholar] [CrossRef] [PubMed]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef]
- Schwab, R.; Palatnik, J.F.; Riester, M.; Schommer, C.; Schmid, M.; Weigel, D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell 2005, 8, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.W.; Slattery, M.; Mann, R.S. Transcription factor choice in the Hippo signaling pathway: Homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes. Dev. 2009, 23, 2307–2319. [Google Scholar] [CrossRef]
- Luo, J.; Zhao, S.; Ren, Q.; Wang, Q.; Chen, Z.; Cui, J.; Jing, Y.; Liu, P.; Yan, R.; Song, X.; et al. Dynamic Analysis of microRNAs from Different Life Stages of Rhipicephalus microplus (Acari: Ixodidae) by High-Throughput Sequencing. Pathogens 2022, 11, 1148. [Google Scholar] [CrossRef]
- McGurran, H.; Kumbol, V.; Krüger, C.; Wallach, T.; Lehnardt, S. miR-154-5p Is a Novel Endogenous Ligand for TLR7 Inducing Microglial Activation and Neuronal Injury. Cells 2024, 13, 407. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Ren, Q.; Liu, W.; Qiu, X.; Zhang, G.; Tan, Y.; Cao, R.; Yin, H.; Luo, J.; Li, X.; et al. MicroRNA-1 Expression and Function in Hyalomma Anatolicum anatolicum (Acari: Ixodidae) Ticks. Front. Physiol. 2021, 12, 596289. [Google Scholar] [CrossRef]
- Mishra, R.; Krishnamoorthy, P.; Kumar, H. MicroRNA-30e-5p Regulates SOCS1 and SOCS3 During Bacterial Infection. Front. Cell Infect. Microbiol. 2021, 10, 604016. [Google Scholar] [CrossRef]
- Omondi, D.; Zweygarth, E.; Murungi, E.; Jongejan, F.; Nijhof, A.M. De novo assembly and annotation of the Amblyomma hebraeum tick midgut transcriptome response to Ehrlichia ruminantium infection. PLoS. Negl. Trop. Dis. 2023, 17, e0011554. [Google Scholar] [CrossRef]
- Luo, J.; Shen, H.; Ren, Q.; Guan, G.; Zhao, B.; Yin, H.; Chen, R.; Zhao, H.; Luo, J.; Li, X.; et al. Characterization of an MLP Homologue from Haemaphysalis longicornis (Acari: Ixodidae) Ticks. Pathogens 2020, 9, 284. [Google Scholar] [CrossRef]

| Primers Name | Primer Sequence (5′–3′) |
|---|---|
| Stem–loop primer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTTCGGGG |
| β-Actin sense primer | GTTCCTGGGTATGGAATCG |
| β-Actin antisense primer | TCCACGTCGCACTTCATGAT |
| miR5309 F | ACACTCCAGCTGGCAATGCCCATGGAA |
| miR5309 R | GTCGTATCCAGTGCAGGGTCC |
| longipain-qPCR-F | CAACTCCTGGAACACCGAATGGG |
| longipain-qPCR-R | TCGTCTTCAATGCCGCACTCATC |
| si-hlo-longipain | GGCGTATTACAATCCGAAA |
| miRNA Name | Count (HLTor) | Count (HLNC) | TPM (HLTor) | TPM (HLNC) | log2 Ratio (NC/Tor) | Up- or Downregulation | p Value |
|---|---|---|---|---|---|---|---|
| miR-5309 | 130 | 1804 | 22.88 | 180.57 | 1.8161 | Up | 8.84 × 10−193 |
| miR-2001 | 1324 | 5229 | 233.01 | 523.98 | 1.1691 | Up | 1.69 × 10−176 |
| miR-96 | 99 | 1305 | 17.42 | 130.77 | 2.9082 | Up | 1.82 × 10−145 |
| miR-184 | 67,803 | 138,893 | 11,932.56 | 13,917.91 | 0.2220 | Up | 1.17 × 10−140 |
| miR-5312 | 63 | 1090 | 11.09 | 109.22 | 3.2999 | Up | 8.67 × 10−137 |
| miR-96 | 906 | 5238 | 155.75 | 343.04 | 1.1391 | Up | 5.64 × 10−127 |
| miR-5315 | 207 | 1329 | 36.43 | 133.17 | 1.8701 | Up | 1.72 × 10−89 |
| miR-5310 | 218 | 1199 | 38.37 | 120.15 | 1.6467 | Up | 2.21 × 10−68 |
| miR-993 | 1839 | 1429 | 240.88 | 97.05 | −1.3115 | Down | 3.84 × 10−147 |
| miR-2a | 1339 | 1180 | 235.65 | 118.24 | −0.9949 | Down | 1.58 × 10−66 |
| miRNA | Site | Free Energy | Binding Site |
|---|---|---|---|
| miR-5309 | 1122 | −27.8 kcal/mol | Target 5′ C U GAUGA C A 3′ UUCG GGG UCC GUG GGCAUUG AAGC CCC AGG UAC CCGUAAC miRNA 3′ AA 5′ |
| Cellular Components | Molecular Functions | Biological Processes |
|---|---|---|
| caspase complex | cysteine-type endopeptidase inhibitor activity | negative regulation of cysteine-type endopeptidase activity |
| cysteine-type endopeptidase activity involved in apoptotic process | positive regulation of cysteine-type endopeptidase activity | |
| GPI-anchor transamidase activity | regulation of cysteine-type endopeptidase activity | |
| cysteine-type endopeptidase activator activity | / | |
| calcium-dependent cysteine-type endopeptidase activity | / | |
| ubiquitin-like protein-specific endopeptidase activity | / |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, J.; Zhao, S.; Ren, Q.; Guan, G.; Luo, J.; Yin, H. Role of Recognition MicroRNAs in Hemaphysalis longicornis and Theileria orientalis Interactions. Pathogens 2024, 13, 288. https://doi.org/10.3390/pathogens13040288
Luo J, Zhao S, Ren Q, Guan G, Luo J, Yin H. Role of Recognition MicroRNAs in Hemaphysalis longicornis and Theileria orientalis Interactions. Pathogens. 2024; 13(4):288. https://doi.org/10.3390/pathogens13040288
Chicago/Turabian StyleLuo, Jin, Shuaiyang Zhao, Qiaoyun Ren, Guiquan Guan, Jianxun Luo, and Hong Yin. 2024. "Role of Recognition MicroRNAs in Hemaphysalis longicornis and Theileria orientalis Interactions" Pathogens 13, no. 4: 288. https://doi.org/10.3390/pathogens13040288







