A Set of Multiresistant Isolates of Mycoplasma bovis Subtype ST-1 with a Variable Susceptibility to Quinolones Are Also Circulating in Spain
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Presence of Mycoplasmas, Identification and Subtyping
2.3. MIC Assays with M. bovis Isolates
3. Results
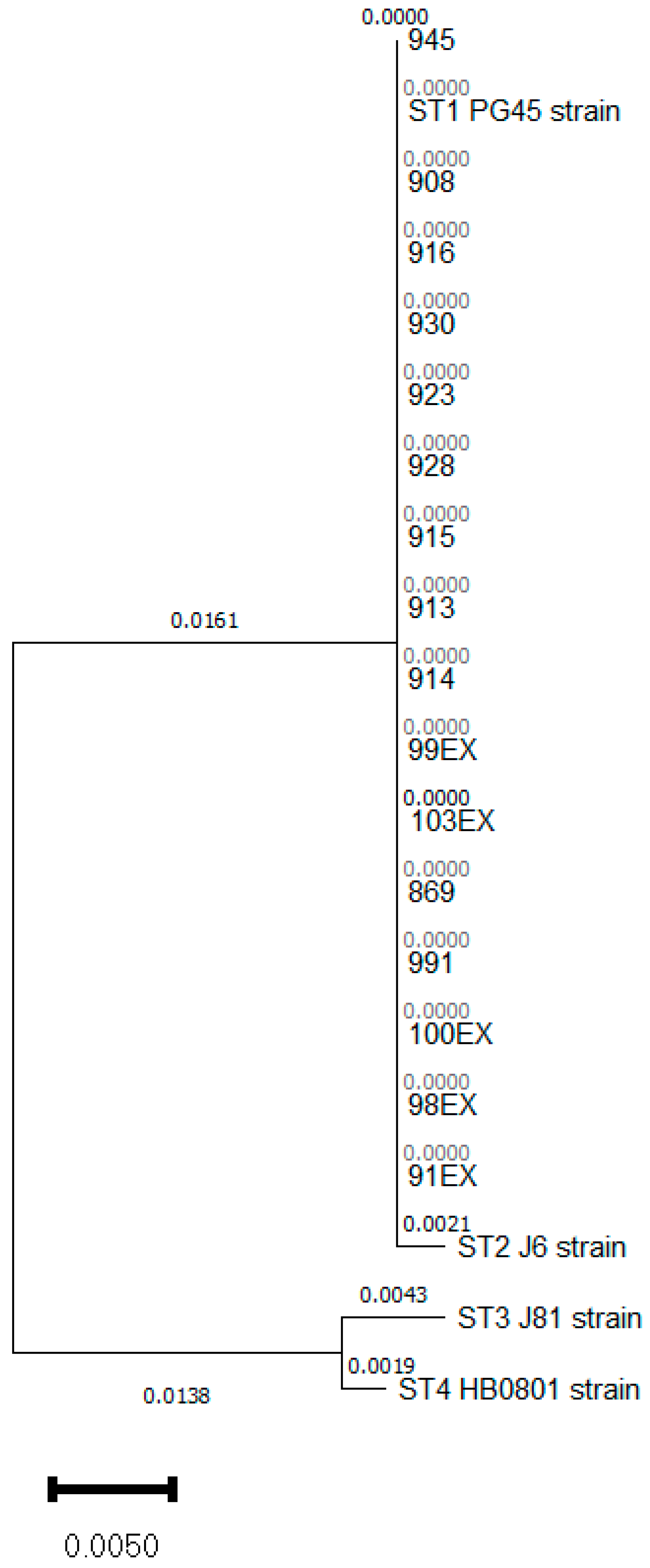
3.1. Mycoplasma bovis ST 1 Is Also Circulating in Spain with a Variable Antimicrobial Susceptibility to FLQ
3.2. Analysis of Point Mutations Conferring Resistance to Quinolones
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Penterman, P.M.; Holzhauer, M.; van Engelen, E.; Smits, D.; Velthuis, A.G.J. Dynamics of Mycoplasma bovis in Dutch dairy herds during acute clinical outbreaks. Vet. J. 2022, 283–284, 105841. [Google Scholar] [CrossRef] [PubMed]
- Torres, F.D.; Madureira, K.M.; Nascimento da Silva, K.; Azevedo, C.; de Moura, T.M.; Skorei, M.R.; Gomes, V. Influence of Mycoplasma bovis infection on milk production and quality of Holstein dairy cows. J. Dairy Res. 2022, 89, 416–418. [Google Scholar] [CrossRef] [PubMed]
- Hasoon, M.F.; Jarocki, V.M.; Mohammed, M.H.; Djordjevic, S.P.; Yip, H.Y.E.; Carr, M.; Khabiri, A.; Azari, A.A.; Amanollahi, R.; Jozani, R.J.; et al. Antimicrobial susceptibility and molecular characteristics of Mycoplasma bovis isolated from cases of bovine respiratory disease in Australian feedlot cattle. Vet. Microbiol. 2023, 283, 109779. [Google Scholar] [CrossRef] [PubMed]
- Thézé, J.; Ambroset, C.; Barry, S.; Masseglia, S.; Colin, A.; Tricot, A.; Tardy, F.; Bailly, X. Genome-wide phylodynamic approach reveals the epidemic dynamics of the main Mycoplasma bovis subtype circulating in France. Microb. Genom. 2023, 9, mgen001067. [Google Scholar] [CrossRef] [PubMed]
- García-Galán, A.; Nouvel, L.X.; Baranowski, E.; Gómez-Martín, Á.; Sánchez, A.; Citti, C.; de la Fe, C. Mycoplasma bovis in Spanish Cattle Herds: Two Groups of Multiresistant Isolates Predominate, with One Remaining Susceptible to Fluoroquinolones. Pathogens 2020, 9, 545. [Google Scholar] [CrossRef] [PubMed]
- García-Galán, A.; Seva, J.; Gómez-Martín, Á.; Ortega, J.; Rodríguez, F.; García-Muñoz, Á.; De la Fe, C. Importance and Antimicrobial Resistance of Mycoplasma bovis in Clinical Respiratory Disease in Feedlot Calves. Animals 2021, 11, 1470. [Google Scholar] [CrossRef]
- Becker, C.A.M.; Thibault, F.M.; Arcangioli, M.-A.; Tardy, F. Loss of diversity within Mycoplasma bovis isolates collected in France from bovines with respiratory diseases over the last 35 years. Infect. Genet. Evol. 2015, 33, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Klein, U.; de Jong, A.; Moyaert, H.; El Garch, F.; Leon, R.; Richard-Mazet, A.; Rose, M.; Maes, D.; Pridmore, A.; Thomson, J.R.; et al. Antimicrobial susceptibility monitoring of Mycoplasma hyopneumoniae and Mycoplasma bovis isolated in Europe. Vet. Microbiol. 2017, 204, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Klein, U.; de Jong, A.; Youala, M.; El Garch, F.; Stevenin, C.; Moyaert, H.; Rose, M.; Catania, S.; Gyuranecz, M.; Pridmore, A.; et al. New antimicrobial susceptibility data from monitoring of Mycoplasma bovis isolated in Europe. Vet. Microbiol. 2019, 238, 108432. [Google Scholar] [CrossRef]
- Dudek, K.; Nicholas, R.A.J.; Szacawa, E.; Bednarek, D. Mycoplasma bovis Infections-Occurrence, Diagnosis and Control. Pathogens 2020, 9, 640. [Google Scholar] [CrossRef]
- Khalil, D.; Becker, C.A.M.; Tardy, F. Alterations in the Quinolone Resistance-Determining Regions and Fluoroquinolone Resistance in Clinical Isolates and Laboratory-Derived Mutants of Mycoplasma bovis: Not All Genotypes May Be Equal. Appl. Environ. Microbiol. 2016, 82, 1060–1068. [Google Scholar] [CrossRef] [PubMed]
- Waites, K.B.; Bébéar, C.M.; Robertson, J.A.; Talkington, D.F.; Kenny, G.E. Cumitech 34: Laboratory Diagnosis of Mycoplasmal Infections; American Society for Microbiology: Washington, DC, USA, 2001. [Google Scholar]
- Tola, S.; Angioi, A.; Rocchigiani, A.M.; Idini, G.; Manunta, D.; Galleri, G.; Leori, G. Detection of Mycoplasma agalactiae in sheep milk samples by polymerase chain reaction. Vet. Microbiol. 1997, 54, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Foddai, A.; Idini, G.; Fusco, M.; Rosa, N.; de la Fe, C.; Zinellu, S.; Corona, L.; Tola, S. Rapid differential diagnosis of Mycoplasma agalactiae and Mycoplasma bovis based on a multiplex-PCR and a PCR-RFLP. Mol. Cell. Probes 2005, 19, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Albers, A.C.; Fletcher, R.D. Simple method for quantitation of viable mycoplasmas. Appl. Environ. Microbiol. 1982, 43, 958–960. [Google Scholar] [CrossRef] [PubMed]
- Hannan, P.C. Guidelines and recommendations for antimicrobial minimum inhibitory concentration (MIC) testing against veterinary mycoplasma species. Vet. Res. 2000, 31, 373–395. [Google Scholar] [CrossRef] [PubMed]
- Gautier-Bouchardon, A.V.; Ferré, S.; Le Grand, D.; Paoli, A.; Gay, E.; Poumarat, F. Overall decrease in the susceptibility of Mycoplasma bovis to antimicrobials over the past 30 years in France. PLoS ONE 2014, 9, e87672. [Google Scholar] [CrossRef] [PubMed]
- Khalil, D.; Becker, C.A.M.; Tardy, F. Monitoring the Decrease in Susceptibility to Ribosomal RNAs Targeting Antimicrobials and Its Molecular Basis in Clinical Mycoplasma bovis Isolates over Time. Microb. Drug Resist. 2017, 23, 799–811. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.C.; Gao, D.; Jia, B.Y.; Wang, Z.; Gao, Y.-H.; Pei, Z.-H.; Liu, S.-M.; Xin, J.-Q.; Ma, H.-X. Antimicrobial susceptibility and molecular characterization of macrolide resistance of Mycoplasma bovis isolates from multiple provinces in China. J. Vet. Med. Sci. 2016, 78, 293–296. [Google Scholar] [CrossRef]
- Sultana, R.; Cordeiro, R.P.; Timsit, E.; McAllister, T.A.; Alexander, T.W. Prevalence and antimicrobial susceptibility of Mycoplasma bovis from the upper and lower respiratory tracts of healthy feedlot cattle and those diagnosed with bovine respiratory disease. Vet. Microbiol. 2023, 285, 109838. [Google Scholar] [CrossRef]
- Stipkovits, L.; Ripley, P.H.; Tenk, M.; Glávits, R.; Molnár, T.; Fodor, L. The efficacy of valnemulin (Econor) in the control of disease caused by experimental infection of calves with Mycoplasma bovis. Res. Vet. Sci. 2005, 78, 207–215. [Google Scholar] [CrossRef] [PubMed]
- García-Galán, A.; Baranowski, E.; Hygonenq, M.C.; Walch, M.; Croville, G.; Citti, C.; De la Fe, C.; Nouvel, L.X. Genome Mosaicism in Field Strains of Mycoplasma bovis as Footprints of In-Host Horizontal Chromosomal Transfer. Appl. Environ. Microbiol. 2022, 88, e0166121. [Google Scholar] [CrossRef] [PubMed]
| Isolates | Animal | Age | Area | Symptoms |
|---|---|---|---|---|
| 991 | Calf | 6–8 months | Murcia | Pneumonia |
| 100EX | Calf | 6–8 months | Aragón | Pneumonia |
| 103EX | Calf | 6–8 months | Castile and León | Pneumonia |
| 98EX | Calf | 6–8 months | Castile La -Mancha | Pneumonia |
| 91EX | Calf | 1 month | Catalunya | Pneumonia |
| 908 | Calf | 6–8 months | Murcia | Pneumonia |
| 916 | Calf | 6–8 months | Murcia | Pneumonia |
| 913 | Calf | 6–8 months | Murcia | Pneumonia |
| 914 | Calf | 6–8 months | Murcia | Pneumonia |
| 923 | Calf | 6–8 months | Murcia | Pneumonia |
| 915 | Calf | 6–8 months | Murcia | Pneumonia |
| 928 | Calf | 6–8 months | Murcia | Pneumonia |
| 930 | Calf | 6–8 months | Murcia | Pneumonia |
| 99EX | Calf | 6–8 months | Murcia | Pneumonia |
| 869 | Calf | 6–8 months | Murcia | Pneumonia |
| 945 | Calf | 6–8 months | Murcia | Pneumonia |
| Macrolides | FLQ | TET | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC Parameter | Tul | Gam | Eri | Tyl | Spy | Lin | Enr | Marb | Dan | Dox | Oxy | Val |
| MIC Range | 2–>16 | >16 | >16 | 8–>16 | 0.25–>16 | 0.5–>16 | 0.0625–>16 | 0.25–>16 | 0.125–8 | 0.5–8 | 2–>16 | 0.0312 |
| MIC50 | >16 | >16 | >16 | >16 | >16 | >16 | 0.25 | 0.5 | 0.25 | 1 | 4 | 0.0312 |
| MIC90 | >16 | >16 | >16 | >16 | >16 | >16 | 0.5 | 1 | 0.5 | 4 | >16 | 0.0312 |
| Isolate | gyrA | gyrB | parE | parC | MIC (µg/mL) a | ||
|---|---|---|---|---|---|---|---|
| 83 b | 105 | 109 | 278 | Enr | Marb | Dan | |
| PG45 | Ser | Asp | Gln | Ser | 0.125 | 0.5 | 0.125 |
| 991 | - | - | - | - | 2 | 0.5 | 1 |
| 100EX | - | - | - | - | <0.063 | 0.5 | 0.5 |
| 103EX | - | - | - | - | <0.063 | 1 | 0.25 |
| 98EX | - | - | - | - | <0.063 | 0.25 | 0.125 |
| 91EX | - | - | - | - | <0.063 | 0.5 | 0.25 |
| 908 | - | - | - | - | 0.25 | 0.5 | 0.125 |
| 916 | - | - | - | - | 0.25 | 0.5 | 8 |
| 913 | - | - | - | - | <0.063 | 0.25 | 0.125 |
| 914 | - | - | - | - | <0.063 | 0.25 | 0.125 |
| 923 | - | - | - | - | 8 | +8 | 4 |
| 915 | - | - | - | - | <0.063 | 0.5 | 0.125 |
| 928 | - | - | - | - | 8 | +8 | 4 |
| 930 | - | - | - | - | 8 | +8 | 4 |
| 99EX | - | - | - | - | 0.25 | 0.5 | 0.125 |
| 869 | Phe | Asn | - | Ile | 8 | +8 | 4 |
| 945 | - | - | Asn | - | +8 | +8 | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corrales, J.C.; Sánchez, A.; Hernández, X.; Amores-Iniesta, J.; Esnal, A.; de la Fe, C. A Set of Multiresistant Isolates of Mycoplasma bovis Subtype ST-1 with a Variable Susceptibility to Quinolones Are Also Circulating in Spain. Pathogens 2024, 13, 329. https://doi.org/10.3390/pathogens13040329
Corrales JC, Sánchez A, Hernández X, Amores-Iniesta J, Esnal A, de la Fe C. A Set of Multiresistant Isolates of Mycoplasma bovis Subtype ST-1 with a Variable Susceptibility to Quinolones Are Also Circulating in Spain. Pathogens. 2024; 13(4):329. https://doi.org/10.3390/pathogens13040329
Chicago/Turabian StyleCorrales, Juan Carlos, Antonio Sánchez, Xóchitl Hernández, Joaquín Amores-Iniesta, Antón Esnal, and Christian de la Fe. 2024. "A Set of Multiresistant Isolates of Mycoplasma bovis Subtype ST-1 with a Variable Susceptibility to Quinolones Are Also Circulating in Spain" Pathogens 13, no. 4: 329. https://doi.org/10.3390/pathogens13040329
APA StyleCorrales, J. C., Sánchez, A., Hernández, X., Amores-Iniesta, J., Esnal, A., & de la Fe, C. (2024). A Set of Multiresistant Isolates of Mycoplasma bovis Subtype ST-1 with a Variable Susceptibility to Quinolones Are Also Circulating in Spain. Pathogens, 13(4), 329. https://doi.org/10.3390/pathogens13040329







