Parvoviruses of Aquatic Animals
Abstract
1. Introduction
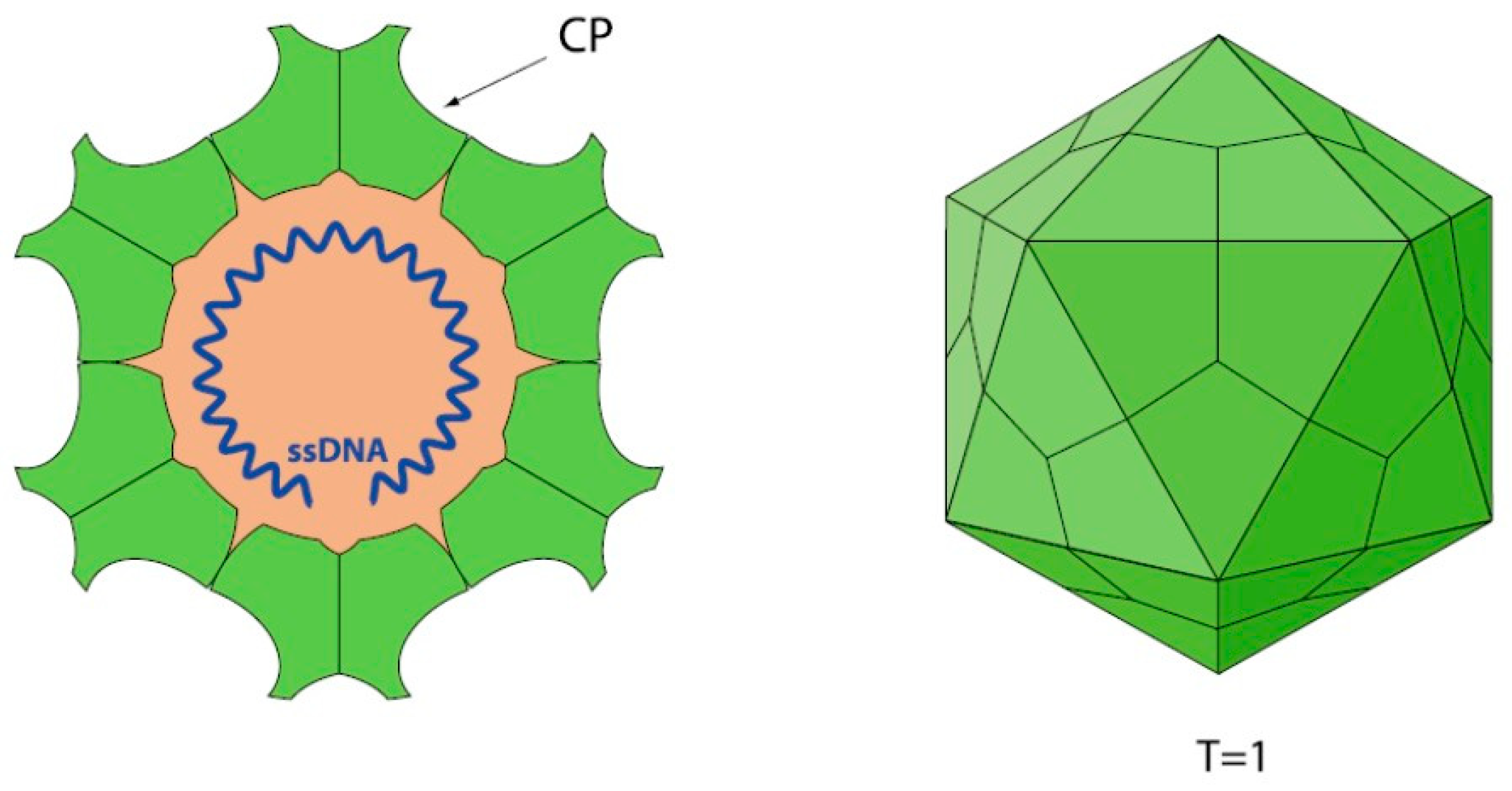
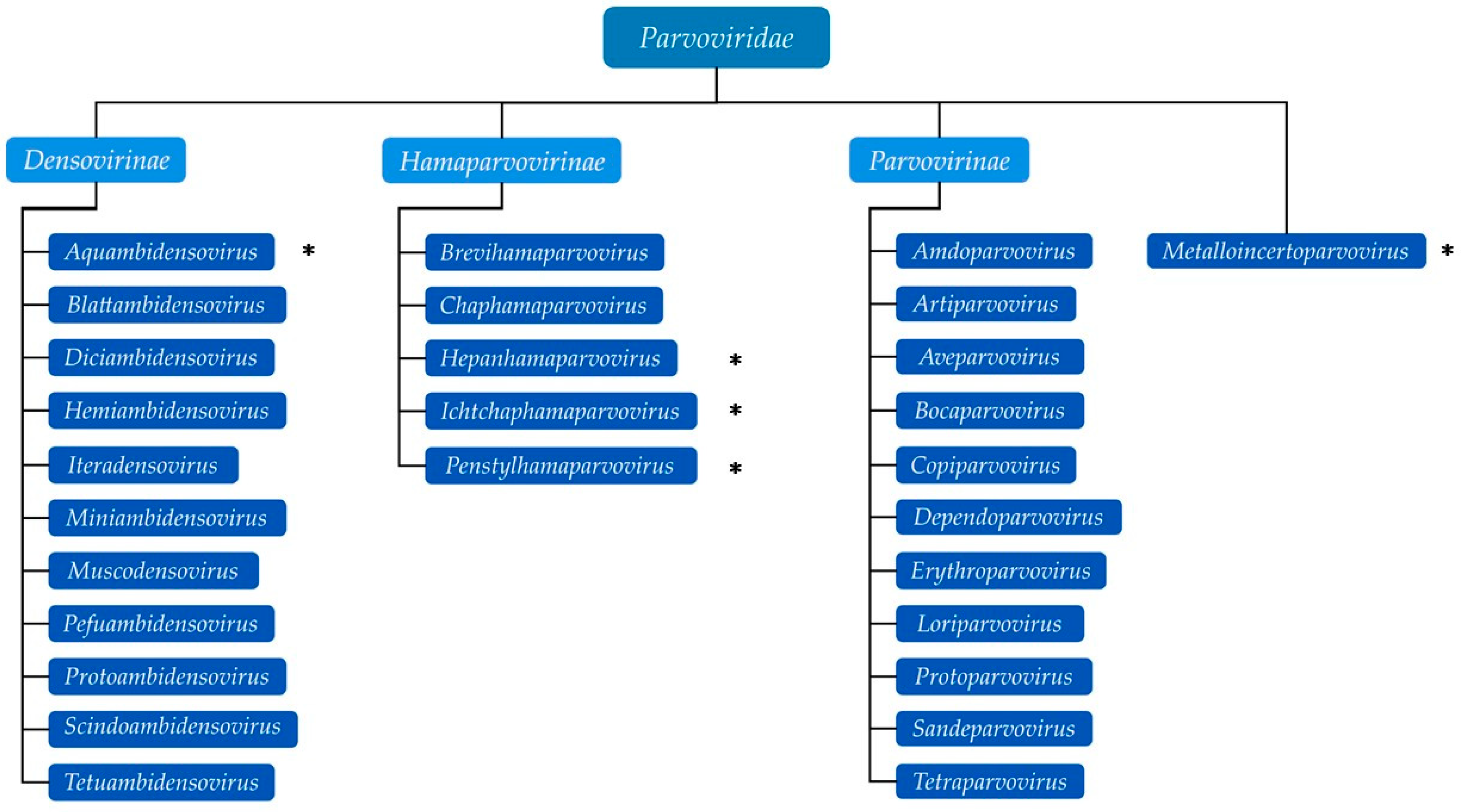
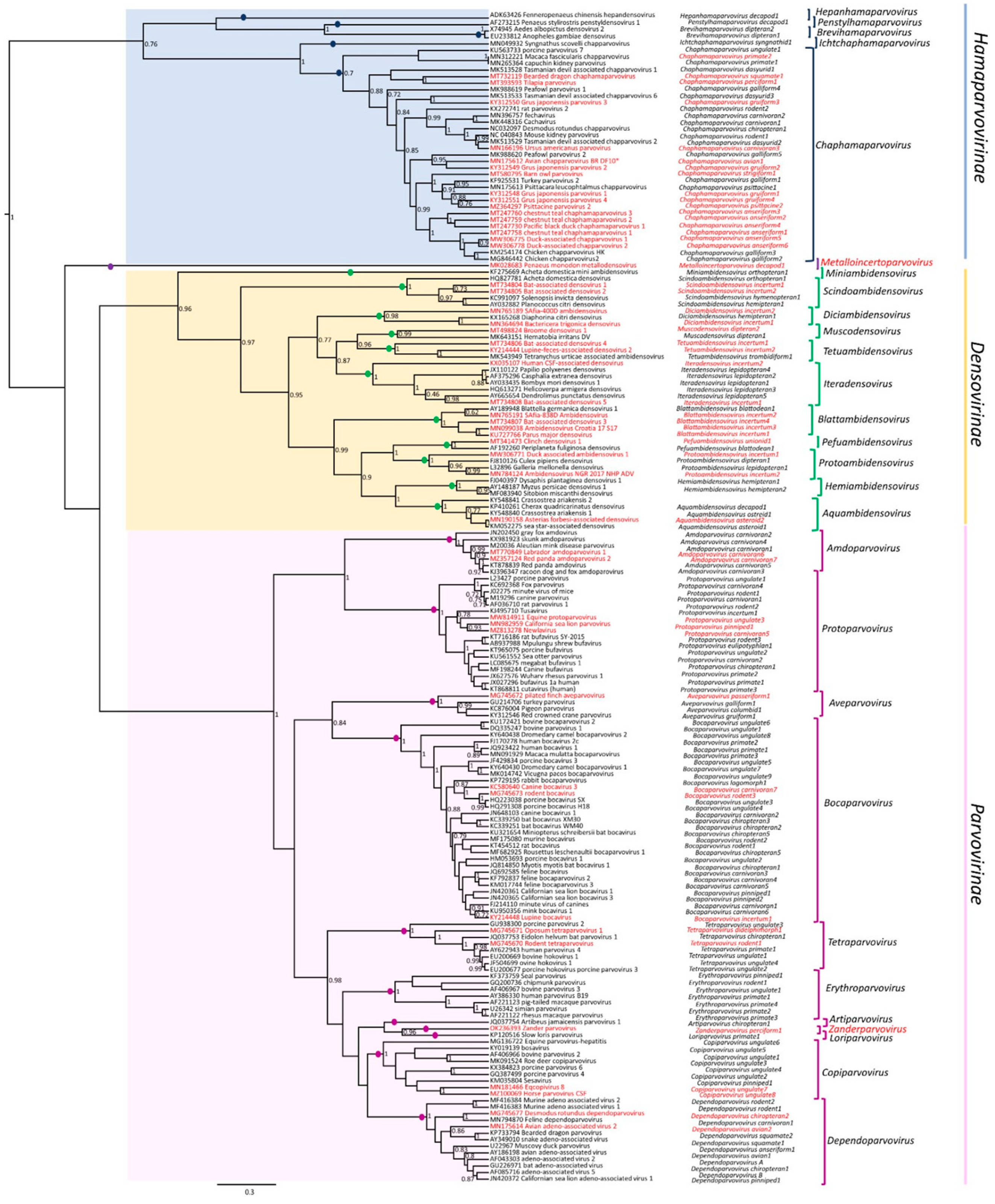
1.1. Parvovirus Classification and Virion Properties
1.1.1. Parvovirus Genome Organization
1.1.2. Virus Replication
1.2. General Pathogenesis of Parvoviruses
| Subfamily and Genus | Species 1 | Virus Common Name (Abbreviation) | Sampled Host | Geographical Area 2 | Clinical Disease and Common Clinical Signs | Reference |
|---|---|---|---|---|---|---|
| Densovirinae, Aquambidensovirus | Decapod aquambidensovirus 1 | Cherax quadricarinatus densovirus [previously known as Cherax quadricarinatus parvo-like virus (CqPlV)]; Cherax destructor systemic parvo-like virus (CdSPV) | redclaw crayfish Cherax quadricarinatus and Cherax destructor | Queensland, Australia | Decrease in stress resistance manifested as chronic mortality or mass mortality of up to 96% cumulative mortality. In experimental infection, inoculated crayfish showed gross signs of malaise, anorexia, and disorientation before dying. Tissues of endodermal, ectodermal, and mesodermal origin have enlarged nuclei containing intranuclear inclusions. Transmission electron microscopy showed the inclusions to consist of viral particles 19.5 nm in diameter. | [9,10] |
| Asteroid aquambidensovirus 1 | Sea star-associated densovirus (SSaDV) | sea stars and sea urchins | Pacific coast of USA and Atlantic coast of North America | Originally associated with sea-star wasting disease (SSWD) causing mass mortality in asteroids. Affected animals show behavioral changes, various lesions such as white lesions along the arms progressing to large areas of exposed underlying white ossicles, loss of turgor, limb autonomy (“dropping” of the limbs), and death characterized by rapid degradation (“melting”). Histologically, pink round to irregular inclusions were noted in the vacuolated nuclei of affected epithelial cells in some samples. | [11,54] | |
| Clinch densovirus 1 | Freshwater mussels | Clinch River, Virginia, and Tennessee, USA | Episodic mass mortality with moribund mussels appearing on the surface of the substrate, gaping, slow to respond to tactile stimuli, and able to close their valves only weakly. Histologic lesions consisted of pervasive necrosis of the internal organs. | [12] | ||
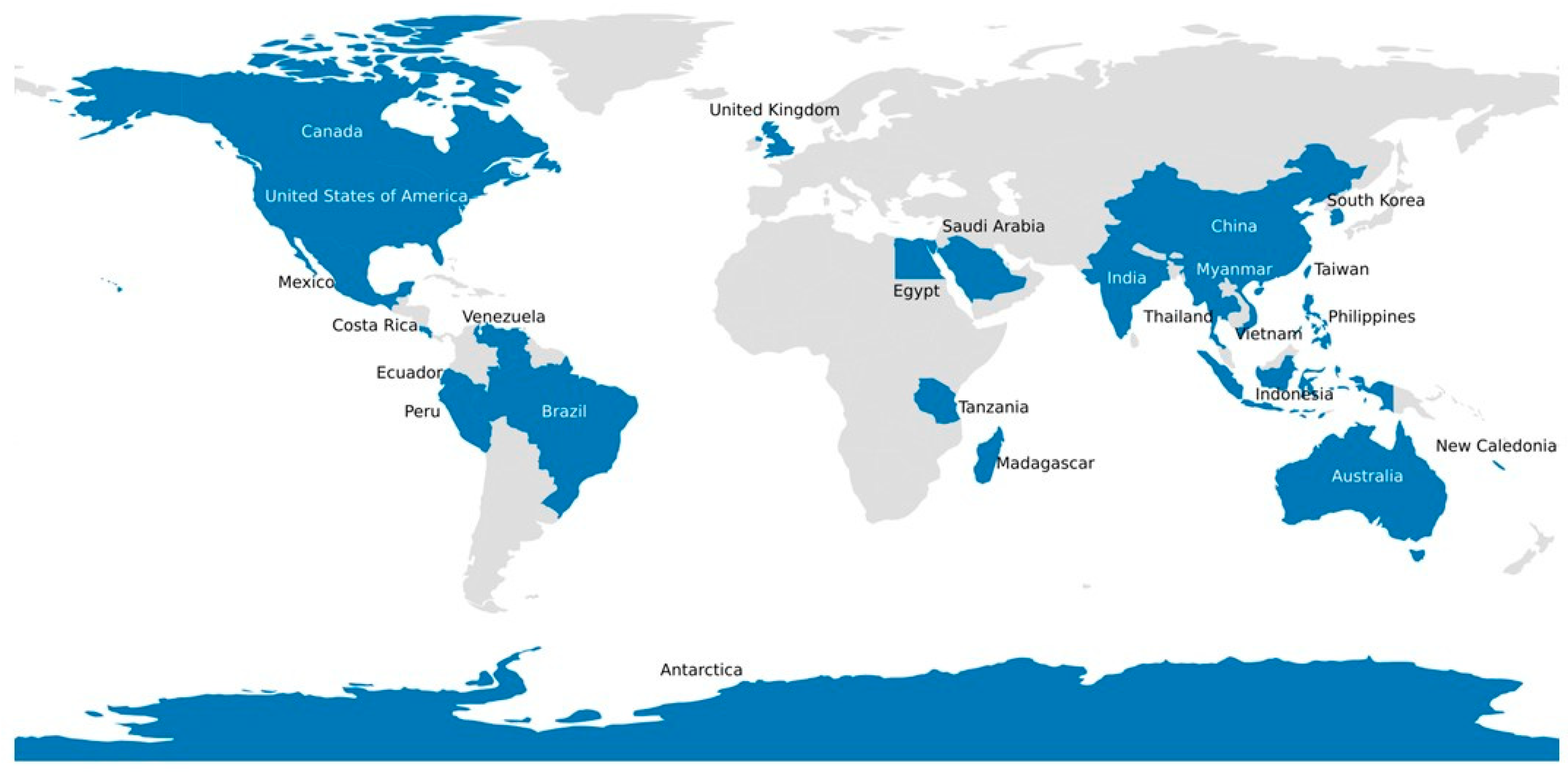
| Hamaparvovirinae, Hepanhamaparvovirus | Decapod hepanhamaparvovirus 1 | Fenneropenaeus chinensis hepatopancreatic densovirus [previously known as Hepatopancreatic parvovirus (HPV)] | Prawns and Shrimp | Australia, Brazil, China, El Salvador, India, Indonesia, Israel, Kenya, Madagascar, Malaysia, Mexico, New Caledonia, Philippines, Singapore, South Korea, Taiwan, Tanzania, Thailand, and the United States of America | Reduced growth in juvenile shrimp, anorexia, reduced preening with a concurrent increase in surface, and gill fouling by epicommensal organisms. Increased mortalities during the larval stages with signs of necrosis and atrophy of the hepatopancreas. Hepatopancreatic cells and epigastric caecal epithelial cells have hypertrophied nuclei with basophilic inclusion bodies. Transmission electron microscopy showed the intranuclear inclusion bodies to consist of viral particles 22–24 nm in diameter. | [45,50,55,56,57] |
| Hamaparvovirinae, Ichtahamaparvovirus | Syngnathid ichthamaparvovirus 1 | Syngnathus scovelli chapparvovirus | gulf pipefish | Gulf of Mexico and parts of South America | Not known. | [58] |
| Tilapia parvovirus (TiPV) | Nile tilapia | China, India, and Thailand | Mass morbidity and 60–70% mortality in adult Nile tilapia (500–600 g); mass mortality of 50–75% in juvenile red hybrid tilapia (10–30 g to 300–800 g). Clinical signs in affected fish include lethargy, reduced feeding, scale loss, redness on body with hemorrhages on the operculum, base of fins and ventral part, opaqueness of eyes, swimming near the pond edge or abnormal swimming patterns before death. TiPV targets the pancreas and the presence of Cowdry type A inclusion bodies in acinar cells of the pancreas is pathognomonic of TiPV infection | [25,26,27,28] | ||
| unclassified Ichthamaparvovirus | ||||||
| Hamaparvovirinae, Penstylhamaparvovirus | Decapod penstylhamaparvovirus 1 | Penaeus stylirostris penstyldensovirus (PstDV 1&2); Penaeus monodon penstyldensovirus (PmoPDV 1&2) [previously known as infectious hypodermal and hematopoietic necrosis virus (IHHNV)] | Prawns and Shrimp | Australia, Brazil, Caribbean, Central America, Ecuador, Indonesia, Israel, Malaysia, New Caledonia, Peru, Philippines, Singapore, Tahiti, Taiwan, Thailand, and United States of America | IHHN; 80–100% mortality in postlarvae and juveniles of Penaeus (Litopenaeus) stylirostris and postlarvae of Macrobrachium rosenbergii; runt deformity syndrome (RDS) in juvenile of P. vannamei and P. monodon; >80% mortality in postlarvae and chronic mortality in juveniles of Penaeus esculentus X Penaeus monodon hybrids. The animals become weak and stop moving prior to death, and the gills have many small melanized foci. There are white or buff colored spots in the cuticular epidermis, especially at the junction of the tergal plates of the abdomen, giving the shrimp a mottled appearance. Histologically, in acute infections, the gills, lymphoid organ, other organs, and connective tissue show intranuclear Cowdrey’s type A inclusions in hypertrophied nuclei. Transmission electron microscopy showed the inclusions to consist of viral particles 17–26 nm in diameter. | [46,59,60,61,62] |
| Unassigned genus | Metalloincertoparvovirus decapod 1 | Penaeus monodon metallodensovirus (PmMDV) | Vietnamese P. monodon | Vietnam | Mass mortality in P. monodon. Clinical signs include a red telson, uropodia, and pleopods, and discoloration of the cephalothorax. | [30] |
| Unassigned | Novel salmon parvovirus | Sockeye salmon | BC, Canada | Not known | [6] | |
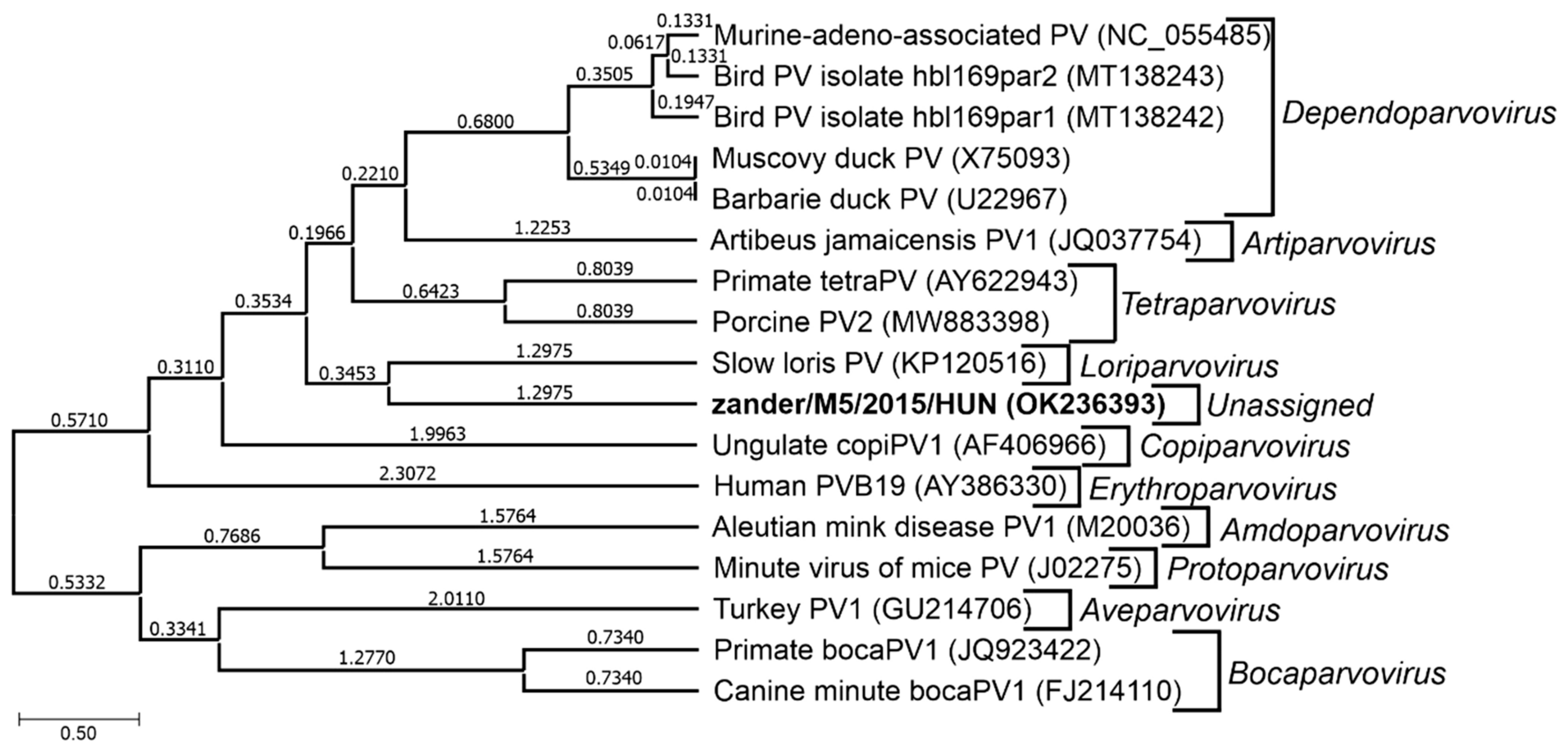
| Unassigned in subfamily Parvovirinae | Novel zander parvovirus (zander/M5/2015/HUN, OK236393) | Zander or pikeperch (Sander lucioperca) | Hungary | Not known | [5] | |
| Unassigned | Crangon crangon parvo-like virus 1 (CcPaLV 1) | Molluscs | Europe | Not known | [7] |
1.3. Challenges Related to Controlling Parvoviruses in Aquatic Animals
2. Genus Aquambidensovirus
2.1. Decapod Aquambidensovirus 1 (Cherax Quadricarinatus Densovirus (CqDV))
2.2. Asteroid Aquambidensovirus 1 (Sea Star-Associated Densovirus (SSaDV))
2.3. Clinch Densovirus 1
3. Genus Hepanhamaparvovirus
Decapod Hepanhamaparvovirus 1 (DHPV-1) (Hepatopancreatic Parvovirus (HPV))
4. Genus Penstylhamaparvovirus
5. Genus Ichthamaparvovirus
5.1. Syngnathid Ichthamaparvovirus 1
5.2. Tilapia Parvovirus (TiPV)
6. Unassigned
6.1. Metalloincertoparvovirus
6.2. Crangon Crangon Parvo-Like Virus (CcPaLV)
6.3. Novel Salmon Parvovirus from Sockeye Salmon
6.4. Novel Zander Parvovirus
6.5. Spawner-Isolated Mortality Virus (SMV)
7. Concluding Remarks
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Pénzes, J.J.; Söderlund-Venermo, M.; Canuti, M.; Eis-Hübinger, A.M.; Hughes, J.; Cotmore, S.F.; Harrach, B. Reorganizing the family Parvoviridae: A revised taxonomy independent of the canonical approach based on host association. Arch. Virol. 2020, 165, 2133–2146. [Google Scholar] [CrossRef]
- Cotmore, S.F.; Agbandje-McKenna, M.; Canuti, M.; Chiorini, J.A.; Eis-Hubinger, A.-M.; Hughes, J.; Mietzsch, M.; Modha, S.; Ogliastro, M.; Pénzes, J.J.; et al. ICTV Virus Taxonomy Profile: Parvoviridae. J. Gen. Virol. 2019, 100, 367–368. [Google Scholar] [CrossRef] [PubMed]
- MacLachlan, N.J.; Dubovi, E.J. Chapter 12, Parvoviridae. In Fenner’s Veterinary Virology, 5th ed.; MacLachlan, N.J., Dubovi, E.J., Eds.; Academic Press: Cambridge, MA, USA, 2017; pp. 245–257. [Google Scholar] [CrossRef]
- Qiu, J.; Soderlund-Venermo, M.; Young, N.S. Human Parvoviruses. Clin. Microbiol. Rev. 2017, 30, 43–113. [Google Scholar] [CrossRef] [PubMed]
- Reuter, G.; Boros, Á.; Mátics, R.; Altan, E.; Delwart, E.; Pankovics, P. A novel parvovirus (family Parvoviridae) in a freshwater fish, zander (Sander lucioperca). Arch. Virol. 2022, 167, 1163–1167. [Google Scholar] [CrossRef]
- Miller, K.M.; Trudel, M.; Patterson, D.A.; Schulze, A.; Kaukinen, K.; Li, S.; Ginther, N.; Ming, T.; Tabata, A. Are Smolts Healthier in Years of Good Ocean Productivity? Technical Report No. 9; North Pacific Anadromous Fish Commission: Vancouver, BC, Canada, 2013; pp. 165–168. Available online: https://www.npafc.org/wp-content/uploads/TechReport9.pdf (accessed on 21 December 2023).
- Van Eynde, B.; Christiaens, O.; Delbare, D.; Shi, C.; Vanhulle, E.; Yinda, C.K.; Matthijnssens, J.; Smagghe, G. Exploration of the virome of the European brown shrimp (Crangon crangon). J. Gen. Virol. 2020, 101, 651–666. [Google Scholar] [CrossRef] [PubMed]
- Fédière, G. Epidemiology and pathology of Densovirinae. In Parvoviruses. From Molecular Biology to Pathology and Therapeutic Uses; Faisst, S., Rommelaere, J., Eds.; Karger: Basel, Switzerland, 2000; pp. 1–11. Available online: https://horizon.documentation.ird.fr/exl-doc/pleins_textes/pleins_textes_7/b_fdi_59-60/010025973.pdf (accessed on 21 December 2023).
- Edgerton, B.F.; Webb, R.; Anderson, I.G.; Kulpa, E.C. Description of a presumptive hepatopancreatic reovirus, and a putative gill parvovirus, in the freshwater crayfish Cherax quadricarinatus. Dis. Aquat. Org. 2000, 41, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Bowater, R.O.; Wingfield, M.; Fisk, A.; Condon, K.M.L.; Reid, A.; Prior, H.; Kulpa, E.C. A parvo-like virus in cultured redclaw crayfish Cherax quadricarinatus from Queensland, Australia. Dis. Aquat. Org. 2002, 50, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Hewson, I.; Button, J.B.; Gudenkauf, B.M.; Miner, B.; Newton, A.L.; Gaydos, J.K.; Wynne, J.; Groves, J.L.; Hendler, G.; Murray, M.; et al. Densovirus associated with sea-star wasting disease and mass mortality. Proc. Natl. Acad. Sci. USA 2014, 111, 17276–17283. [Google Scholar] [CrossRef] [PubMed]
- Richard, J.C.; Leis, E.; Dunn, C.D.; Agbalog, R.; Waller, D.; Knowles, S.; Putnam, J.; Goldberg, T.L. Mass mortality in freshwater mussels (Actinonaias pectorosa) in the Clinch River, USA, linked to a novel densovirus. Sci. Rep. 2020, 10, 14498. [Google Scholar] [CrossRef]
- Kalagayan, H.; Godin, D.; Kanna, R.; Hagino, G.; Sweeney, J.; Wyban, J.; Brock, J. IHHN Virus as an Etiological Factor in Runt-Deformity Syndrome (RDS) of Juvenile Penaeus vannamei Cultured in Hawaii. J. World Aquac. Soc. 1991, 22, 235–243. [Google Scholar] [CrossRef]
- Lee, C.; Jeon, H.J.; Kim, B.; Suh, S.; Piamsomboon, P.; Kim, J.H.; Han, J.E. Cultured Penaeus vannamei in Korea co-infected with white spot syndrome virus and decapod hepanhamaparvovirus. J. World Aquac. Soc. 2024, 55, 373–385. [Google Scholar] [CrossRef]
- Lightner, D.V.; Redman, R.M.; Bell, T.A. Infectious hypodermal and hematopoietic necrosis, a newly recognized virus disease of penaeid shrimp. J. Invertebr. Pathol. 1983, 42, 62–70. [Google Scholar] [CrossRef]
- Lightner, D.V.; Redman, R.M.; Bell, T.A.; Brock, J.A. DETECTION OF IHHN VIRUS IN Penaeus stylirostris AND P. vannamei IMPORTED INTO HAWAII. J. World Maric. Soc. 1983, 14, 212–225. [Google Scholar] [CrossRef]
- Tang, K.F.; Durand, S.V.; White, B.L.; Redman, R.M.; Pantoja, C.R.; Lightner, D.V. Postlarvae and juveniles of a selected line of Penaeus stylirostris are resistant to infectious hypodermal and hematopoietic necrosis virus infection. Aquaculture 2000, 190, 203–210. [Google Scholar] [CrossRef]
- Alday-Sanz, V.; Brock, J.; Flegel, T.W.; McIntosh, R.; Bondad-Reantaso, M.G.; Salazar, M.; Subasinghe, R. Facts, truths and myths about SPF shrimp in Aquaculture. Rev. Aquac. 2018, 12, 76–84. [Google Scholar] [CrossRef]
- Wyban, J. Selective Breeding of Penaeus vannamei: Impact on World Aquaculture and Lessons for Future. J. Coast. Res. 2019, 86, 1–5. [Google Scholar] [CrossRef]
- Briggs, M. Introductions and Movement of Two Penaeid Shrimp Species in Asia and the Pacific; FAO Fisheries Technical Paper; FAO: Rome, Italy, 2005; Volume 476, p. 99. [Google Scholar] [CrossRef]
- Subramaniam, K. Commercial Production of SPF Penaeus Monodon Brood Stock in Malaysia. 2009. Available online: https://s9f9e7d2357d9daa5.jimcontent.com/download/version/1446562656/module/9553567799/name/SPF%20Black%20Tiger%20Broodstock%20production.pdf (accessed on 6 July 2024).
- Dhar, A.K.; Cruz-Flores, R.; Caro, L.F.A.; Siewiora, H.M.; Jory, D. Diversity of single-stranded DNA containing viruses in shrimp. Virus Dis. 2019, 30, 43–57. [Google Scholar] [CrossRef]
- Cruz-Flores, R.; Siewiora, H.M.; Kanrar, S.; Le Groumellec, M.; Dhar, A.K.; Cruz-Flores, R.; Siewiora, H.M.; Kanrar, S.; Le Groumellec, M.; Dhar, A.K.; et al. Identification and characterization of a novel strain of Decapod hepanhamaparvovirus in black tiger shrimp (Penaeus monodon) from Madagascar that does not cause histological lesions. Aquaculture 2024, 588, 740904. [Google Scholar] [CrossRef]
- Sellars, M.J.; Cowley, J.A.; Musson, D.; Rao, M.; Menzies, M.L.; Coman, G.J.; Murphy, B.S. Reduced growth performance of Black Tiger shrimp (Penaeus monodon) infected with infectious hypodermal and hematopoietic necrosis virus. Aquaculture 2019, 499, 160–166. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, Y.; Ma, J.; Jiang, N.; Fan, Y.; Zhou, Y.; Cain, K.; Yi, M.; Jia, K.; Wen, H.; et al. Determination of a novel parvovirus pathogen associated with massive mortality in adult tilapia. PLoS Pathog. 2020, 16, e1008765. [Google Scholar] [CrossRef]
- Yamkasem, J.; Tattiyapong, P.; Gorgoglione, B.; Surachetpong, W. Uncovering the first occurrence of Tilapia parvovirus in Thailand in tilapia during co-infection with Tilapia tilapinevirus. Transbound. Emerg. Dis. 2021, 68, 3136–3144. [Google Scholar] [CrossRef] [PubMed]
- Piewbang, C.; Tattiyapong, P.; Khemthong, M.; Lachroje, S.; Boonrungsiman, S.; Kasantikul, T.; Surachetpong, W.; Techangamsuwan, S. Dual infections of tilapia parvovirus (TiPV) and tilapia lake virus (TiLV) in multiple tilapia farms: Their impacts, genetic diversity, viral tropism, and pathological effects. Aquaculture 2022, 550, 737887. [Google Scholar] [CrossRef]
- Paul, A.; Mohanty, S.; Parida, S.; Biswal, S.; Sandha, B.K.; Sahoo, P.K. Tilapia parvovirus disease: An emerging threat for the tilapia aquaculture industry. Aquac. Asia Mag. 2023, 27, 17–18. Available online: https://enaca.org/enclosure/?id=1304 (accessed on 26 May 2024).
- Du, J.; Wang, W.; Chan, J.F.-W.; Wang, G.; Huang, Y.; Yi, Y.; Zhu, Z.; Peng, R.; Hu, X.; Wu, Y.; et al. Identification of a Novel Ichthyic Parvovirus in Marine Species in Hainan Island, China. Front. Microbiol. 2019, 10, 2815. [Google Scholar] [CrossRef] [PubMed]
- Pénzes, J.J.; Pham, H.T.; Chipman, P.; Bhattacharya, N.; McKenna, R.; Agbandje-McKenna, M.; Tijssen, P. Molecular biology and structure of a novel penaeid shrimp densovirus elucidate convergent parvoviral host capsid evolution. Proc. Natl. Acad. Sci. USA 2020, 117, 20211–20222. [Google Scholar] [CrossRef] [PubMed]
- Pénzes, J.J.; Canuti, M.; Söderlund-Venermo, M.; François, S.; Eis-Hübinger, A.M. Parvoviridae: Introduction of the Binomial Nomenclature, Establishment of Two New Genera and the Classification Eligibility of Parvoviruses Derived from Ambiguous Host Origin. 2021. 2022.005D. Available online: https://ictv.global/ictv/proposals/2022.005D.Parvoviridae_2ngen_49nsp_125rensp.zip (accessed on 20 July 2024).
- Gordon, J.C.; Angrick, E.J. Canine parvovirus: Environmental effects on infectivity. Am. J. Vet. Res. 1986, 47, 1464–1467. [Google Scholar] [PubMed]
- Viral Zone Parvoviridae. Available online: https://viralzone.expasy.org/103?outline=all_by_species (accessed on 27 May 2024).
- Pénzes, J.J.; Pham, H.T.; Chipman, P.; Smith, E.W.; McKenna, R.; Tijssen, P. Bipartite genome and structural organization of the parvovirus Acheta domesticus segmented densovirus. Nat. Commun. 2023, 14, 3515. [Google Scholar] [CrossRef] [PubMed]
- Pénzes, J.J.; Canuti, M.; Francois, S.; Söderlund-Venermo, M.; Tijssen, P.; Tattersall, P. Creating 13 New Species in Family Parvoviridae. 2023. 2023.007D. Available online: https://ictv.global/filebrowser/download/16005 (accessed on 19 July 2024).
- Cotmore, S.F.; Tattersall, P. Parvoviruses: Small Does Not Mean Simple. Annu. Rev. Virol. 2014, 1, 517–537. [Google Scholar] [CrossRef] [PubMed]
- Cotmore, S.F.; Tattersall, P. Genome Packaging Sense Is Controlled by the Efficiency of the Nick Site in the Right-End Replication Origin of Parvoviruses Minute Virus of Mice and LuIII. J. Virol. 2005, 79, 2287–2300. [Google Scholar] [CrossRef]
- Jager, M.C.; Tomlinson, J.E.; Lopez-Astacio, R.A.; Parrish, C.R.; Van de Walle, G.R. Small but mighty: Old and new parvoviruses of veterinary significance. Virol. J. 2021, 18, 210. [Google Scholar] [CrossRef]
- Chen, A.Y.; Qiu, J. Parvovirus Infection-Induced Cell Death and Cell Cycle Arrest. Future Virol. 2010, 5, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Söderlund-Venermo, M. Emerging Human Parvoviruses: The Rocky Road to Fame. Annu. Rev. Virol. 2019, 6, 71–91. [Google Scholar] [CrossRef] [PubMed]
- Kailasan, S.; Agbandje-McKenna, M.; Parrish, C.R. Parvovirus Family Conundrum: What Makes a Killer? Annu. Rev. Virol. 2015, 2, 425–450. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Yan, Z.; Cheng, F.; Engelhardt, J.F.; Qiu, J. Replication of an Autonomous Human Parvovirus in Non-dividing Human Airway Epithelium Is Facilitated through the DNA Damage and Repair Pathways. PLoS Pathog. 2016, 12, e1005399. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Xu, P.; Zou, W.; Shen, W.; Peng, J.; Liu, K.; Engelhardt, J.F.; Yan, Z.; Qiu, J. DNA Damage Signaling Is Required for Replication of Human Bocavirus 1 DNA in Dividing HEK293 Cells. J. Virol. 2017, 91, e01831-16. [Google Scholar] [CrossRef] [PubMed]
- Eisenlord, M.E.; Groner, M.L.; Yoshioka, R.M.; Elliott, J.; Maynard, J.; Fradkin, S.; Turner, M.; Pyne, K.; Rivlin, N.; van Hooidonk, R.; et al. Ochre star mortality during the 2014 wasting disease epizootic: Role of population size structure and temperature. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20150212. [Google Scholar] [CrossRef] [PubMed]
- Bower, S.M. Synopsis of Infectious Diseases and Parasites of Commercially Exploited Shellfish: Hepatopancreatic Parvovirus (HPV) Disease of Shrimp and Prawns. Available online: https://www.dfo-mpo.gc.ca/science/aah-saa/diseases-maladies/hpvsp-eng.html (accessed on 2 July 2024).
- Hsieh, C.Y.; Chuang, P.C.; Chen, L.C.; Tu, C.; Chien, M.S.; Huang, K.C.; Kao, H.F.; Tung, M.C.; Tsai, S.S. Infectious hypodermal and haematopoietic necrosis virus (IHHNV) infections in giant freshwater prawn, Macrobrachium rosenbergii. Aquaculture 2006, 258, 73–79. [Google Scholar] [CrossRef]
- Phromjai, J.; Boonsaeng, V.; Withyachumnarnkul, B.; Flegel, T.W. Detection of hepatopancreatic parvovirus in Thai shrimp Penaeus monodon by in situ hybridization, dot blot hybridization and PCR amplification. Dis. Aquat. Org. 2002, 51, 227–232. [Google Scholar] [CrossRef]
- Hewson, I.; Bistolas, K.S.I.; Cardé, E.M.Q.; Button, J.B.; Foster, P.J.; Flanzenbaum, J.M.; Kocian, J.; Lewis, C.K. Investigating the Complex Association Between Viral Ecology, Environment, and Northeast Pacific Sea Star Wasting. Front. Mar. Sci. 2018, 5, 77. [Google Scholar] [CrossRef]
- Aquino, C.A.; Besemer, R.M.; DeRito, C.M.; Kocian, J.; Porter, I.R.; Raimondi, P.T.; Rede, J.E.; Schiebelhut, L.M.; Sparks, J.P.; Wares, J.P.; et al. Evidence That Microorganisms at the Animal-Water Interface Drive Sea Star Wasting Disease. Front. Microbiol. 2021, 11, 610009. [Google Scholar] [CrossRef]
- Manivannan, S.; Otta, S.K.; Karunasagar, I. Multiple viral infection in Penaeus monodon shrimp postlarvae in an Indian hatchery. Dis. Aquat. Org. 2002, 48, 233–236. [Google Scholar] [CrossRef] [PubMed]
- Safeena, M.P.; Rai, P.; Karunasagar, I. Molecular Biology and Epidemiology of Hepatopancreatic parvovirus of Penaeid Shrimp. Indian J. Virol. 2012, 23, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.-Y.; Yang, N.; Hou, Z.-H.; Wang, J.-J.; Li, T.; Chang, L.-R.; Fang, Y.; Yan, D.-C. Research progress on hosts and carriers, prevalence, virulence of infectious hypodermal and hematopoietic necrosis virus (IHHNV). J. Invertebr. Pathol. 2021, 183, 107556. [Google Scholar] [CrossRef] [PubMed]
- Rusaini; La Fauce, K.A.; Elliman, J.; Bowater, R.O.; Owens, L. Endogenous Brevidensovirus-like elements in Cherax quadricarinatus: Friend or foe? Aquaculture 2013, 396–399, 136–145. [Google Scholar] [CrossRef]
- Bucci, C.; Francoeur, M.; McGreal, J.; Smolowitz, R.; Zazueta-Novoa, V.; Wessel, G.M.; Gomez-Chiarri, M. Sea Star Wasting Disease in Asterias forbesi along the Atlantic Coast of North America. PLoS ONE 2017, 12, e0188523. [Google Scholar] [CrossRef] [PubMed]
- Flegel, T.W. Detection of major penaeid shrimp viruses in Asia, a historical perspective with emphasis on Thailand. Aquaculture 2006, 258, 1–33. [Google Scholar] [CrossRef]
- Spann, K.M.; Adlard, R.D.; Hudson, D.A.D.; Pyecroft, S.B.; Jones, T.C.; Voigt, M.O.C. Hepatopancreatic parvo-like virus (HPV) of Penaeus japonicus cultured in Australia. Dis. Aquat. Org. 1997, 31, 239–241. [Google Scholar] [CrossRef]
- Madan, N.; Raj, N.S.; Farook, M.A.; Vimal, S.; Venkatesan, C.; Majeed, S.A.; Nambi, K.S.N.; Hameed, A.S. Partial cloning and production of polyclonal antiserum against recombinant capsid protein of hepatopancreatic parvovirus (HPV) and its application for diagnostics in penaeid shrimp. Process Biochem. 2013, 48, 1893–1898. [Google Scholar] [CrossRef]
- Pénzes, J.J.; de Souza, W.M.; Agbandje-McKenna, M.; Gifford, R.J. An Ancient Lineage of Highly Divergent Parvoviruses Infects both Vertebrate and Invertebrate Hosts. Viruses 2019, 11, 525. [Google Scholar] [CrossRef] [PubMed]
- Bower, S.M. Synopsis of Infectious Diseases and Parasites of Commercially Exploited Shellfish: Infectious Hypodermal and Haematopoietic Necrosis Virus (IHHNV) of Penaeid Shrimp. Available online: https://www.dfo-mpo.gc.ca/science/aah-saa/diseases-maladies/ihnvsp-eng.html (accessed on 2 July 2024).
- Vega-Heredia, S.; Mendoza-Cano, F.; Sánchez-Paz, A. The Infectious Hypodermal and Haematopoietic Necrosis Virus: A Brief Review of What We Do and Do Not Know. Transbound. Emerg. Dis. 2012, 59, 95–105. [Google Scholar] [CrossRef]
- WOAH (World Organization for Animal Health). Infection with infectious hypodermal and haematopoietic necrosis virus. In Manual of Diagnostic Tests for Aquatic Animals; WOAH (World Organization for Animal Health): Paris, France, 2021; Chapter 2.2; pp. 138–153. Available online: https://www.woah.org/fileadmin/Home/eng/Health_standards/aahm/current/2.2.04_IHHN.pdf (accessed on 21 December 2023).
- Owens, L.; Anderson, I.; Kenway, M.; Trort, L.; Benzie, J. Infectious hypodermal and haematopoietic necrosis virus (IHHNV) in a hybrid penaeid prawn from tropical Australia. Dis. Aquat. Org. 1992, 14, 219–228. [Google Scholar] [CrossRef]
- NCBI Taxonomy Browser. Available online: https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi (accessed on 9 July 2024).
- NCBI Virus Sequences for Discovery Portal. Available online: https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/ (accessed on 9 July 2024).
- Kibenge, F.S.B.; Strange, R. Introduction to the anatomy and physiology of the major aquatic animal species in aquaculture. In Aquaculture Pharmacology; Kibenge, F.S.B., Baldisserotto, B., Chong, R.S.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; Chapter 1; pp. 1–111. [Google Scholar]
- Evensen, Ø. Immunology and vaccinology of farmed aquatic animals. In Aquaculture Pharmacology; Kibenge, F.S.B., Baldisserotto, B., Chong, R.S.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; Chapter 1; pp. 117–135. [Google Scholar]
- Wang, B.; Thompson, K.D.; Wangkahart, E.; Yamkasem, J.; Bondad-Reantaso, M.G.; Tattiyapong, P.; Jian, J.; Surachetpong, W. Strategies to enhance tilapia immunity to improve their health in aquaculture. Rev. Aquac. 2023, 15, 41–56. [Google Scholar] [CrossRef]
- Seibert, C.H.; Pinto, A.R. Challenges in shrimp aquaculture due to viral diseases: Distribution and biology of the five major penaeid viruses and interventions to avoid viral incidence and dispersion. Braz. J. Microbiol. 2012, 43, 857–864. [Google Scholar] [CrossRef] [PubMed]
- MacKinnon, B.; Debnath, P.P.; Bondad-Reantaso, M.G.; Fridman, S.; Bin, H.; Nekouei, O. Improving tilapia biosecurity through a value chain approach. Rev. Aquac. 2023, 15, 57–91. [Google Scholar] [CrossRef]
- Global Tilapia Market Published by Research and Markets. Global Tilapia Market Forecast by Production, Import, Export Countries, Company Analysis. Available online: https://www.researchandmarkets.com/reports/5317038/global-tilapia-market-forecast-by-production#product--toc (accessed on 18 July 2024).
- Thompson, K.D.; Rodkhum, C.; Bunnoy, A.; Thangsunan, P.; Kitiyodom, S.; Sukkarun, P.; Yostawornkul, J.; Yata, T.; Pirarat, N. Addressing Nanovaccine Strategies for Tilapia. Vaccines 2023, 11, 1356. [Google Scholar] [CrossRef]
- Attasart, P.; Kaewkhaw, R.; Chimwai, C.; Kongphom, U.; Panyim, S. Clearance of Penaeus monodon densovirus in naturally pre-infected shrimp by combined ns1 and vp dsRNAs. Virus Res. 2011, 159, 79–82. [Google Scholar] [CrossRef] [PubMed]
- Itsathitphaisarn, O.; Thitamadee, S.; Weerachatyanukul, W.; Sritunyalucksana, K. Potential of RNAi applications to control viral diseases of farmed shrimp. J. Invertebr. Pathol. 2017, 147, 76–85. [Google Scholar] [CrossRef] [PubMed]
- WOAH (World Organization for Animal Health). Diseases listed by WOAH. Chapter 1. In Aquatic Animal Health Code; WOAH (World Organization for Animal Health): Paris, France, 2024; pp. 138–153. Available online: https://www.woah.org/fileadmin/Home/eng/Health_standards/aahc/current/chapitre_diseases_listed.pdf (accessed on 26 May 2024).
- Stentiford, G.D.; Neil, D.M.; Peeler, E.J.; Shields, J.D.; Small, H.J.; Flegel, T.W.; Vlak, J.M.; Jones, B.; Morado, F.; Moss, S.; et al. Disease will limit future food supply from the global crustacean fishery and aquaculture sectors. J. Invertebr. Pathol. 2012, 110, 141–157. [Google Scholar] [CrossRef]
- Dhar, A.K.; Robles-Sikisaka, R.; Saksmerprome, V.; Lakshman, D.K. Biology, genome organization, and evolution of parvoviruses in marine shrimp. Adv. Virus Res. 2014, 89, 85–139. [Google Scholar] [CrossRef]
- Thitamadee, S.; Prachumwat, A.; Srisala, J.; Jaroenlak, P.; Salachan, P.V.; Sritunyalucksana, K.; Flegel, T.W.; Itsathitphaisarn, O. Review of current disease threats for cultivated penaeid shrimp in Asia. Aquaculture 2016, 452, 69–87. [Google Scholar] [CrossRef]
- Tang, K.F.; Lightner, D.V. Infectious hypodermal and hematopoietic necrosis virus (IHHNV)-related sequences in the genome of the black tiger prawn Penaeus monodon from Africa and Australia. Virus Res. 2006, 118, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Srisala, J.; Thaiue, D.; Sanguanrut, P.; Aldama-Cano, D.J.; Flegel, T.W.; Sritunyalucksana, K. Potential universal PCR method to detect decapod hepanhamaparvovirus (DHPV) in crustaceans. Aquaculture 2021, 541, 736782. [Google Scholar] [CrossRef]
- Lee, C.; Chang, Y.; Chiou, H.; Chang, H. Concurrent infection of a novel genotype of hepatopancreatic parvovirus and Enterocytozoon hepatopenaei in Penaeus vannamei in Taiwan. J. Fish Dis. 2022, 45, 1201–1210. [Google Scholar] [CrossRef] [PubMed]
- Singaravel, V.; Gopalakrishnan, A.; Martin, G.G. Multiple infections of Entrerocytozoon hepatopenaei and Hepatopancreatic parvovirus in pond-reared Penaeus vannamei in India. Aquaculture 2021, 545, 737232. [Google Scholar] [CrossRef]
- Joseph, T.C.; James, R.; Rajan, L.A.; Surendran, P.k.; Lalitha, K.v. Occurrence of viral pathogens in Penaeus monodon post-larvae from aquaculture hatcheries. Data Brief 2015, 4, 170–176. [Google Scholar] [CrossRef]
- Lee, C.; Jeon, H.J.; Kim, B.; Choi, S.-K.; Kim, J.H.; Han, J.E. Multiple infections of a new-type decapod hepanhamaparvovirus (DHPV) and Enterocytozoon hepatopenaei in Korea and DHPV infectivity in Penaeus vannamei. Aquaculture 2023, 563, 738922. [Google Scholar] [CrossRef]
- Flegel, T.; Thamavit, V.; Pasharawipas, T.; Alday-Sanz, V. Statistical correlation between severity of hepatopancreatic parvovirus infection and stunting of farmed black tiger shrimp (Penaeus monodon). Aquaculture 1999, 174, 197–206. [Google Scholar] [CrossRef]
- Chayaburakul, K.; Nash, G.; Pratanpipat, P.; Sriurairatana, S.; Withyachumnarnkul, B. Multiple pathogens found in growth-retarded black tiger shrimp Penaeus monodon cultivated in Thailand. Dis. Aquat. Org. 2004, 60, 89–96. [Google Scholar] [CrossRef]
- Chantanachookin, C.; Boonyaratpalin, S.; Kasornchandra, J.; Direkbusarakom, S.; Ekpanithanpong, U.; Supamataya, K.; Sriurairatana, S.; Flegel, T. Histology and ultrastructure reveal a new granulosis-like virus in Penaeus monodon affected by yellow-head disease. Dis. Aquat. Org. 1993, 17, 145–157. [Google Scholar] [CrossRef]
- Cotmore, S.F.; Agbandje-McKenna, M.; Chiorini, J.A.; Mukha, D.V.; Pintel, D.J.; Qiu, J.; Soderlund-Venermo, M.; Tattersall, P.; Tijssen, P.; Gatherer, D.; et al. The family Parvoviridae. Arch. Virol. 2014, 159, 1239–1247. [Google Scholar] [CrossRef]
- Edgerton, B.; Webb, R.; Wingfield, M. A systemic parvo-like virus in the freshwater crayfish Cherax destructor. Dis. Aquat. Org. 1997, 29, 73–78. [Google Scholar] [CrossRef]
- Bochow, S. Characterisation of Cherax quadricarinatus Densovirus: The First Virus Characterised from Australian Freshwater Crayfish. Ph.D. Thesis, James Cook University, Douglas, Australia, 2016. Available online: https://researchonline.jcu.edu.au/48889/ (accessed on 30 November 2023).
- Bochow, S.; Condon, K.; Elliman, J.; Owens, L. First complete genome of an Ambidensovirus; Cherax quadricarinatus densovirus, from freshwater crayfish Cherax quadricarinatus. Mar. Genom. 2015, 24, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Jackson, E.W.; Wilhelm, R.C.; Johnson, M.R.; Lutz, H.L.; Danforth, I.; Gaydos, J.K.; Hart, M.W.; Hewson, I. Diversity of sea star-associated densoviruses and transcribed endogenized viral elements of densovirus origin. J. Virol. 2020, 95, e01594-20. [Google Scholar] [CrossRef] [PubMed]
- Nunan, L.; Poulos, B.; Lightner, D. Use of Polymerase Chain Reaction for the Detection of Infectious Hypodermal and Hematopoietic Necrosis Virus in Penaeid Shrimp. Mar. Biotechnol. 2000, 2, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Dhar, A.K.; Roux, M.M.; Klimpel, K.R. Detection and Quantification of Infectious Hypodermal and Hematopoietic Necrosis Virus and White Spot Virus in Shrimp Using Real-Time Quantitative PCR and SYBR Green Chemistry. J. Clin. Microbiol. 2001, 39, 2835–2845. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.F.J.; Navarro, S.A.; Lightner, D.V. PCR assay for discriminating between infectious hypodermal and hematopoietic necrosis virus (IHHNV) and virus-related sequences in the genome of Penaeus monodon. Dis. Aquat. Org. 2007, 74, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Rai, P.; Pradeep, B.; Karunasagar, I.; Karunasagar, I. Detection of viruses in Penaeus monodon from India showing signs of slow growth syndrome. Aquaculture 2009, 289, 231–235. [Google Scholar] [CrossRef]
- Cowley, J.A.; Rao, M.; Coman, G. Real-time PCR tests to specifically detect IHHNV lineages and an IHHNV EVE integrated in the genome of Penaeus monodon. Dis. Aquat. Org. 2018, 129, 145–158. [Google Scholar] [CrossRef] [PubMed]
- Madan, N.; Nambi, K.; Majeed, S.A.; Taju, G.; Raj, N.S.; Farook, M.; Vimal, S.; Hameed, A.S. In vitro propagation of hepatopancreatic parvo-like virus (HPV) of shrimp in C6/36 (Aedes albopictus) cell line. J. Invertebr. Pathol. 2013, 112, 229–235. [Google Scholar] [CrossRef]
- Pantoja, C.R.; Lightner, D.V. Detection of hepatopancreatic parvovirus (HPV) of penaeid shrimp by in situ hybridization at the electron microscope level. Dis. Aquat. Org. 2001, 44, 87–96. [Google Scholar] [CrossRef][Green Version]
- Umesha, K.R.; Dass, B.K.M.; Naik, B.M.; Venugopal, M.N.; Karunasagar, I.; Karunasagar, I. High prevalence of dual and triple viral infections in black tiger shrimp ponds in India. Aquaculture 2006, 258, 91–96. [Google Scholar] [CrossRef]
- Yamkasem, J.; Tattiyapong, P.; Surachetpong, W. Development and application of TaqMan probe-based quantitative PCR assays for the detection of tilapia parvovirus. J. Fish Dis. 2022, 45, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhou, Y.; Fan, Y.; Jiang, N.; Meng, Y.; Li, Y.; Xue, M.; Xu, C.; Guo, W.; Liu, W. Development and application of a sensitive droplet digital PCR-based method to detect tilapia parvovirus. J. Fish Dis. 2023, 46, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Gudenkauf, B.M.; Eaglesham, J.B.; Aragundi, W.M.; Hewson, I. Discovery of urchin-associated densoviruses (family Parvoviridae) in coastal waters of the Big Island, Hawaii. J. Gen. Virol. 2014, 95, 652–658. [Google Scholar] [CrossRef] [PubMed]
- Pham, H.T. Molecular Biology of Single-Stranded DNA Viruses in Shrimps and Crickets. Ph.D. Thesis, Université du Québec, Quebec City, QC, Canada, 2015. [Google Scholar]
- La Fauce, K.A.; Elliman, J.; Owens, L. Molecular characterisation of hepatopancreatic parvovirus (PmergDNV) from Australian Penaeus merguiensis. Virology 2007, 362, 397–403. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tang, K.F.J.; Pantoja, C.R.; Lightner, D.V. Nucleotide sequence of a Madagascar hepatopancreatic parvovirus (HPV) and comparison of genetic variation among geographic isolates. Dis. Aquat. Org. 2008, 80, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Jeeva, S.; Kang, S.-W.; Lee, Y.-S.; Jang, I.K.; Seo, H.C.; Choi, T.-J. Complete nucleotide sequence analysis of a Korean strain of hepatopancreatic parvovirus (HPV) from Fenneropenaeus chinensis. Virus Genes 2012, 44, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Gangnonngiw, W.; Kiatpathomchai, W.; Sriurairatana, S.; Laisutisan, K.; Chuchird, N.; Limsuwan, C.; Flegel, T.W. Parvo-like virus in the hepatopancreas of freshwater prawns Macrobrachium rosenbergii cultivated in Thailand. Dis. Aquat. Org. 2009, 85, 167–173. [Google Scholar] [CrossRef]
- Anderson, I.G.; Law, A.; Shariff, M.; Nash, G. A parvo-like virus in the giant freshwater prawn, Macrobrachium rosenbergii. J. Invertebr. Pathol. 1990, 55, 447–449. [Google Scholar] [CrossRef]
- Lightner, D.V.; Redman, R.M.; Poulos, B.T.; Mari, J.L.; Bonami, J.R.; Shariff, M. Distinction of HPV-Type Viruses in Penaeus chinensis and Macrobrachium rosenbergii Using a DNA Probe. Asian Fish. Sci. 1994, 7, 267–272. [Google Scholar] [CrossRef]
- Chong, Y.C.; Loh, H. Hepatopancreas chlamydial and parvoviral infections of farmed marine prawns in Singapore. Singap. Vet. J. 1984, 9, 51–56. [Google Scholar]
- Lightner, D.V. A Handbook of Pathology and Diagnostic Procedures for Diseases of Penaeid Shrimp; World Aquaculture Society: Baton Rouge, LA, USA, 1996; Available online: https://lib.ugent.be/catalog/rug01:002170818 (accessed on 11 November 2023).
- Flegel, T.W.; Nielsen, L.; Thamavit, V.; Kongtim, S.; Pasharawipas, T. Presence of multiple viruses in non-diseased, cultivated shrimp at harvest. Aquaculture 2004, 240, 55–68. [Google Scholar] [CrossRef]
- Sukhumsirichart, W.; Attasart, P.; Boonsaeng, V.; Panyim, S. Complete nucleotide sequence and genomic organization of hepatopancreatic parvovirus (HPV) of Penaeus monodon. Virology 2006, 346, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Cervellione, F.; McGurk, C.; Eriksen, T.B.; Broeck, W.V.D. Use of computer-assisted image analysis for semi-quantitative histology of the hepatopancreas in whiteleg shrimp Penaeus vannamei (Boone). J. Fish Dis. 2017, 40, 1223–1234. [Google Scholar] [CrossRef] [PubMed]
- Lightner, D.V.; Redman, R.M. Shrimp diseases and current diagnostic methods. Aquaculture 1998, 164, 201–220. [Google Scholar] [CrossRef]
- Tang, K.F.J.; Poulos, B.T.; Wang, J.; Redman, R.M.; Shih, H.-H.; Lightner, D.V. Geographic variations among infectious hypodermal and hematopoietic necrosis virus (IHHNV) isolates and characteristics of their infection. Dis. Aquat. Org. 2003, 53, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Rai, P.; Pradeep, B.; Safeena, M.P.; Karunasagar, I.; Karunasagar, I. Simultaneous presence of infectious hypodermal and hematopoietic necrosis virus (IHHNV) and Type A virus-related sequence in Penaeus monodon from India. Aquaculture 2009, 295, 168–174. [Google Scholar] [CrossRef]
- Saksmerprome, V.; Jitrakorn, S.; Chayaburakul, K.; Laiphrom, S.; Boonsua, K.; Flegel, T.W. Additional random, single to multiple genome fragments of Penaeus stylirostris densovirus in the giant tiger shrimp genome have implications for viral disease diagnosis. Virus Res. 2011, 160, 180–190. [Google Scholar] [CrossRef]
- Alcivar-Warren, A.; Bao, W.; Tang, K.F. Endogenous viral elements (EVE) of Decapod penstylhamaparvovirus 1 (Infectious hypodermal and hematopoietic necrosis virus, IHHNV)—Implications for shrimp diagnosis. In Proceedings of the Aquaculture 2022, San Diego, CA, USA, 28 February–4 March 2022; Available online: https://www.was.org/Meeting/Program/PaperDetail/158479 (accessed on 7 June 2022).
- Sritunyalucksana-Dangtip, K. Progress on understanding the development of shrimp vaccines. In Proceedings of the FAO International Conference (Hybrid Event) Titled “Aquatic Organism Health: To Vaccinate or Not to Vaccinate, Rome, Italy, 20 June 2024. [Google Scholar]
- Campbell, M.A.; Loncar, S.; Kotin, R.M.; Gifford, R.J. Comparative analysis reveals the long-term coevolutionary history of parvoviruses and vertebrates. PLoS Biol. 2022, 20, e3001867. [Google Scholar] [CrossRef]
- Salcedo-Mejía, L.A.; Durán-Ramirez, Y.; Velazco-Peña, R.Z.; Pinto, J.A.; Rebaza-Caballero, A. Near-Complete Genome Sequences of 12 Peruvian Strains of Infectious Hypodermal and Hematopoietic Necrosis Virus Infecting the Shrimp Penaeus vannamei. Genome Announc. 2021, 10, e00169-21. [Google Scholar] [CrossRef]
- CEFAS (Centre for Environment Fisheries and Aquaculture Science). Infectious Hypodermal and Hematopoietic Necrosis Virus. Available online: https://idaad.cefas.co.uk/diseases/39 (accessed on 29 June 2022).
- Bajaña, L.; Betancourt, I.; Bayot, B. Complete coding genome sequence of infectious hypodermal and hematopoietic necrosis virus isolated from Penaeus (Litopenaeus) vannamei shrimp in Ecuador. Microbiol. Resour. Announc. 2022, 11, 4. [Google Scholar] [CrossRef]
- Bell, T.A.; Lightner, D.V. IHHN virus: Infectivity and pathogenicity studies in Penaeus stylirostris and Penaeus vannamei. Aquaculture 1984, 38, 185–194. [Google Scholar] [CrossRef]
- Chai, C.; Liu, Y.; Xia, X.; Wang, H.; Pan, Y.; Yan, S.; Wang, Y. Prevalence and genomic analysis of infectious hypodermal and hematopoietic necrosis virus (IHHNV) in Litopenaeus vannamei shrimp farmed in Shanghai, China. Arch. Virol. 2016, 161, 3189–3201. [Google Scholar] [CrossRef] [PubMed]
- Lightner, D.V.; Redman, R.M.; Poulos, B.T.; Nunan, L.M.; Mari, J.L.; Hasson, K.W. Risk of spread of peneid shrimp viruses in the Americas by the international movement of live and frozen shrimp. Rev. Sci. Tech. Off. Int. Epiz. 1997, 16, 146–160. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.D.; Hill, J.M.; Smith, P.R.K.; Kendrick, M.R.; Gooding, E.L.; Fuchs, C.; Whelan, N.V.; Bullard, S.A. First detection of white spot syndrome virus (WSSV) and infectious hypodermal and hematopoietic necrosis virus (IHHNV) from wild-caught giant tiger prawn, Penaeus monodon Fabricius, 1798 (Penaeoidea: Penaeidae) from the Gulf of Mexico and Northwestern Atlantic Ocean. BioInvasions Rec. 2024, 13, 121–140. [Google Scholar] [CrossRef]
- Hazreen-Nita, M.K.; Kua, B.C.; Bhassu, S.; Othman, R.Y. Detection and genetic profiling of infectious hypodermal and haematopoietic necrosis virus (IHHNV) infections in wild berried freshwater prawn, Macrobrachium rosenbergii collected for hatchery production. Mol. Biol. Rep. 2012, 39, 3785–3790. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.-K.; Dong, Z.; Liu, D.-P.; Yan, Y.-B.; Pang, N.-Y.; Nian, Y.-Y.; Yan, D.-C. Infectious hypodermal and haematopoietic necrosis virus (IHHNV) infection in freshwater crayfish Procambarus clarkii. Aquaculture 2017, 477, 76–79. [Google Scholar] [CrossRef]
- Chen, B.-K.; Dong, Z.; Pang, N.-Y.; Nian, Y.-Y.; Yan, D.-C. A novel real-time PCR approach for detection of infectious hypodermal and haematopoietic necrosis virus (IHHNV) in the freshwater crayfish Procambarus clarkii. J. Invertebr. Pathol. 2018, 157, 100–103. [Google Scholar] [CrossRef]
- Lee, C.; Choi, S.-K.; Jeon, H.J.; Lee, S.H.; Kim, Y.K.; Park, S.; Park, J.-K.; Han, S.-H.; Bae, S.; Kim, J.H.; et al. Detection of Infectious Hypodermal and Hematopoietic Necrosis Virus (IHHNV, Decapod Penstylhamaparvovirus 1) in Commodity Red Claw Crayfish (Cherax quadricarinatus) Imported into South Korea. J. Mar. Sci. Eng. 2021, 9, 856. [Google Scholar] [CrossRef]
- Yang, B.; Song, X.-L.; Huang, J.; Shi, C.-Y.; Liu, L. Evidence of existence of infectious hypodermal and hematopoietic necrosis virus in penaeid shrimp cultured in China. Veter. Microbiol. 2007, 120, 63–70. [Google Scholar] [CrossRef]
- Cavalli, L.S.; Batista, C.R.; Nornberg, B.F.S.; Mayer, F.Q.; Seixas, F.K.; Romano, L.A.; Marins, L.F.; Abreu, P.C. Natural occurrence of White spot syndrome virus and Infectious hypodermal and hematopoietic necrosis virus in Neohelice granulata crab. J. Invertebr. Pathol. 2013, 114, 86–88. [Google Scholar] [CrossRef]
- Macías-Rodríguez, N.A.; Mañón-Ríos, N.; Romero-Romero, J.L.; Camacho-Beltrán, E.; Magallanes-Tapia, M.A.; Leyva-López, N.E.; Hernández-López, J.; Magallón-Barajas, F.J.; Perez-Enriquez, R.; Sánchez-González, S.; et al. Prevalence of viral pathogens WSSV and IHHNV in wild organisms at the Pacific Coast of Mexico. J. Invertebr. Pathol. 2014, 116, 8–12. [Google Scholar] [CrossRef]
- Wei, Y.-W.; Fan, D.-D.; Chen, J. The mussel Mytilus edulis L. as an important reservoir of infectious hypodermal and hematopoietic necrosis virus (IHHNV). Aquac. Res. 2015, 48, 758–759. [Google Scholar] [CrossRef]
- Owens, L.; De Beer, S.; Smith, J. Lymphoidal parvovirus-like particles in Australian penaeid prawns. Dis. Aquat. Org. 1991, 11, 129–134. [Google Scholar] [CrossRef]
- Owens, L. Abolishment of spawner-isolated mortality virus and where the remaining science leads. J. Gen. Virol. 2023, 104, 001878. [Google Scholar] [CrossRef]
- Owens, L.; McElnea, C. Natural infection of the redclaw crayfish Cherax quadricarinatus with presumptive spawner-isolated mortality virus. Dis. Aquat. Org. 2000, 40, 219–223. [Google Scholar] [CrossRef]
- Rai, P.; Safeena, M.P.; Krabsetsve, K.; La Fauce, K.; Owens, L.; Karunasagar, I. Genomics, Molecular Epidemiology and Diagnostics of Infectious hypodermal and hematopoietic necrosis virus. Indian J. Virol. 2012, 23, 203–214. [Google Scholar] [CrossRef]
- DAWE (Australian Government Department of Agriculture, Water and the Environment). Infection with Infectious Hypodermal and Haematopoietic Necrosis Virus (IHHNV). 2020. Available online: https://www.agriculture.gov.au/sites/default/files/documents/infection-infectious-hypodermal-haematopoietic-necrosis-virus.pdf (accessed on 29 June 2022).
- Lightner, D. Virus diseases of farmed shrimp in the Western Hemisphere (the Americas): A review. J. Invertebr. Pathol. 2011, 106, 110–130. [Google Scholar] [CrossRef]
- Jagadeesan, V.; Praveena, P.E.; Otta, S.K.; Jithendran, K.P. Classical Runt Deformity Syndrome Cases in Farmed Penaeus vannamei Along the East Coast of India. J. Coast. Res. 2019, 86, 107–111. [Google Scholar] [CrossRef]
- Zhang, C.; Yuan, J.-F.; Shi, Z.-L. Molecular epidemiological investigation of infectious hypodermal and hematopoietic necrosis virus and Taura syndrome virus in Penaeus Vannamei cultured in China. Virol. Sin. 2007, 22, 380–388. [Google Scholar] [CrossRef]
- Nian, Y.-Y.; Chen, B.-K.; Wang, J.-J.; Zhong, W.-T.; Fang, Y.; Li, Z.; Zhang, Q.-S.; Yan, D.-C. Transcriptome analysis of Procambarus clarkii infected with infectious hypodermal and haematopoietic necrosis virus. Fish Shellfish. Immunol. 2020, 98, 766–772. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Yu, Y.-B.; Choi, J.-H.; Jo, A.-H.; Hong, S.-M.; Kang, J.-C.; Kim, J.-H. Viral Shrimp Diseases Listed by the OIE: A Review. Viruses 2022, 14, 585. [Google Scholar] [CrossRef]
- Bonnichon, V.; Lightner, D.V.; Bonami, J.-R. Viral interference between infectious hypodermal and hematopoietic necrosis virus and white spot syndrome virus in Litopenaeus vannamei. Dis. Aquat. Org. 2006, 72, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Molthathong, S.; Jitrakorn, S.; Joyjinda, Y.; Boonchird, C.; Witchayachamnarnkul, B.; Pongtippatee, P.; Flegel, T.; Saksmerprome, V. Persistence of Penaeus stylirostris densovirus delays mortality caused by white spot syndrome virus infection in black tiger shrimp (Penaeus monodon). BMC Veter. Res. 2013, 9, 33. [Google Scholar] [CrossRef]
- Robles-Sikisaka, R.; Bohonak, A.J.; McClenaghan, L.R., Jr.; Dhar, A.K. Genetic Signature of Rapid IHHNV (Infectious Hypodermal and Hematopoietic Necrosis Virus) Expansion in Wild Penaeus Shrimp Populations. PLoS ONE 2010, 5, e11799. [Google Scholar] [CrossRef] [PubMed]
- Arbon, P.M.; Condon, K.; Martinez, M.A.; Jerry, D.R. Molecular detection of six viral pathogens from Australian wild sourced giant black tiger shrimp (Penaeus monodon) broodstock. Aquaculture 2021, 548, 737651. [Google Scholar] [CrossRef]
- Silva, D.C.D.; Nunes, A.R.D.; Teixeira, D.I.A.; Lima, J.P.M.S.; Lanza, D.C.F. Infectious hypodermal and hematopoietic necrosis virus from Brazil: Sequencing, comparative analysis and PCR detection. Virus Res. 2014, 189, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Jaroenram, W.; Owens, L. Recombinase polymerase amplification combined with a lateral flow dipstick for discriminating between infectious Penaeus stylirostris densovirus and virus-related sequences in shrimp genome. J. Virol. Methods 2014, 208, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, K.; Liu, M.; Hu, J.; Bao, Z.; Wang, M. Development of a real-time enzymatic recombinase amplification assay for rapid detection of infectious hypodermal and hematopoietic necrosis virus (IHHNV) in shrimp Penaeus vannamei. J. Invertebr. Pathol. 2023, 201, 108024. [Google Scholar] [CrossRef]
- Clyde, C.W.; Tan, J.P.; Yeap, S.K.; Yong, C.Y. Current updates on viral infections affecting tilapia. Aquac. Fish. 2024, in press. [CrossRef]
- Dong, H.T.; Sangpo, P.; Dien, L.T.; Mai, T.T.; Linh, N.V.; Del-Pozo, J.; Salin, K.R.; Senapin, S. Usefulness of the pancreas as a prime target for histopathological diagnosis of Tilapia parvovirus (TiPV) infection in Nile tilapia, Oreochromis niloticus. J. Fish Dis. 2022, 45, 1323–1331. [Google Scholar] [CrossRef]
- Rajendran, K.V.; Sood, N.; Rao, B.M.; Valsalam, A.; Bedekar, M.K.; Jeena, K.; Pradhan, P.K.; Paria, A.; Swaminathan, T.R.; Verma, D.K.; et al. Widespread occurrence of Tilapia parvovirus in farmed Nile tilapia Oreochromis niloticus from India. J. Fish Dis. 2023. online ahead of print. [Google Scholar] [CrossRef]
- Prasartset, T.; Surachetpong, W. Simultaneous detection of three important viruses affecting tilapia using a multiplex PCR assay. J. Fish Dis. 2023, 46, 459–464. [Google Scholar] [CrossRef]
- Phusantisampan, T.; Yamkasem, J.; Tattiyapong, P.; Sriariyanun, M.; Surachetpong, W. Specific and rapid detection of tilapia parvovirus using loop-mediated isothermal amplification (LAMP) method. J. Fish Dis. 2022, 45, 1893–1898. [Google Scholar] [CrossRef] [PubMed]
- Hargitai, R.; Pankovics, P.; Boros, Á.; Mátics, R.; Altan, E.; Delwart, E.; Reuter, G. Novel picornavirus (family Picornaviridae) from freshwater fishes (Perca fluviatilis, Sander lucioperca, and Ameiurus melas) in Hungary. Arch. Virol. 2021, 166, 2627–2632. [Google Scholar] [CrossRef] [PubMed]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef]
- Owens, L.; McElnea, C.; Snape, N.; Harris, L.; Smith, M. Prevalence and effect of spawner-isolated mortality virus on the hatchery phases of Penaeus monodon and P. merguiensis in Australia. Dis. Aquat. Org. 2003, 53, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Fraser, C.A.; Owens, L. Spawner-isolated mortality virus from Australian Penaeus monodon. Dis. Aquat. Org. 1996, 27, 141–148. [Google Scholar] [CrossRef][Green Version]
- Owens, L.; Haqshenas, G.; McElnea, C.; Coelen, R. Putative spawner-isolated mortality virus associated with mid-crop mortality syndrome in farmed Penaeus monodon from northern Australia. Dis. Aquat. Org. 1998, 34, 177–185. [Google Scholar] [CrossRef]
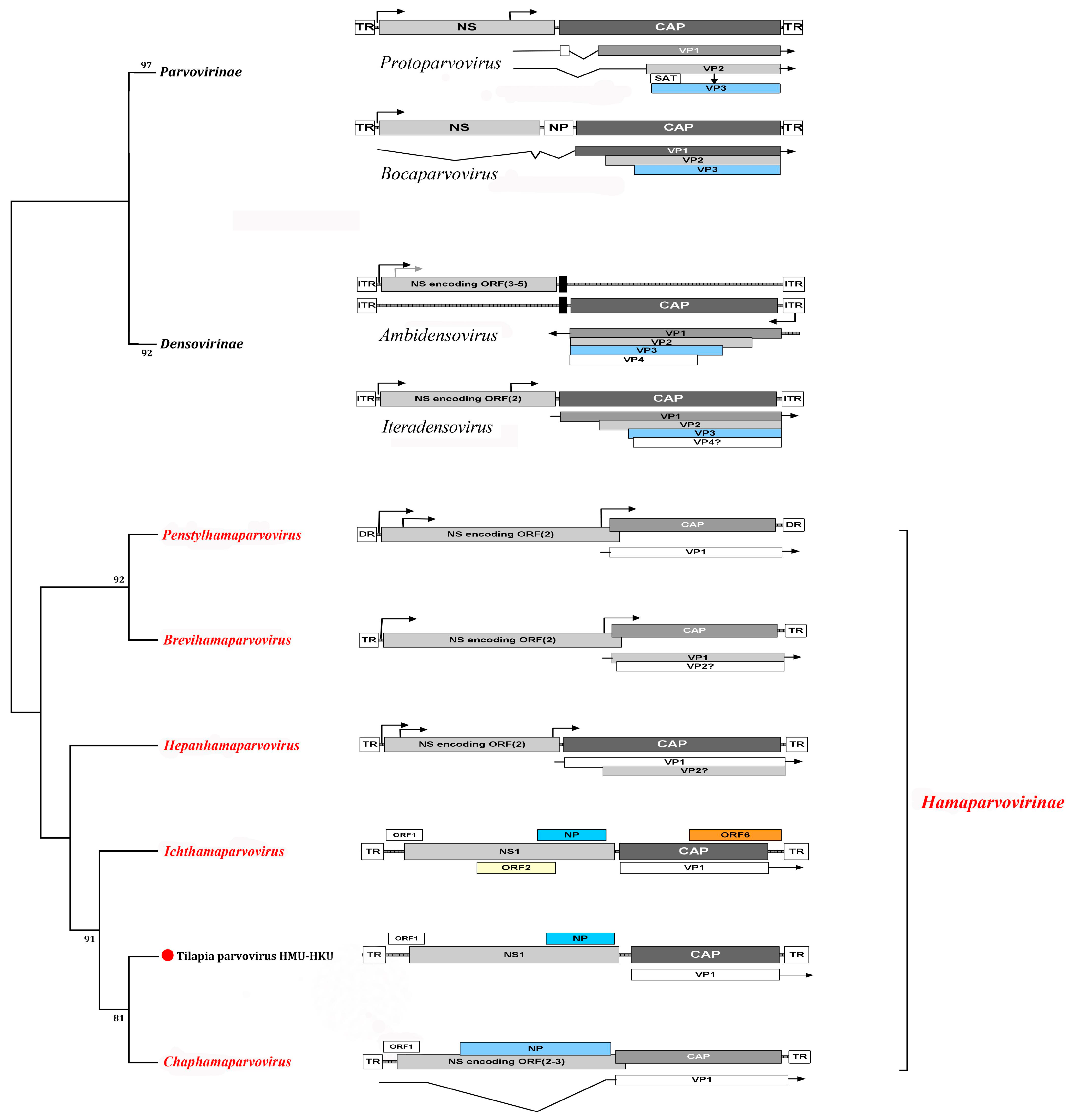
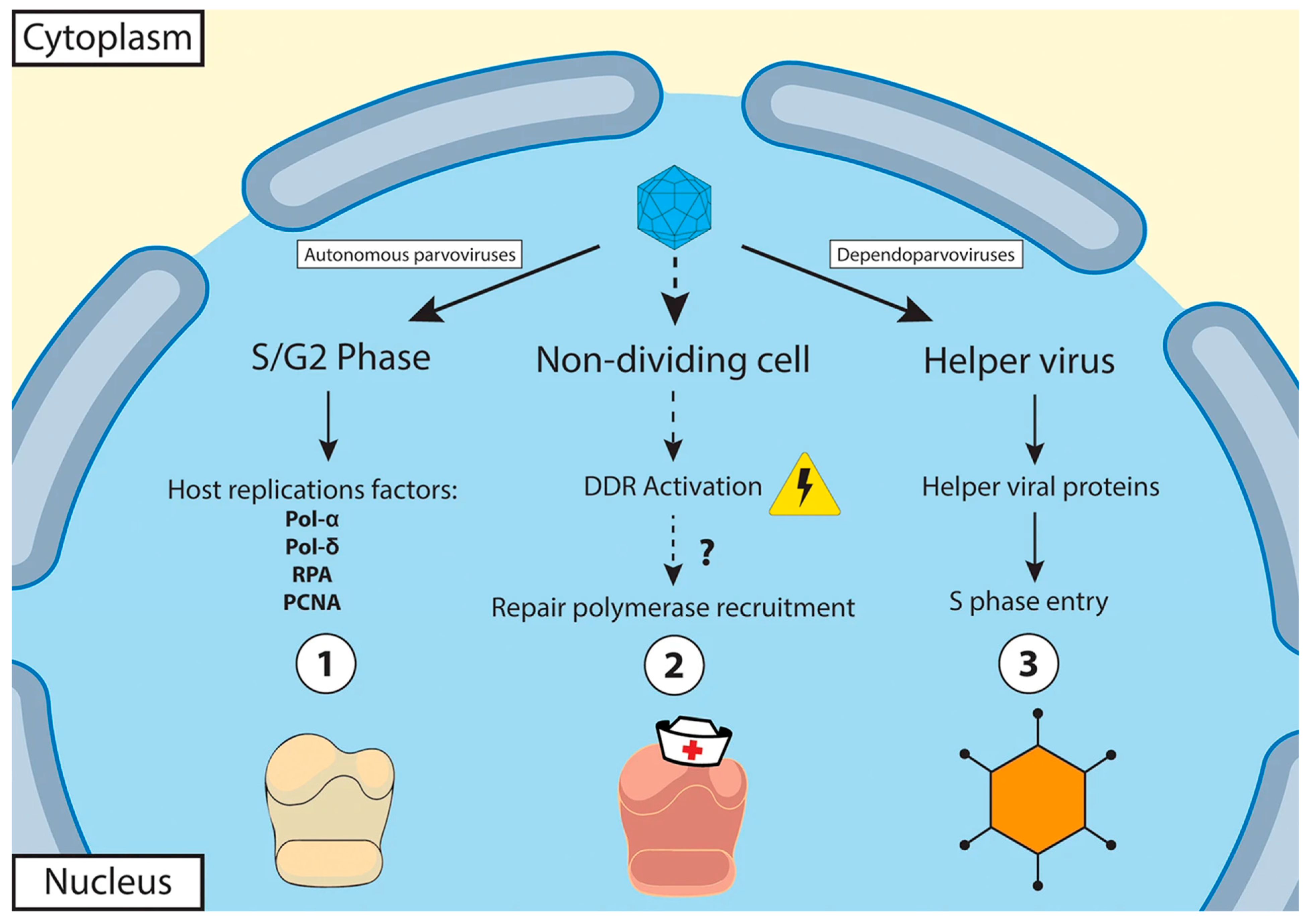
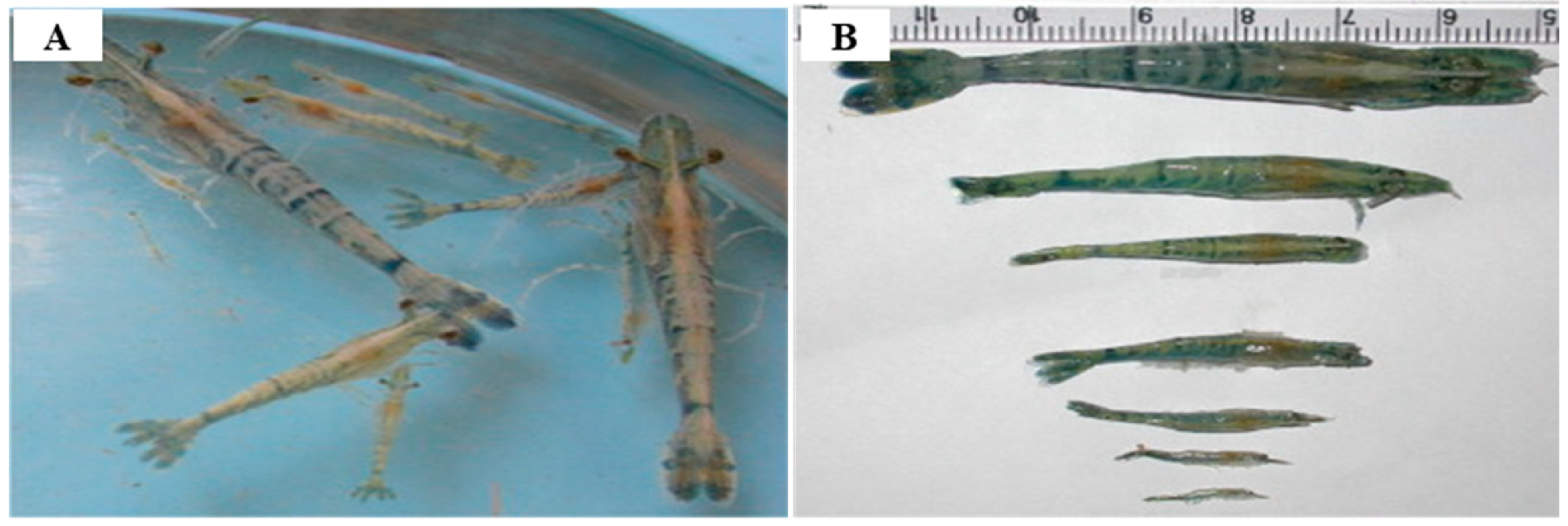
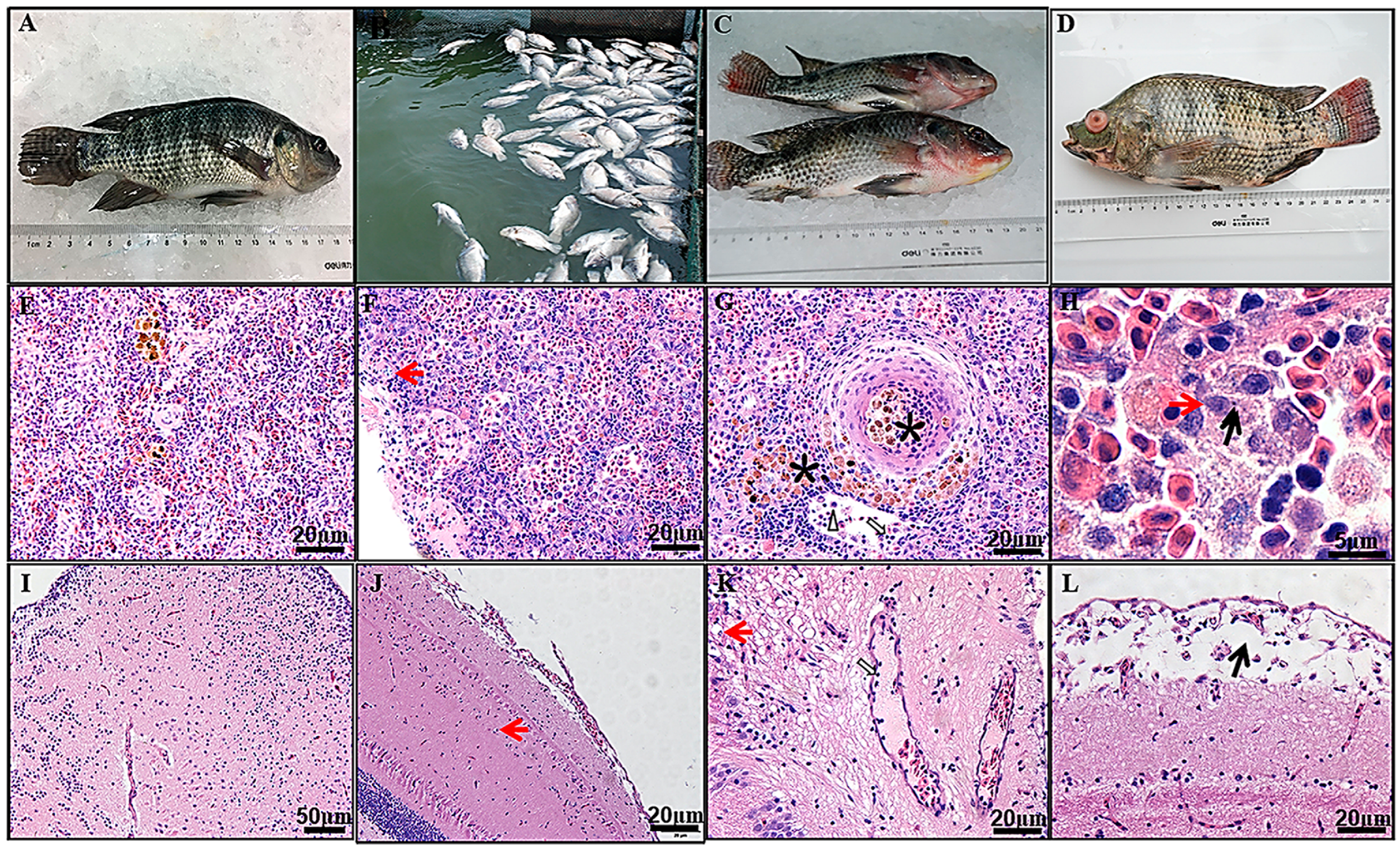
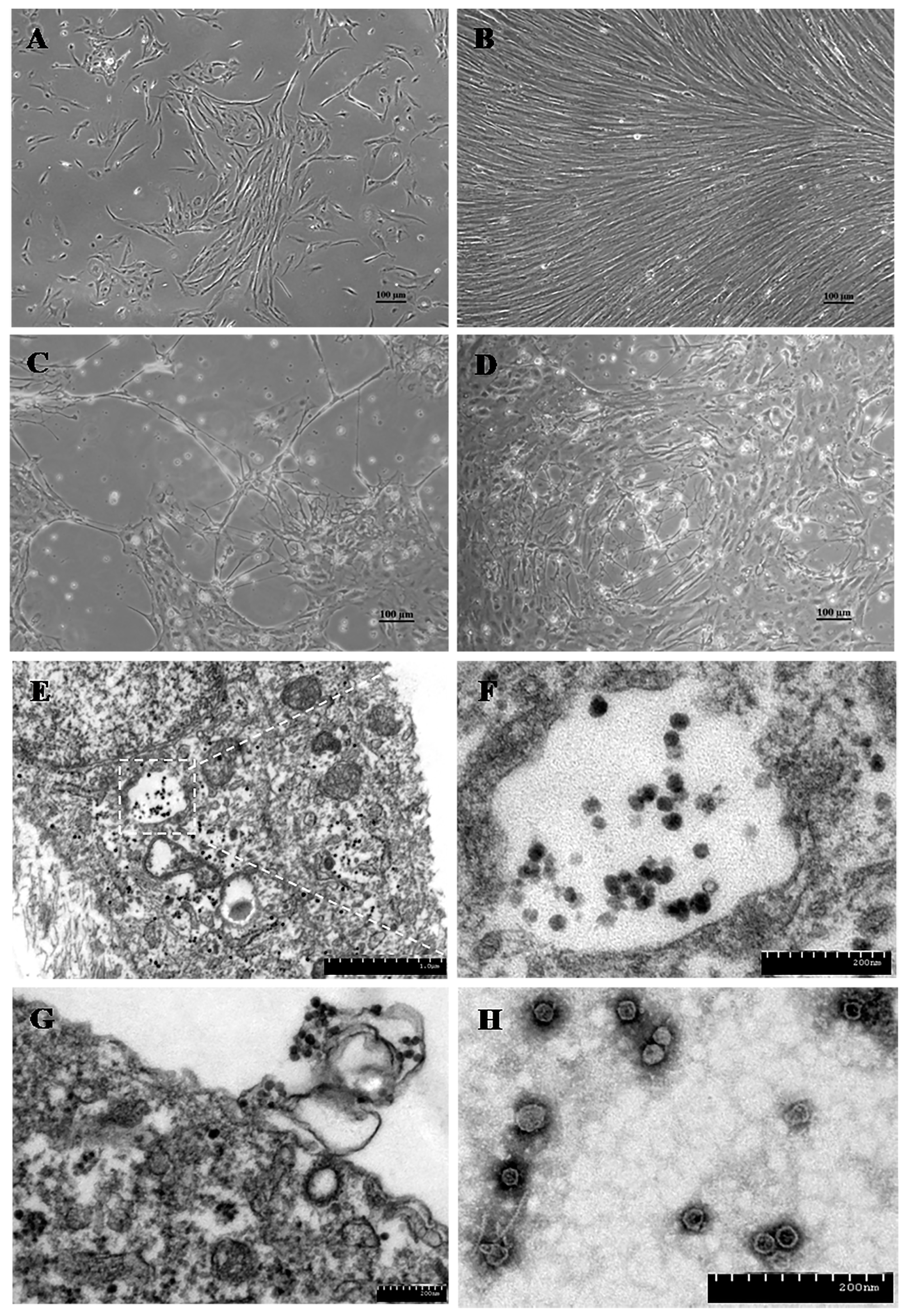
| Virus Subfamily | Virus Genus | Virus Species or Isolate 1 in Host | Virus Isolation and Propagation | Conventional PCR/RT-PCR or Quantitative (q)PCR/RT-PCR | |||
|---|---|---|---|---|---|---|---|
| Fish | Crustaceans | Cell Lines 2 | Incubation Temp. 3 | Virus-Specific Primers/Probes | Amplicon Size, PCR Chemistry 4, and Reference | ||
| Densovirinae | Aquambidensovirus | CqDV SSaDV | - - | - - | CqDV 5 Fq 5′-CGCTGTGGAGAGTGCACTAGAGGC-3′ 2A Rq 5′-TCTGAATCAATCTCCTCACGATCGC-3′ VP1 forward primer 5′-TGGCCACTCATCATGTCTCT-3′ VP1 reverse primer 5′-CTTGGGGTCCTTCATGAGC-3′ | 283 bp, qPCR-SYBR Green [89] 534 bp, cPCR [91] | |
| Hamaparvovirinae Parvovirinae | Penstylhamaparvovirus Hepanhamaparvovirus Ichthamaparvovirus Unassigned | TiPV Novel zander parvovirus | IHHNV HPV/DHPV-1 | - C6/36 6 E-11; TiB | - 28 °C 25 °C 28 °C | 77012F 5′-ATCGGTGCACTACTCGGA-3′ 77353R 5′-TCGTACTGGCTGTTCATC-3′ 313F Forward 5′-AGGAGACAACCGACGACATCA-3′ 363R Reverse 5′-CGATTTCCATTGCTTCCATGA-3′ IHHNV309F 5′-TCCAACACTTAGTCAAAACCAA-3′ IHHNV309R 5′-TGTCTGCTACGATGATTATCCA-3′ IHHNV648F 5′-GAACGGCTTTCGTATTTTGG-3′ IHHNV648R 5′-AGCGTAGGACTTGCCGATTA-3′ 77112F 5′-ATCGGTGCACTACTCGGA-3′ 77012R 5′-′TCGTACTGGCTGTTCATC-3′ 389F 5′-CGGAACACAACCCGACTTTA-3′ 389R 5′-CGCCAAGACCAAAATACGAA-3′ IHHNV-q309F1 5′-CCTAAAGAAAACAGTGCAGAATAT-3′ IHHNV-q309R1 5′-TCATCGTCAAGTTTATTGACAAGTTC-3′ IHHNV-q309Pr1 5′-6FAM-CTCCAACACTTAGTCAAA-TAMRA-3′ HPVF 5′-GCATTACAAGAGCCAAGCAG-3′ HPVR 5′- ACACTCAGCCTCTACCTTGT-3′ qHPVF 5′- CGCGGATCCAGGTAGAGGCTGAGTGTAA-3′ qHPVR 5′-CGCGAATTCCAGGTAGTGACGCCGAAA-3′ 1120F 5′-GGTGATGTGGAGGAGAGA-3′ 1120R 5′-GTAACTATCGCCGCCAAC-3′ HPVnF 5′-ATAGAACGCATAGAAAACGCT-3′ HPVnR 5′-CAGCGATTCATTCCAGCGCCACC-3′ DHPV-U 1538 F 5′-CCTCTTGTTACATTTTACTC-3′ DHPV-U 1887 R 5′-GATGTCTTCTGTAGTCC-3′ DHPV-U 1622 F 5′-AAGTTTGCACAGTGGTTGT-3′ HPV F 5′- CCACAACATAAGTGCTGCAGT-3′ HPV R 5′- TAGCCGCGGAATAAAACCCT-3′ TaqMan probe for HPV Madagascar 5′- 6FAM -TGAATGTTGTAAAGACTCAGCCA- TAMRA-3 HPV F 5′-AAGCCTGTGTTTCTGACTAACCA-3′ HPV R 5′- TGAGTTTACCGCCTCACTTCC-3′ TiPV-Fq 5′-GAGATGGTGTGAAAATGAACGGG-3′ TiPV-Rq 5′-CTATCTCCTCGTTGCTCGGTGTATC-3′ TiPV-F 5′-GAGATGGTGTGAAAATGAACGGG-3′ TiPV-R 5′-CTATCTCCTCGTTGCTCGGTGTATC-3′ VP734F 5′-TGGCTTTATGGACTTTGCTGAT-3′ VP734R 5′-CATCCCTCCTGCTCTTGGTT-3′ TiPV-NS124F 5′-AAAGTACCTAAGGCGGAGCG-3′ TiPV-NS124R 5′- TTTCAATCACCTTCCCGCCA-3′ TiPV-NS124Probe 5′-6FAM-CCGAAGAGCATAGTGG-BHQ1-3′ TiPV-VP205F 5′-CCAGATTGAAAGGGGCACGA-3′ TiPV-VP205R 5′-TTGGTGTTGGTGGTACGCAT-3′ TiPV-VP205Probe 5′-6FAM-CCAGTCCCGACCTACTCAAA-BHQ1-3′ TiPV-NS965F 5′-TGGCTACCGAGAAGGGGTTA-3′ TiPV-NS965R 5′-GCTCTTCCCGCTTGAGTCTT-3′ TiPV Forward 5′-GTATTAGTGGCGTCATTGCAGAG-3′ TiPV Reverse 5′-GGCAGGTTCCCCACTTCAC-3′ TiPV Probe 5′-6FAM-CCCTTCTCGGTAGCCAC-MGB-3′ ZanderParvoscreen-F 5′-GGCTAATCATCAAACAGGAAAGA-3′ ZanderParvo-screen-R 5′-AGCTCC CACCACTTAATATCTT-3′ | 1681 bp, cPCR [92] 50 bp, qPCR-SYBR Green [93] 309 bp, cPCR [94] 648 bp, cPCR-1st step 5 [95] 356 bp, cPCR [82] 389 bp, cPCR [82] 98 bp, qPCR -TaqMan [96] 441 bp, cPCR [47,97] 147 bp, qPCR-SYBR Green [97] 592 bp, cPCR [98] 265 bp, cPCR-2nd step 7 [99] 350 bp and 266 bp, cPCR-semi nested [51] 165 bp, cPCR and TaqMan [23] 120 bp, cPCR [23] 134 bp, qPCR-SYBR Green [25] 534 bp, cPCR [25] 734 bp, cPCR [26] 124 bp, qPCR-TaqMan [100] 205 bp, qPCR-TaqMan [100] 965 bp, cPCR [100] ddPCR [101] 492 bp, cPCR [5]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kibenge, F.; Kibenge, M.; Montes de Oca, M.; Godoy, M. Parvoviruses of Aquatic Animals. Pathogens 2024, 13, 625. https://doi.org/10.3390/pathogens13080625
Kibenge F, Kibenge M, Montes de Oca M, Godoy M. Parvoviruses of Aquatic Animals. Pathogens. 2024; 13(8):625. https://doi.org/10.3390/pathogens13080625
Chicago/Turabian StyleKibenge, Frederick, Molly Kibenge, Marco Montes de Oca, and Marcos Godoy. 2024. "Parvoviruses of Aquatic Animals" Pathogens 13, no. 8: 625. https://doi.org/10.3390/pathogens13080625
APA StyleKibenge, F., Kibenge, M., Montes de Oca, M., & Godoy, M. (2024). Parvoviruses of Aquatic Animals. Pathogens, 13(8), 625. https://doi.org/10.3390/pathogens13080625







