Phylogenetics and Mobilization of Genomic Traits of Cephalosporin-Resistant Escherichia coli Originated from Retail Meat
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection and Selection of Samples
2.2. DNA Extraction
2.3. Library Preparation and Sequencing
2.4. Data Analysis
3. Results
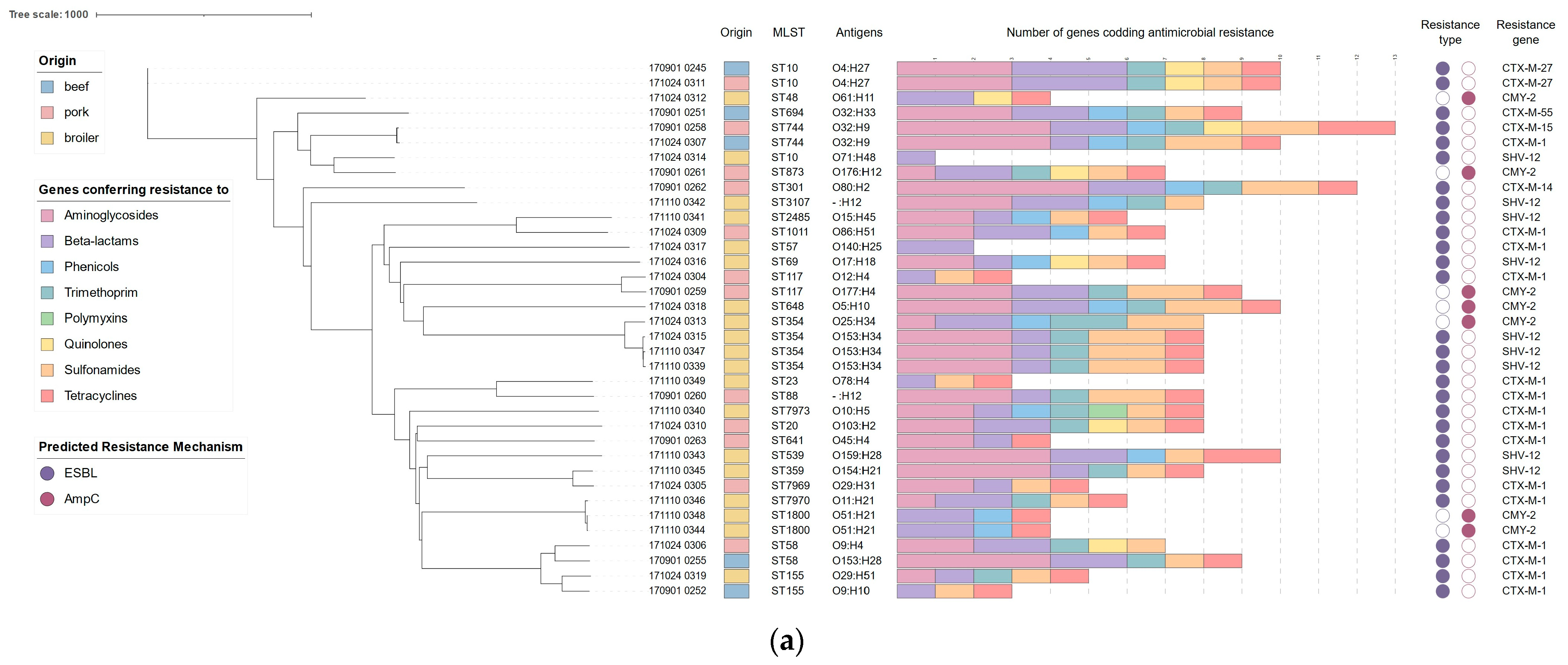
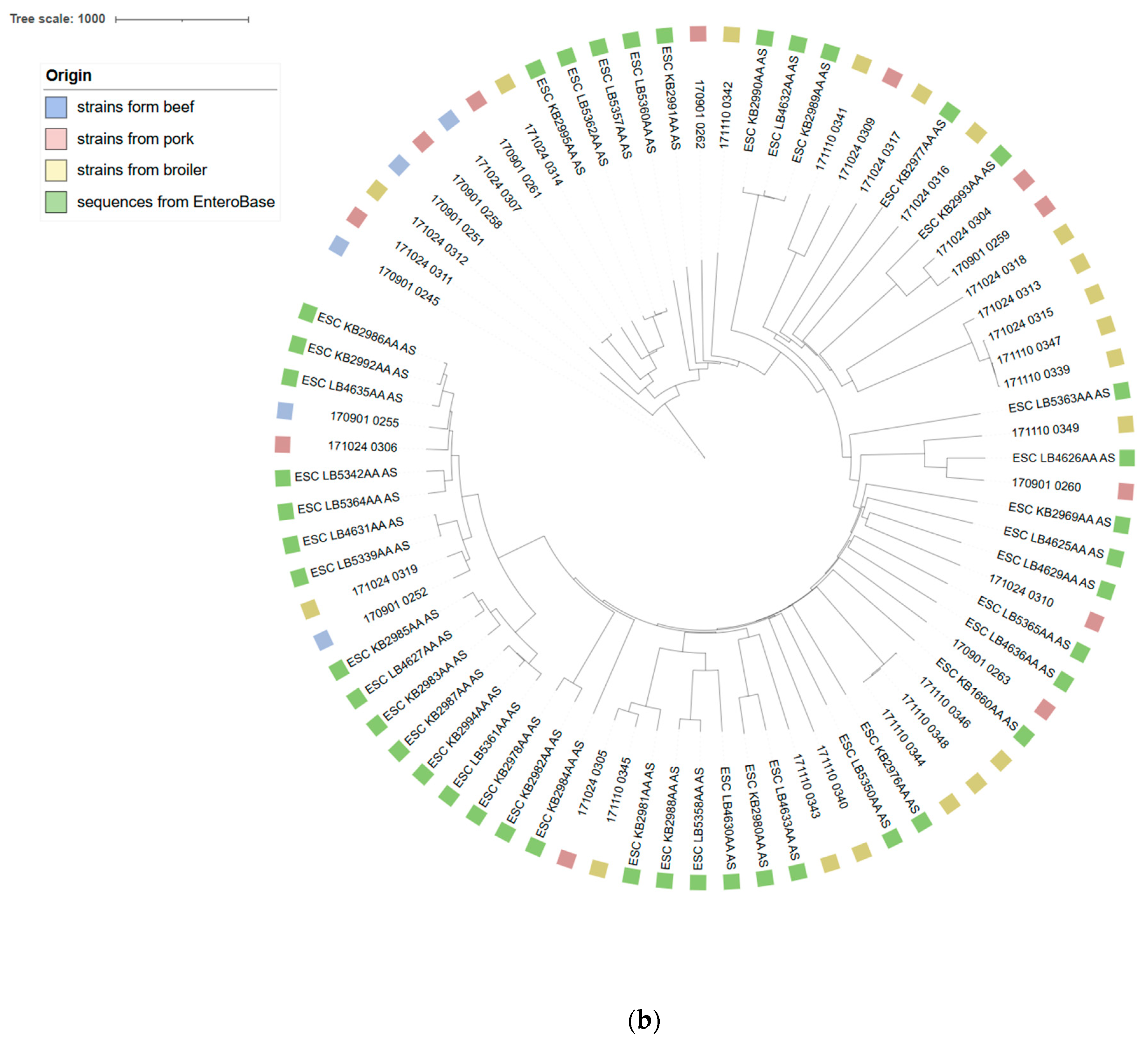
3.1. Molecular Typing
3.2. Acquired Resistance
3.3. Virulence Factors
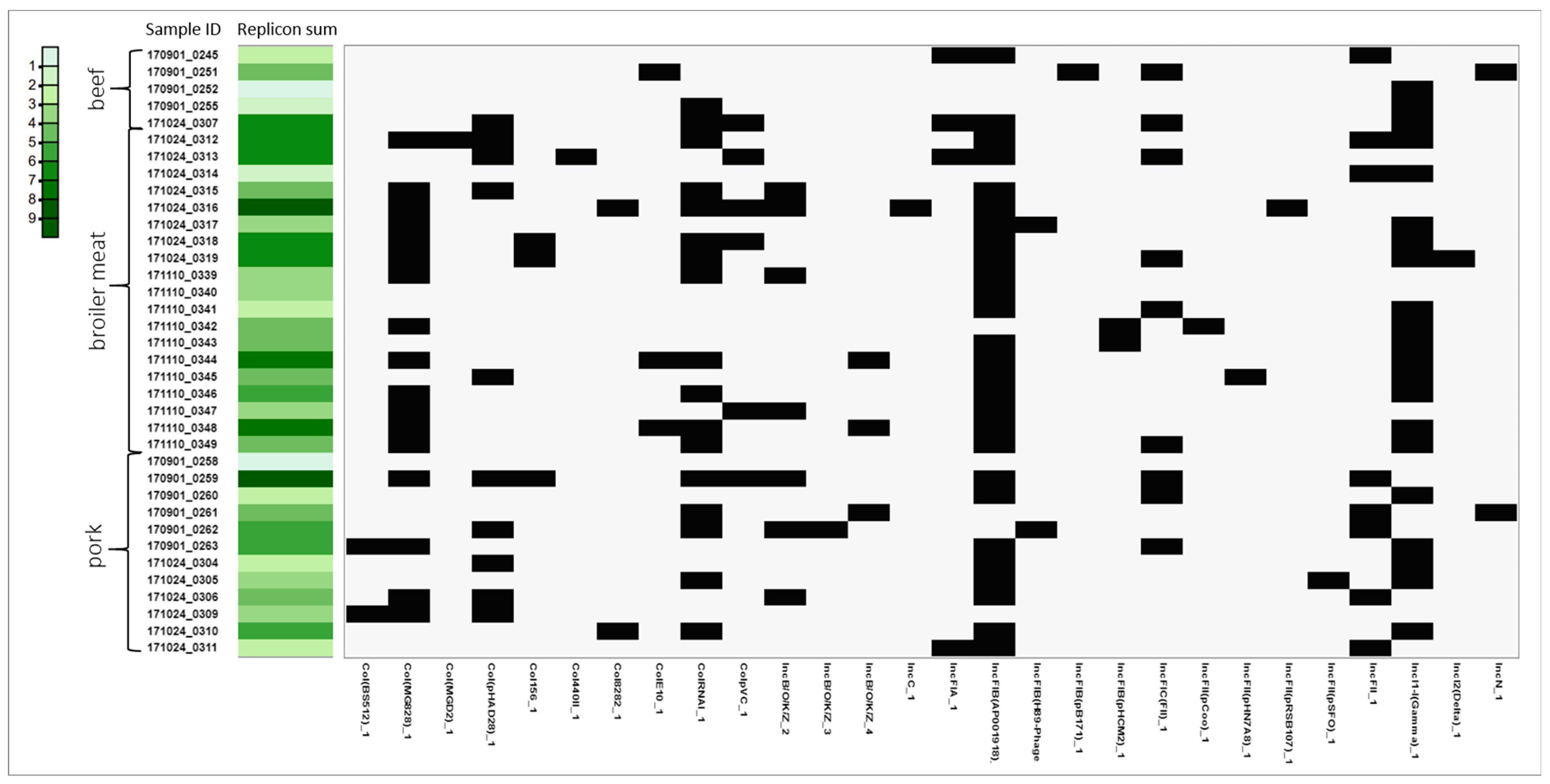
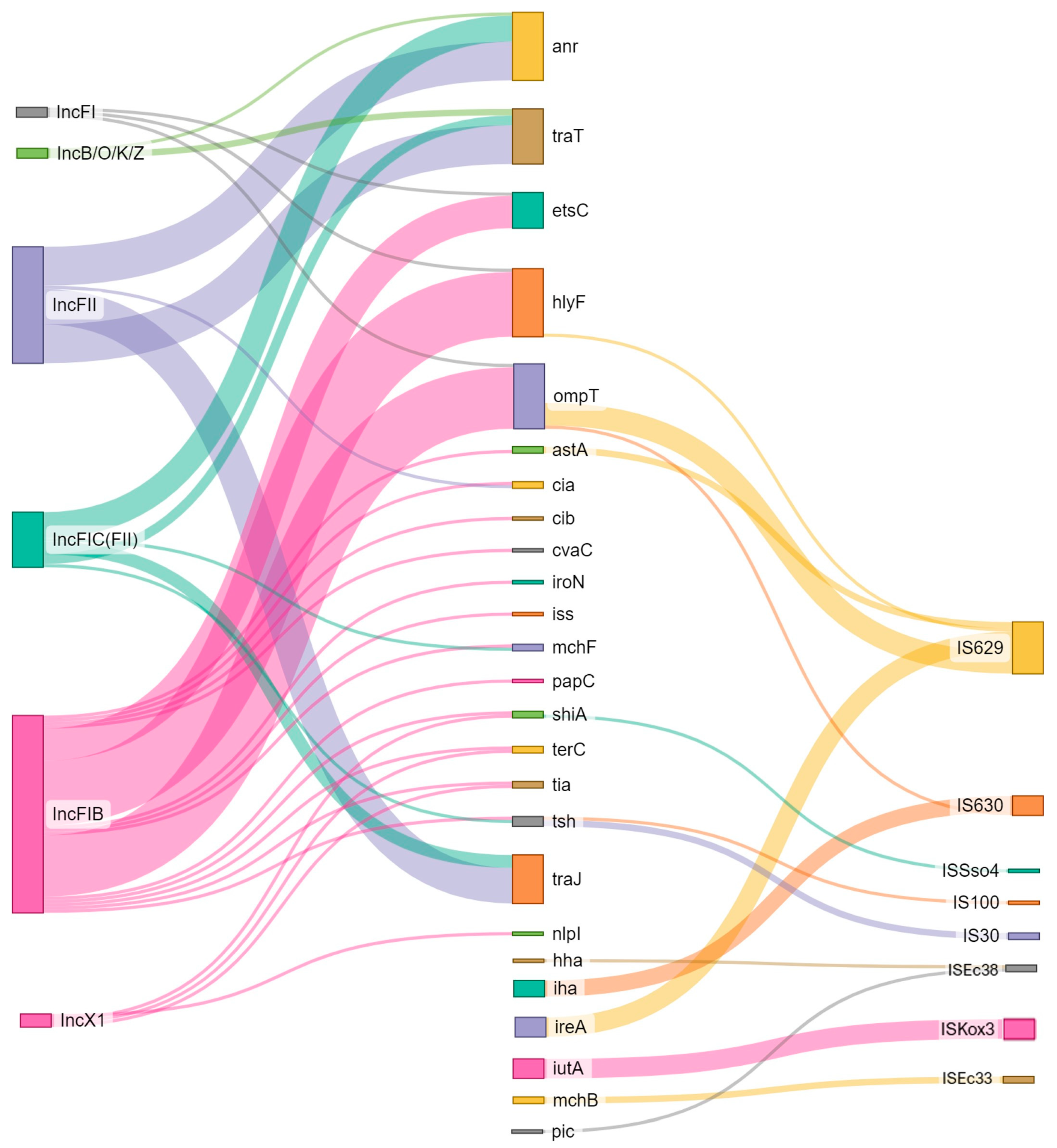
3.4. Plasmids
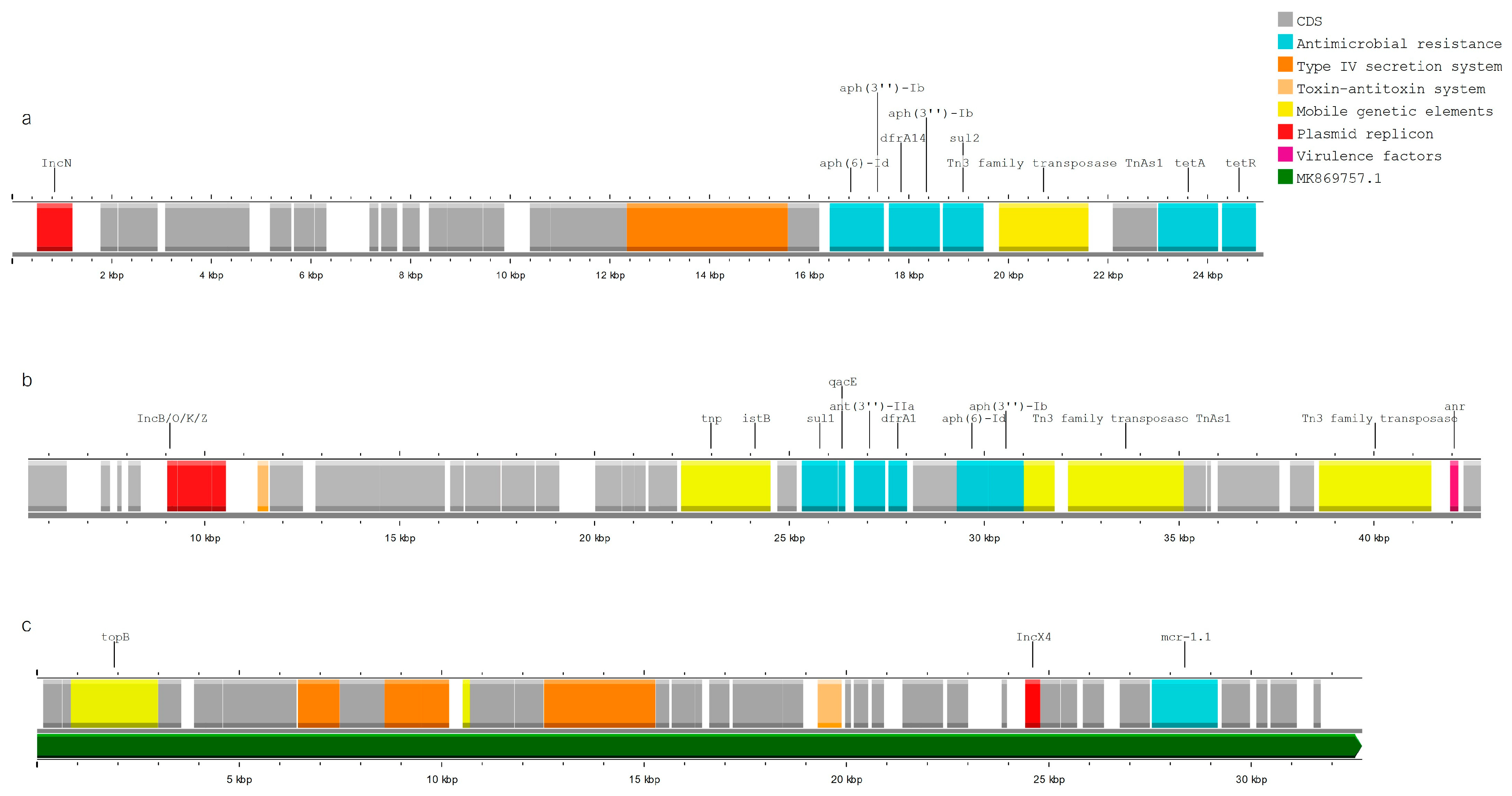
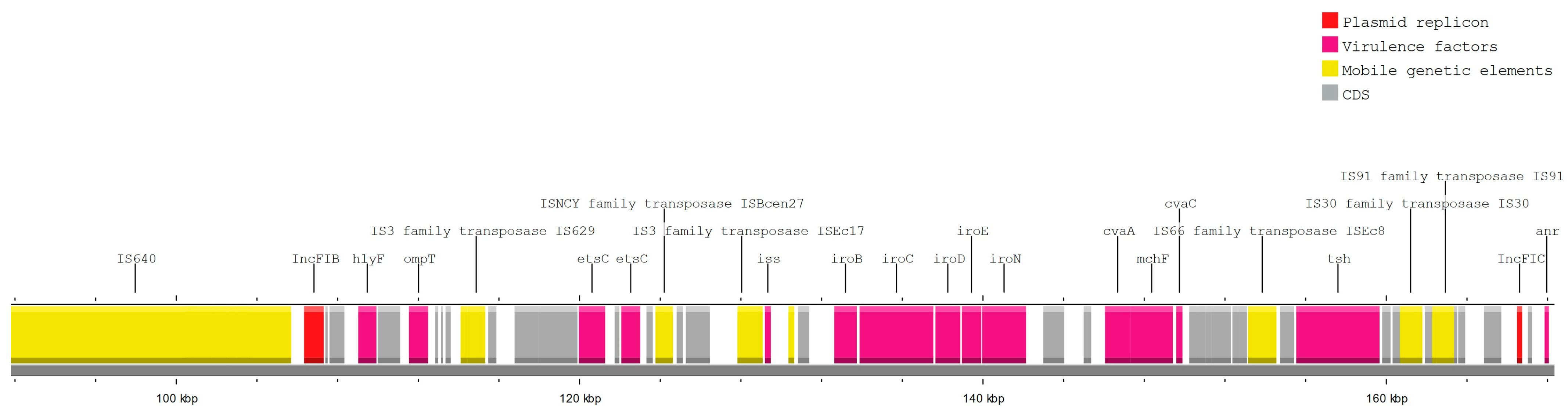
3.5. MGE Associated with Resistance and Virulence Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Haley, B.J.; Kim, S.W.; Salaheen, S.; Hovingh, E.; Van Kessel, J.A.S. Virulome and genome analyses identify associations between antimicrobial resistance genes and virulence factors in highly drug resistant Escherichia coli isolated from veal calves. PLoS ONE 2022, 17, e0265445. [Google Scholar] [CrossRef]
- Poirel, L.; Madec, J.-Y.; Lupo, A.; Schink, A.-K.; Kieffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial Resistance in Escherichia coli. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Stokes, H.W.; Gillings, M.R. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol. Rev. 2011, 35, 790–819. [Google Scholar] [CrossRef]
- Lam, M.M.C.; Wick, R.R.; Wyres, K.L.; Gorrie, C.L.; Judd, L.M.; Jenney, A.W.J.; Brisse, S.; Holt, K.E. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb. Genom. 2018, 4, e000196. [Google Scholar] [CrossRef] [PubMed]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile genetic elements associated with antimicrobial resistance. Am. Soc. Microbiol. 2018, 31, 10–128. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J. 2019, 17, 136–160. [Google Scholar] [CrossRef]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2019–2020. EFSA J. 2022, 20, 68–88. [Google Scholar] [CrossRef]
- Florez-Cuadrado, D.; Moreno, M.A.; Ugarte-Ruíz, M.; Domínguez, L. Antimicrobial Resistance in the Food Chain in the European Union. Adv. Food Nutr. Res. 2018, 86, 115–136. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2020/2021. EFSA J. 2023, 21, e07867. [Google Scholar] [CrossRef]
- WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Im-Portance to Guide Research, Development and Strategies to Prevent and Control Antimicro-Bial Resistance; World Health Organization: Geneva, Switzerland, 2024.
- Food Processing Sector Report—Poland USDA’S Global Agriculture Information Network (GAIN), PL2022-0028. 2023. Available online: https://apps.fas.usda.gov/newgainapi/api/Report/DownloadReportByFileName?fileName=Food%20Processing%20Sector%20Report_Warsaw_Poland_PL2022-0028.pdf (accessed on 5 August 2024).
- Volkov, S. WHO List of Medically Important Antimicrobials A Risk Management Tool for Mitigating Antimicrobial Resistance Due to Non-Human Use. 2024. Available online: https://cdn.who.int/media/docs/default-source/gcp/who-mia-list-2024-lv.pdf?sfvrsn=3320dd3d_2 (accessed on 5 August 2024).
- European Medicines Agency. Categorisation of Antibiotics for Use in Animals. Available online: https://www.ema.europa.eu/en/documents/report/infographic-categorisation-antibiotics-use-animals-prudent-and-responsible-use_en.pdf (accessed on 5 August 2024).
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4, 481–511. [Google Scholar] [CrossRef]
- Van Der Steen, M.; Leenstra, T.; Kluytmans, J.A.J.W.; Van Der Bij, A.K. Trends in expanded-spectrum cephalosporin-resistant Escherichia coli and Klebsiella pneumoniae among Dutch clinical isolates, from 2008 to 2012. PLoS ONE 2015, 10, e0138088. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bush, K. Past and Present Perspective on B-Lactamase Inhibitors. Antimicrob. Agents Chemother. 2018, 62, e01076-18. [Google Scholar] [CrossRef] [PubMed]
- Tamma, P.D.; Doi, Y.; Bonomo, R.A.; Johnson, J.K.; Simner, P.J. A Primer on AmpC β-Lactamases: Necessary Knowledge for an Increasingly Multidrug-Resistant World; Oxford University Press: Oxford, UK, 2019; Volume 69, pp. 1446–1455. [Google Scholar] [CrossRef]
- Irrgang, A.; Roschanski, N.; Brenner Michael, G.; Hamprecht, A.; Rieber, H.; Käsbohrer, A.; Schwarz, S.; Rösler, U.; Kreienbrock, L.; Pfeifer, Y.; et al. Whole genome analyses of CMY-2-producing Escherichia coli isolates from humans, animals and food in Germany. BMC Genom. 2018, 19, 601. [Google Scholar] [CrossRef]
- Lalak, A.; Wasyl, D.; Zając, M.; Skarżyńska, M.; Hoszowski, A.; Samcik, I.; Woźniakowski, G.; Szulowski, K. Mechanisms of cephalosporin resistance in indicator Escherichia coli isolated from food animals. Vet. Microbiol. 2016, 194, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Wasyl, D.; Zając, M.; Skarżyńska, M. Simple and efficient screening method for detection of cephalosporin resistant Escherichia coli. Bull. Vet. Inst. Pulawy 2010, 54, 147–151. [Google Scholar]
- de Been, M.; Lanza, V.F.; de Toro, M.; Scharringa, J.; Dohmen, W.; Du, Y.; Hu, J.; Lei, Y.; Li, N.; Tooming-Klunderud, A.; et al. Dissemination of Cephalosporin Resistance Genes between Escherichia coli Strains from Farm Animals and Humans by Specific Plasmid Lineages. PLoS Genet. 2014, 10, e1004776. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, D.; Kant, A.; Van Essen-Zandbergen, A.; Dierikx, C.; Hordijk, J.; Wit, B.; Mevius, D.J.; Veldman, K.T. Diversity of plasmids and genes encoding resistance to extended spectrum cephalosporins in commensal Escherichia coli From Dutch livestock in 2007–2017. Front. Microbiol. 2019, 10, 76. [Google Scholar] [CrossRef]
- Braz, V.S.; Melchior, K.; Moreira, C.G. Escherichia coli as a Multifaceted Pathogenic and Versatile Bacterium. Front. Cell. Infect. Microbiol. 2020, 10, 548492. [Google Scholar] [CrossRef]
- Denamur, E.; Clermont, O.; Bonacorsi, S.; Gordon, D. The population genetics of pathogenic Escherichia coli. Nat. Res. 2021, 19, 37–54. [Google Scholar] [CrossRef]
- Ranasinghe, R.A.S.S.; Satharasinghe, D.A.; Anwarama, P.S.; Parakatawella, P.M.S.D.K.; Jayasooriya, L.J.P.A.P.; Ranasinghe, R.M.S.B.K.; Rajapakse, R.P.V.J.; Huat, J.T.Y.; Rukayadi, Y.; Nakaguchi, Y.; et al. Prevalence and Antimicrobial Resistance of Escherichia coli in Chicken Meat and Edible Poultry Organs Collected from Retail Shops and Supermarkets of North Western Province in Sri Lanka. J. Food Qual. 2022, 2022, 8962698. [Google Scholar] [CrossRef]
- Omer, M.K.; Álvarez-Ordoñez, A.; Prieto, M.; Skjerve, E.; Asehun, T.; Alvseike, O.A. A Systematic Review of Bacterial Foodborne Outbreaks Related to Red Meat and Meat Products. Foodborne Pathog. Dis. 2018, 15, 598–611. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, B.; Rood, J.; Singer, E. BBMerge—Accurate paired shotgun read merging via overlap. PLoS ONE 2017, 12, e0185056. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.; Korobeynikov, A.; Lapidus, A.; Prjibelsky, A.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling Genomes and Mini-metagenomes from Highly Chimeric Reads. In Research in Computational Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Clausen, P.T.L.C.; Aarestrup, F.M.; Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinform. 2018, 19, 307. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Alikhan, N.F.; Mohamed, K.; Fan, Y.; Achtman, M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Ingle, D.J.; Valcanis, M.; Kuzevski, A.; Tauschek, M.; Inouye, M.; Stinear, T.; Levine, M.M.; Robins-Browne, R.M.; Holt, K.E. In silico serotyping of E. Coli from short read data identifies limited novel o-loci but extensive diversity of O:H serotype combinations within and between pathogenic lineages. Microb. Genom. 2016, 2, e000064. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Johansson, M.H.K.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genom. 2021, 7, 000685. [Google Scholar] [CrossRef] [PubMed]
- Soncini, J.G.M.; Cerdeira, L.; Sano, E.; Koga, V.L.; Tizura, A.T.; Tano, Z.N.; Nakazato, G.; Kobayashi, R.K.T.; Aires, C.A.M.; Lincopan, N.; et al. Genomic insights of high-risk clones of ESBL-producing Escherichia coli isolated from community infections and commercial meat in southern Brazil. Sci. Rep. 2022, 12, 9354. [Google Scholar] [CrossRef]
- Belas, A.; Marques, C.; Menezes, J.; Da Gama, L.T.; Cavaco-Silva, P.; Pomba, C. ESBL/pAmpC-Producing Escherichia coli Causing Urinary Tract Infections in Non-Related Companion Animals and Humans. Antibiotics 2022, 11, 559. [Google Scholar] [CrossRef]
- Cameron, A.; Mangat, R.; Mostafa, H.H.; Taffner, S.; Wang, J.; Dumyati, G.; Stanton, R.A.; Daniels, J.B.; Campbell, D.; Lutgring, J.D.; et al. Detection of CTX-M-27 b-lactamase genes on two distinct plasmid types in ST38 Escherichia coli from three U.S. States. Antimicrob. Agents Chemother. 2021, 65, e00825-21. [Google Scholar] [CrossRef]
- Antimicrobial consumption and resistance in bacteria from humans and food-producing animals: Fourth joint inter-agency report on integrated analysis of antimicrobial agent consumption and occurrence of antimicrobial resistance in bacteria from humans and food-producing animals in the EU/EEA JIACRA IV—2019–2021. EFSA J. 2024, 22, 1–141. [CrossRef]
- Irrgang, A.; Falgenhauer, L.; Fischer, J.; Ghosh, H.; Guiral, E.; Guerra, B.; Schmoger, S.; Imirzalioglu, C.; Chakraborty, T.; Hammerl, J.A.; et al. CTX-M-15-producing E. coli isolates from food products in Germany are mainly associated with an IncF-type plasmid and belong to two predominant clonal E. coli lineages. Front. Microbiol. 2017, 8, 2318. [Google Scholar] [CrossRef]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-Spectrum β-Lactamases: An Update on Their Characteristics, Epidemiology and Detection; Oxford University Press: Oxford, UK, 2021. [Google Scholar] [CrossRef]
- Bevan, E.R.; Jones, A.M.; Hawkey, P.M. Global epidemiology of CTX-M β-lactamases: Temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 2017, 72, 2145–2155. [Google Scholar] [CrossRef] [PubMed]
- Birgy, A.; Levy, C.; Nicolas-Chanoine, M.H.; Cointe, A.; Hobson, C.A.; Magnan, M.; Bechet, S.; Bidet, P.; Cohen, R.; Bonacorsi, S. Independent host factors and bacterial genetic determinants of the emergence and dominance of Escherichia coli sequence type 131 CTX-M-27 in a community pediatric cohort study. Antimicrob. Agents Chemother. 2019, 63, e00382-19. [Google Scholar] [CrossRef]
- Matsumura, Y.; Yamamoto, M.; Nagao, M.; Komori, T.; Fujita, N.; Hayashi, A.; Shimizu, T.; Watanabe, H.; Doi, S.; Tanaka, M.; et al. Multicenter retrospective study of cefmetazole and flomoxef for treatment of extended-spectrum-β-lactamase-producing Escherichia coli bacteremia. Antimicrob. Agents Chemother. 2015, 59, 5107–5113. [Google Scholar] [CrossRef] [PubMed]
- Lupo, A.; Saras, E.; Madec, J.Y.; Haenni, M. Emergence of bla CTX-M-55 associated with fosA, rmtB and mcr gene variants in Escherichia coli from various animal species in France. J. Antimicrob. Chemother. 2018, 73, 867–872. [Google Scholar] [CrossRef] [PubMed]
- Bourély, C.; Coeffic, T.; Caillon, J.; Thibaut, S.; Cazeau, G.; Jouy, E.; Jarrige, N.; Chauvin, C.; Madec, J.Y.; Haenni, M.; et al. Trends in antimicrobial resistance among Escherichia coli from defined infections in humans and animals. J. Antimicrob. Chemother. 2021, 75, 1525–1529. [Google Scholar] [CrossRef]
- Koga, V.L.; Maluta, R.P.; Da Silveira, W.D.; Ribeiro, R.A.; Hungria, M.; Vespero, E.C.; Nakazato, G.; Kobayashi, R.K.T. Characterization of CMY-2-type beta-lactamase-producing Escherichia coli isolated from chicken carcasses and human infection in a city of South Brazil. BMC Microbiol. 2019, 19, 174. [Google Scholar] [CrossRef]
- Carattoli, A. Plasmids and the spread of resistance. Int. J. Med. Microbiol. 2013, 303, 298–304. [Google Scholar] [CrossRef]
- Hammerum, A.M.; Larsen, J.; Andersen, V.D.; Lester, C.H.; Skytte, T.S.S.; Hansen, F.; Olsen, S.S.; Mordhorst, H.; Skov, R.L.; Aarestrup, F.M.; et al. Characterization of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli obtained from Danish pigs, pig farmers and their families from farms with high or no consumption of third- or fourth-generation cephalosporins. J. Antimicrob. Chemother. 2014, 69, 2650–2657. [Google Scholar] [CrossRef]
- Abraham, S.; Kirkwood, R.N.; Laird, T.; Saputra, S.; Mitchell, T.; Singh, M.; Linn, B.; Abraham, R.J.; Pang, S.; Gordon, D.M.; et al. Dissemination and persistence of extended-spectrum cephalosporin-resistance encoding IncI1-blaCTXM-1 plasmid among Escherichia coli in pigs. ISME J. 2018, 12, 2352–2362. [Google Scholar] [CrossRef] [PubMed]
- Rozwandowicz, M.; Brouwer, M.S.M.; Fischer, J.; Wagenaar, J.A.; Gonzalez-Zorn, B.; Guerra, B.; Mevius, D.J.; Hordijk, J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 1121–1137. [Google Scholar] [CrossRef]
- Liakopoulos, A.; Van Der Goot, J.; Bossers, A.l.; Betts, J.; Brouwer, M.S.M.; Kant, A.; Smith, H.; Ceccarelli, D.; Mevius, D. Genomic and functional characterisation of IncX3 plasmids encoding blaSHV-12 in Escherichia coli from human and animal origin. Sci. Rep. 2018, 8, 7674. [Google Scholar] [CrossRef]
- Alonso, C.A.; Michael, G.B.; Li, J.; Somalo, S.; Simón, C.; Wang, Y.; Kaspar, H.; Kadlec, K.; Torres, C.; Schwarz, S. Analysis of blaSHV-12-carrying Escherichia coli clones and plasmids from human, animal and food sources. J. Antimicrob. Chemother. 2017, 72, 1589–1596. [Google Scholar] [CrossRef]
- Saliu, E.M.; Vahjen, W.; Zentek, J. Types and Prevalence of Extended-Spectrum Beta-Lactamase Producing Enterobacteriaceae in Poultry; Cambridge University Press: Cambridge, UK, 2017. [Google Scholar] [CrossRef]
- Poirel, L.; Decousser, J.W.; Nordmann, P. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M β-lactamase gene. Antimicrob. Agents Chemother. 2003, 47, 2938–2945. [Google Scholar] [CrossRef]
- Aldea, I.; Gibello, A.; Hernández, M.; Leekitcharoenphon, P.; Bortolaia, V.; Moreno, M.A. Clonal and plasmid-mediated flow of ESBL/AmpC genes in Escherichia coli in a commercial laying hen farm. Vet. Microbiol. 2022, 270, 109453. [Google Scholar] [CrossRef] [PubMed]
- Kurittu, P.; Khakipoor, B.; Brouwer, M.S.M.; Heikinheimo, A. Plasmids conferring resistance to extended-spectrum beta-lactamases including a rare IncN+IncR multireplicon carrying blaCTX-M-1 in Escherichia coli recovered from migrating barnacle geese (Branta leucopsis). Open Res. Eur. 2021, 1, 46. [Google Scholar] [CrossRef] [PubMed]
- Dobiasova, H.; Dolejska, M. Prevalence and diversity of IncX plasmids carrying fluoroquinolone and β-lactam resistance genes in Escherichia coli originating from diverse sources and geographical areas. J. Antimicrob. Chemother. 2016, 71, 2118–2124. [Google Scholar] [CrossRef]
- Nohejl, T.; Valcek, A.; Papousek, I.; Palkovicova, J.; Wailan, A.M.; Pratova, H.; Minoia, M.; Dolejska, M. Genomic analysis of qnr-harbouring IncX plasmids and their transferability within different hosts under induced stress. BMC Microbiol. 2022, 22, 136. [Google Scholar] [CrossRef]
- Abgottspon, H.; Stephan, R.; Bagutti, C.; Brodmann, P.; Hächler, H.; Zurfluh, K. Characteristics of extended-spectrum cephalosporin-resistant Escherichia coli isolated from Swiss and imported poultry meat. J. Food Prot. 2014, 77, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Shirakawa, R.; Ishikawa, K.; Furuta, K.; Kaito, C. Knockout of ribosomal protein RpmJ leads to zinc resistance in Escherichia coli. PLoS ONE 2023, 18, e0277162. [Google Scholar] [CrossRef] [PubMed]
- Salinas, L.; Cárdenas, P.; Graham, J.P.; Trueba, G. IS 26 drives the dissemination of bla CTX-M genes in an Ecuadorian community. Microbiol Spectr. 2024, 12, e02504-23. [Google Scholar] [CrossRef]
- Pires, J.; Huisman, J.S.; Bonhoeffer, S.; Van Boeckel, T.P. Increase in antimicrobial resistance in Escherichia coli in food animals between 1980 and 2018 assessed using genomes from public databases. J. Antimicrob. Chemother. 2022, 77, 646–655. [Google Scholar] [CrossRef]
- Woegerbauer, M.; Kuffner, M.; Domingues, S.; Nielsen, K.M. Involvement of aph(3′)-IIa in the formation of mosaic aminoglycoside resistance genes in natural environments. Front. Microbiol. 2015, 6, 442. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Poirel, L. Plasmid-mediated colistin resistance: An additional antibiotic resistance menace. Clin. Microbiol. Infect. 2016, 22, 398–400. [Google Scholar] [CrossRef]
- Matamoros, S.; Van Hattem, J.M.; Arcilla, M.S.; Willemse, N.; Melles, D.C.; Penders, J.; Vinh, T.N.; Thi Hoa, N.; Bootsma, M.C.J.; Van Genderen, P.J.; et al. Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci. Rep. 2017, 7, 15364. [Google Scholar] [CrossRef]
- Zając, M.; Sztromwasser, P.; Bortolaia, V.; Leekitcharoenphon, P.; Cavaco, L.M.; Ziȩtek-Barszcz, A.; Hendriksen, R.S.; Wasyl, D. Occurrence and Characterization of mcr-1-Positive Escherichia coli Isolated From Food-Producing Animals in Poland, 2011–2016. Front. Microbiol. 2019, 10, 1753. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Wang, Y.; Shi, X.; Wang, S.; Ren, H.; Shen, Z.; Wang, Y.; Lin, J.; Wang, S. Rapid rise of the ESBL and mcr-1 genes in Escherichia coli of chicken origin in China, 2008–2014. Emerg Microbes Infect. 2018, 7, 30. [Google Scholar] [CrossRef] [PubMed]
- Cameron, E.A.; Sperandio, V.; Dunny, G.M. Enterococcus faecalis enhances expression and activity of the enterohemorrhagic Escherichia coli type III secretion system. mBio 2019, 10, e02547-19. [Google Scholar] [CrossRef]
- Sarowska, J.; Futoma-Koloch, B.; Jama-Kmiecik, A.; Frej-Madrzak, M.; Ksiazczyk, M.; Bugla-Ploskonska, G.; Choroszy-Krol, I. Virulence Factors, Prevalence and Potential Transmission of Extraintestinal Pathogenic Escherichia coli Isolated from Different Sources: Recent Reports; BioMed Central Ltd.: London, UK, 2019; Volume 21. [Google Scholar] [CrossRef]
- Hüttener, M.; Dietrich, M.; Paytubi, S.; Juárez, A. HilA-like regulators in Escherichia coli pathotypes: The YgeH protein from the enteroaggregative strain 042. BMC Microbiol. 2014, 14, 268. [Google Scholar] [CrossRef]
- Christenson, J.K.; Gordon, D.M. Evolution of Colicin BM plasmids: The loss of the Colicin B activity gene. Microbiology 2009, 155, 1645–1655. [Google Scholar] [CrossRef][Green Version]
- Noll, L.W.; Worley, J.N.; Yang, X.; Shridhar, P.B.; Ludwig, J.B.; Shi, X.; Bai, J.; Caragea, D.; Meng, J.; Nagaraja, T.G. Comparative genomics reveals differences in mobile virulence genes of Escherichia coli O103 pathotypes of bovine fecal origin. PLoS ONE 2018, 13, e0191362. [Google Scholar] [CrossRef]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L.T. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef]
- Shridhar, P.B.; Patel, I.R.; Gangiredla, J.; Noll, L.W.; Shi, X.; Bai, J.; Elkins, C.A.; Strockbine, N.A.; Nagaraja, T.G. Genetic analysis of virulence potential of Escherichia coli O104 serotypes isolated from cattle feces using whole genome sequencing. Front. Microbiol. 2018, 9, 341. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.D.; Chen, L. The Significance of Epidemic Plasmids in the Success of Multidrug-Resistant Drug Pandemic Extraintestinal Pathogenic Escherichia coli. Infect. Dis. Ther. 2023, 12, 1029–1041. [Google Scholar] [CrossRef] [PubMed]
- Toro, M.; Rump, L.V.; Cao, G.; Meng, J.; Brown, E.W.; Gonzalez-Escalona, N. Simultaneous presence of insertion sequence excision enhancer and insertion sequence is629 correlates with increased diversity and virulence in Shiga toxin-producing Escherichia coli. J. Clin. Microbiol. 2015, 53, 3466–3473. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iwan, E.; Zając, M.; Bomba, A.; Olejnik, M.; Skarżyńska, M.; Wasiński, B.; Wieczorek, K.; Tłuścik, K.; Wasyl, D. Phylogenetics and Mobilization of Genomic Traits of Cephalosporin-Resistant Escherichia coli Originated from Retail Meat. Pathogens 2024, 13, 700. https://doi.org/10.3390/pathogens13080700
Iwan E, Zając M, Bomba A, Olejnik M, Skarżyńska M, Wasiński B, Wieczorek K, Tłuścik K, Wasyl D. Phylogenetics and Mobilization of Genomic Traits of Cephalosporin-Resistant Escherichia coli Originated from Retail Meat. Pathogens. 2024; 13(8):700. https://doi.org/10.3390/pathogens13080700
Chicago/Turabian StyleIwan, Ewelina, Magdalena Zając, Arkadiusz Bomba, Małgorzata Olejnik, Magdalena Skarżyńska, Bernard Wasiński, Kinga Wieczorek, Katarzyna Tłuścik, and Dariusz Wasyl. 2024. "Phylogenetics and Mobilization of Genomic Traits of Cephalosporin-Resistant Escherichia coli Originated from Retail Meat" Pathogens 13, no. 8: 700. https://doi.org/10.3390/pathogens13080700
APA StyleIwan, E., Zając, M., Bomba, A., Olejnik, M., Skarżyńska, M., Wasiński, B., Wieczorek, K., Tłuścik, K., & Wasyl, D. (2024). Phylogenetics and Mobilization of Genomic Traits of Cephalosporin-Resistant Escherichia coli Originated from Retail Meat. Pathogens, 13(8), 700. https://doi.org/10.3390/pathogens13080700






