In the Depths of Wash Water: Isolation of Opportunistic Bacteria from Fresh-Cut Processing Plants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Characteristics of Fresh-Cut Processing Plants
2.2. Wash Water Sampling
2.3. Species Identification
2.4. Evaluation of Phenotypic Antibiotic Susceptibility of Selected Strains
3. Results
3.1. Species Identification
3.2. Antibiotic Susceptibility
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lehto, M.; Sipilä, I.; Alakukku, L.; Kymäläinen, H.-R. Water Consumption and Wastewaters in Fresh-Cut Vegetable Production. AFSci 2014, 23, 246–256. [Google Scholar] [CrossRef]
- Raffo, A.; Paoletti, F. Fresh-Cut Vegetables Processing: Environmental Sustainability and Food Safety Issues in a Comprehensive Perspective. Front. Sustain. Food Syst. 2022, 5, 681459. [Google Scholar] [CrossRef]
- Manzocco, L.; Ignat, A.; Anese, M.; Bot, F.; Calligaris, S.; Valoppi, F.; Nicoli, M.C. Efficient Management of the Water Resource in the Fresh-Cut Industry: Current Status and Perspectives. Trends Food Sci. Technol. 2015, 46, 286–294. [Google Scholar] [CrossRef]
- Nahim-Granados, S.; Rivas-Ibáñez, G.; Antonio Sánchez Pérez, J.; Oller, I.; Malato, S.; Polo-López, M.I. Synthetic Fresh-Cut Wastewater Disinfection and Decontamination by Ozonation at Pilot Scale. Water Res. 2020, 170, 115304. [Google Scholar] [CrossRef] [PubMed]
- López-Gálvez, F.; Allende, A.; Gil, M.I. Recent Progress on the Management of the Industrial Washing of Fresh Produce with a Focus on Microbiological Risks. Curr. Opin. Food Sci. 2021, 38, 46–51. [Google Scholar] [CrossRef]
- Balali, G.I.; Yar, D.D.; Afua Dela, V.G.; Adjei-Kusi, P. Microbial Contamination, an Increasing Threat to the Consumption of Fresh Fruits and Vegetables in Today’s World. Int. J. Microbiol. 2020, 2020, e3029295. [Google Scholar] [CrossRef]
- Kanarek, P.; Breza-Boruta, B.; Poćwiardowski, W.; Szulc, J. Sustainable Water Use in a Fruit Processing Plant: Evaluation of Microbiological and Physicochemical Properties of Wash Water after Application of a Modular Water Recovery System. Sustainability 2024, 16, 2181. [Google Scholar] [CrossRef]
- Gil, M.I.; Selma, M.V.; Suslow, T.; Jacxsens, L.; Uyttendaele, M.; Allende, A. Pre- and Postharvest Preventive Measures and Intervention Strategies to Control Microbial Food Safety Hazards of Fresh Leafy Vegetables. Crit. Rev. Food Sci. Nutr. 2015, 55, 453–468. [Google Scholar] [CrossRef]
- Kanarek, P.; Breza-Boruta, B.; Rolbiecki, R. Microbial Composition and Formation of Biofilms in Agricultural Irrigation Systems- a Review. Ecohydrol. Hydrobiol. 2024, 24, 583–590. [Google Scholar] [CrossRef]
- Gu, G.; Luo, Z.; Cevallos-Cevallos, J.M.; Adams, P.; Vellidis, G.; Wright, A.; van Bruggen, A.H.C. Occurrence and Population Density of Campylobacter Jejuni in Irrigation Ponds on Produce Farms in the Suwannee River Watershed. Can. J. Microbiol. 2013, 59, 339–346. [Google Scholar] [CrossRef]
- Kovačić, A.; Huljev, Ž.; Sušić, E. Ground Water as the Source of an Outbreak of Salmonella Enteritidis. J. Epidemiol. Glob. Health 2017, 7, 181–184. [Google Scholar] [CrossRef]
- Gartley, S.; Anderson-Coughlin, B.; Sharma, M.; Kniel, K.E. Listeria Monocytogenes in Irrigation Water: An Assessment of Outbreaks, Sources, Prevalence, and Persistence. Microorganisms 2022, 10, 1319. [Google Scholar] [CrossRef]
- Blaustein, R.A.; Shelton, D.R.; Van Kessel, J.A.S.; Karns, J.S.; Stocker, M.D.; Pachepsky, Y.A. Irrigation Waters and Pipe-Based Biofilms as Sources for Antibiotic-Resistant Bacteria. Environ. Monit. Assess. 2016, 188, 56. [Google Scholar] [CrossRef]
- Black, Z.; Balta, I.; Black, L.; Naughton, P.J.; Dooley, J.S.G.; Corcionivoschi, N. The Fate of Foodborne Pathogens in Manure Treated Soil. Front. Microbiol. 2021, 12, 781357. [Google Scholar] [CrossRef]
- Oh, S.-S.; Han, S.-J.; Gong, Y.-W.; Nam, H.-J.; Kim, K.-A.; Kim, N.-Y.; Kim, K.-S.; Lee, D.-G.; Shin, H.-K.; Lee, H.-B.; et al. Investigation of Pathogenic Escherichia Coli Contamination of Vegetables Distributed in a Korean Agricultural Wholesale Market. Foodborne Pathog. Dis. 2021, 18, 887–893. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Szonyi, B.; Gautam, R.; Nightingale, K.; Anciso, J.; Ivanek, R. Risk Factors for Microbial Contamination in Fruits and Vegetables at the Preharvest Level: A Systematic Review. J. Food Prot. 2012, 75, 2055–2081. [Google Scholar] [CrossRef] [PubMed]
- Machado, D.C.; Maia, C.M.; Carvalho, I.D.; Silva, N.F.D.; André, M.C.D.P.B.; Serafini, Á.B. Microbiological Quality of Organic Vegetables Produced in Soil Treated with Different Types of Manure and Mineral Fertilizer. Braz. J. Microbiol. 2006, 37, 538–544. [Google Scholar] [CrossRef]
- Yang, Y.; Luo, Y.; Millner, P.; Turner, E.; Feng, H. Assessment of Escherichia Coli O157:H7 Transference from Soil to Iceberg Lettuce via a Contaminated Field Coring Harvesting Knife. Int. J. Food Microbiol. 2012, 153, 345–350. [Google Scholar] [CrossRef]
- Mostafidi, M.; Sanjabi, M.R.; Shirkhan, F.; Zahedi, M.T. A Review of Recent Trends in the Development of the Microbial Safety of Fruits and Vegetables. Trends Food Sci. Technol. 2020, 103, 321–332. [Google Scholar] [CrossRef]
- Kusstatscher, P.; Cernava, T.; Abdelfattah, A.; Gokul, J.; Korsten, L.; Berg, G. Microbiome Approaches Provide the Key to Biologically Control Postharvest Pathogens and Storability of Fruits and Vegetables. FEMS Microbiol. Ecol. 2020, 96, fiaa119. [Google Scholar] [CrossRef]
- Sarno, E.; Pezzutto, D.; Rossi, M.; Liebana, E.; Rizzi, V. A Review of Significant European Foodborne Outbreaks in the Last Decade. J. Food Prot. 2021, 84, 2059–2070. [Google Scholar] [CrossRef] [PubMed]
- Martinović, T.; Andjelković, U.; Gajdošik, M.Š.; Rešetar, D.; Josić, D. Foodborne Pathogens and Their Toxins. J. Proteom. 2016, 147, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Schirone, M.; Visciano, P. Trends of Major Foodborne Outbreaks in the European Union during the Years 2015–2019. Hygiene 2021, 1, 106–119. [Google Scholar] [CrossRef]
- Gourama, H. Foodborne Pathogens. In Food Safety Engineering; Food Engineering Series; Demirci, A., Feng, H., Krishnamurthy, K., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 25–49. ISBN 978-3-030-42660-6. [Google Scholar]
- Morgado, M.E.; Jiang, C.; Zambrana, J.; Upperman, C.R.; Mitchell, C.; Boyle, M.; Sapkota, A.R.; Sapkota, A. Climate Change, Extreme Events, and Increased Risk of Salmonellosis: Foodborne Diseases Active Surveillance Network (FoodNet), 2004–2014. Environ. Health 2021, 20, 105. [Google Scholar] [CrossRef]
- Kunhikannan, S.; Thomas, C.J.; Franks, A.E.; Mahadevaiah, S.; Kumar, S.; Petrovski, S. Environmental Hotspots for Antibiotic Resistance Genes. MicrobiologyOpen 2021, 10, e1197. [Google Scholar] [CrossRef]
- Larsson, D.G.J.; Flach, C.-F. Antibiotic Resistance in the Environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef]
- Denissen, J.; Reyneke, B.; Waso-Reyneke, M.; Havenga, B.; Barnard, T.; Khan, S.; Khan, W. Prevalence of ESKAPE Pathogens in the Environment: Antibiotic Resistance Status, Community-Acquired Infection and Risk to Human Health. Int. J. Hyg. Environ. Health 2022, 244, 114006. [Google Scholar] [CrossRef]
- Reyes, J.; Komarow, L.; Chen, L.; Ge, L.; Hanson, B.M.; Cober, E.; Herc, E.; Alenazi, T.; Kaye, K.S.; Garcia-Diaz, J.; et al. Global Epidemiology and Clinical Outcomes of Carbapenem-Resistant Pseudomonas Aeruginosa and Associated Carbapenemases (POP): A Prospective Cohort Study. Lancet Microbe 2023, 4, e159–e170. [Google Scholar] [CrossRef]
- Tiwari, A.; Krolicka, A.; Tran, T.T.; Räisänen, K.; Ásmundsdóttir, Á.M.; Wikmark, O.-G.; Lood, R.; Pitkänen, T. Antibiotic Resistance Monitoring in Wastewater in the Nordic Countries: A Systematic Review. Environ. Res. 2024, 246, 118052. [Google Scholar] [CrossRef]
- Sun, S.; Wang, Q.; Wang, N.; Yang, S.; Qi, H. High-Risk Antibiotics Positively Correlated with Antibiotic Resistance Genes in Five Typical Urban Wastewater. J. Environ. Manag. 2023, 342, 118296. [Google Scholar] [CrossRef]
- PN-EN ISO 19458:2007; Water Quality—Sampling for Microbiological Analyses. Polish Committee for Standardisation: Warsaw, Poland, 2007.
- EUCAST The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 14.0; EUCAST: Madrid, Spain, 2024; Available online: https://www.eucast.org/ (accessed on 6 August 2024).
- Weng, S.-C.; Jacangelo, J.G.; Schwab, K.J. Sustainable Practice for the Food Industry: Assessment of Selected Treatment Options for Reclamation of Washwater from Vegetable Processing. Int. J. Environ. Sci. Technol. 2019, 16, 1369–1378. [Google Scholar] [CrossRef]
- Barth, M.; Hankinson, T.R.; Zhuang, H.; Breidt, F. Microbiological Spoilage of Fruits and Vegetables. In Compendium of the Microbiological Spoilage of Foods and Beverages; Sperber, W.H., Doyle, M.P., Eds.; Springer: New York, NY, USA, 2009; pp. 135–183. ISBN 978-1-4419-0825-4. [Google Scholar]
- Ölmez, H.; Kretzschmar, U. Potential Alternative Disinfection Methods for Organic Fresh-Cut Industry for Minimizing Water Consumption and Environmental Impact. LWT-Food Sci. Technol. 2009, 42, 686–693. [Google Scholar] [CrossRef]
- Zhou, B.; Luo, Y.; Teng, Z.; Nou, X.; Millner, P. Factors Impacting Chemical and Microbiological Quality of Wash Water during Simulated Dump Tank Wash of Grape Tomatoes. J. Food Prot. 2021, 84, 695–703. [Google Scholar] [CrossRef]
- Liu, N.T.; Lefcourt, A.M.; Nou, X.; Shelton, D.R.; Zhang, G.; Lo, Y.M. Native Microflora in Fresh-Cut Produce Processing Plants and Their Potentials for Biofilm Formation. J. Food Prot. 2013, 76, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhao, X.; Ma, Y.; Ma, Z.; He, Z.; Zhao, W.; Wang, P.; Zhao, S.; Wang, D. Investigation on the Microbial Diversity of Fresh-Cut Lettuce during Processing and Storage Using High Throughput Sequencing and Their Relationship with Quality. Foods 2022, 11, 1683. [Google Scholar] [CrossRef]
- Leroy, S.; Lebert, I.; Chacornac, J.-P.; Chavant, P.; Bernardi, T.; Talon, R. Genetic Diversity and Biofilm Formation of Staphylococcus equorum Isolated from Naturally Fermented Sausages and Their Manufacturing Environment. Int. J. Food Microbiol. 2009, 134, 46–51. [Google Scholar] [CrossRef]
- Chajęcka-Wierzchowska, W.; Gajewska, J.; Wiśniewski, P.; Zadernowska, A. Enterotoxigenic Potential of Coagulase-Negative Staphylococci from Ready-to-Eat Food. Pathogens 2020, 9, 734. [Google Scholar] [CrossRef]
- Meservey, A.; Sullivan, A.; Wu, C.; Lantos, P.M. Staphylococcus Sciuri Peritonitis in a Patient on Peritoneal Dialysis. Zoonoses Public Health 2020, 67, 93–95. [Google Scholar] [CrossRef]
- Santana, J.A.; Colombo, S.A.; Silva, B.A.; Diniz, A.N.; de Almeida, L.R.; Oliveira Junior, C.A.; Lobato, F.C.F.; de Souza Trindade, G.; Paglia, A.P.; Silva, R.O.S. Clostridioides Difficile and Multi-Drug-Resistant Staphylococci in Free-Living Rodents and Marsupials in Parks of Belo Horizonte, Brazil. Braz. J. Microbiol. 2022, 53, 401–410. [Google Scholar] [CrossRef]
- Pintor-Cora, A.; Álvaro-Llorente, L.; Otero, A.; Rodríguez-Calleja, J.M.; Santos, J.A. Extended-Spectrum Beta-Lactamase-Producing Enterobacteriaceae in Fresh Produce. Foods 2021, 10, 2609. [Google Scholar] [CrossRef]
- Wareth, G.; Neubauer, H. The Animal-Foods-Environment Interface of Klebsiella Pneumoniae in Germany: An Observational Study on Pathogenicity, Resistance Development and the Current Situation. Vet. Res. 2021, 52, 16. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Ferrer, S.; Peñaloza, H.F.; Budnick, J.A.; Bain, W.G.; Nordstrom, H.R.; Lee, J.S.; Van Tyne, D. Finding Order in the Chaos: Outstanding Questions in Klebsiella Pneumoniae Pathogenesis. Infect. Immun. 2021, 89, e00693-20. [Google Scholar] [CrossRef]
- Salem, M.S.E.; Mahfouz, A.Y.; Fathy, R.M. The Antibacterial and Antihemolytic Activities Assessment of Zinc Oxide Nanoparticles Synthesized Using Plant Extracts and Gamma Irradiation against the Uro-Pathogenic Multidrug Resistant Proteus Vulgaris. Biometals 2021, 34, 175–196. [Google Scholar] [CrossRef] [PubMed]
- Tavares-Carreon, F.; Anda-Mora, K.D.; Rojas-Barrera, I.C.; Andrade, A. Serratia Marcescens Antibiotic Resistance Mechanisms of an Opportunistic Pathogen: A Literature Review. PeerJ 2023, 11, e14399. [Google Scholar] [CrossRef] [PubMed]
- Staley, C.; Dunny, G.M.; Sadowsky, M.J. Chapter Four—Environmental and Animal-Associated Enterococci. In Advances in Applied Microbiology; Sariaslani, S., Gadd, G.M., Eds.; Academic Press: Cambridge, MA, USA, 2014; Volume 87, pp. 147–186. [Google Scholar]
- Kim, H.J.; Kim, S.M. Occurrence and antibiotic resistance of Enterococcus spp. from retail fresh-cut products in Korea. Korean J. Food Sci. Technol. 2018, 50, 581–586. [Google Scholar] [CrossRef]
- Xie, Y.; Nitin, N.; Harris, L.J. Przeniesienie Enterococcus faecium i Salmonella enterica Podczas Symulowanego Postępowania z Cebulą Żółtą Po Zbiorach (Allium cepa). Food Microbiol. 2023, 115, 104340. [Google Scholar] [CrossRef]
- Rudi, K.; Flateland, S.L.; Hanssen, J.F.; Bengtsson, G.; Nissen, H. Development and Evaluation of a 16S Ribosomal DNA Array-Based Approach for Describing Complex Microbial Communities in Ready-To-Eat Vegetable Salads Packed in a Modified Atmosphere. Appl. Environ. Microbiol. 2002, 68, 1146–1156. [Google Scholar] [CrossRef]
- Mulaosmanovic, E.; Lindblom, T.U.T.; Windstam, S.T.; Bengtsson, M.; Rosberg, A.K.; Mogren, L.; Alsanius, B.W. Processing of Leafy Vegetables Matters: Damage and Microbial Community Structure from Field to Bag. Food Control 2021, 125, 107894. [Google Scholar] [CrossRef]
- Abd-Alla, M.H.; Bashandy, S.R. Production of Quorum Sensing Inhibitors in Growing Onion Bulbs Infected with Pseudomonas Aeruginosa E (HQ324110). ISRN Microbiol. 2012, 2012, 161890. [Google Scholar] [CrossRef]
- Jurado-Martín, I.; Sainz-Mejías, M.; McClean, S. Pseudomonas Aeruginosa: An Audacious Pathogen with an Adaptable Arsenal of Virulence Factors. Int. J. Mol. Sci. 2021, 22, 3128. [Google Scholar] [CrossRef]
- Reynolds, D.; Kollef, M. The Epidemiology and Pathogenesis and Treatment of Pseudomonas Aeruginosa Infections: An Update. Drugs 2021, 81, 2117–2131. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Wang, H.; Pu, S.; Huang, S.; Niu, S. Resistance Trends of Klebsiella Pneumoniae Causing Urinary Tract Infections in Chongqing, 2011–2019. Infect. Drug Resist. 2021, 14, 475–481. [Google Scholar] [CrossRef]
- Okaiyeto, S.A.; Sutar, P.P.; Chen, C.; Ni, J.-B.; Wang, J.; Mujumdar, A.S.; Zhang, J.-S.; Xu, M.-Q.; Fang, X.-M.; Zhang, C.; et al. Antibiotic Resistant Bacteria in Food Systems: Current Status, Resistance Mechanisms, and Mitigation Strategies. Agric. Commun. 2024, 2, 100027. [Google Scholar] [CrossRef]
- Isler, B.; Harris, P.; Stewart, A.G.; Paterson, D.L. An Update on Cefepime and Its Future Role in Combination with Novel β-Lactamase Inhibitors for MDR Enterobacterales and Pseudomonas Aeruginosa. J. Antimicrob. Chemother. 2021, 76, 550–560. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Ucha, J.C.; Lasarte-Monterrubio, C.; Guijarro-Sánchez, P.; Oviaño, M.; Álvarez-Fraga, L.; Alonso-García, I.; Arca-Suárez, J.; Bou, G.; Beceiro, A. GEMARA-SEIMC/Informacje o autorach grupy badawczej REIPI Enterobacterales Assessment of Activity and Resistance Mechanisms to Cefepime in Combination with the Novel β-Lactamase Inhibitors Zidebactam, Taniborbactam, and Enmetazobactam against a Multicenter Collection of Carbapenemase-Producing Enterobacterales. Antimicrob. Agents Chemother. 2022, 66, e01676-21. [Google Scholar] [CrossRef]
- Wang, H.; Feng, Y.; Lu, H. Low-Level Cefepime Exposure Induces High-Level Resistance in Environmental Bacteria: Molecular Mechanism and Evolutionary Dynamics. Environ. Sci. Technol. 2022, 56, 15074–15083. [Google Scholar] [CrossRef]
- Salmanov, A.G.; Ushkalov, V.O.; Shunko, Y.Y.; Piven, N.; Vygovska, L.M.; Verner, O.M.; Kushnirenko, S. One health: Antibiotic-resistant bacteria contamination in fresh vegetables sold at a retail markets in Kyiv, Ukraine. Wiadomości Lek. 2021, 74, 83–89. [Google Scholar] [CrossRef]
- Saksena, R.; Malik, M.; Gaind, R. Bacterial Contamination and Prevalence of Antimicrobial Resistance Phenotypes in Raw Fruits and Vegetables Sold in Delhi, India. J. Food Saf. 2020, 40, e12739. [Google Scholar] [CrossRef]
- Tiedje, J.M.; Fu, Y.; Mei, Z.; Schäffer, A.; Dou, Q.; Amelung, W.; Elsner, M.; Adu-Gyamfi, J.; Heng, L.; Virta, M.; et al. Antibiotic Resistance Genes in Food Production Systems Support One Health Opinions. Curr. Opin. Environ. Sci. Health 2023, 34, 100492. [Google Scholar] [CrossRef]
- Allydice-Francis, K.; Brown, P.D. Diversity of Antimicrobial Resistance and Virulence Determinants in Pseudomonas Aeruginosa Associated with Fresh Vegetables. Int. J. Microbiol. 2012, 2012, 426241. [Google Scholar] [CrossRef]
- Oulahal, N.; Degraeve, P. Phenolic-Rich Plant Extracts With Antimicrobial Activity: An Alternative to Food Preservatives and Biocides? Front. Microbiol. 2022, 12, 753518. [Google Scholar] [CrossRef] [PubMed]

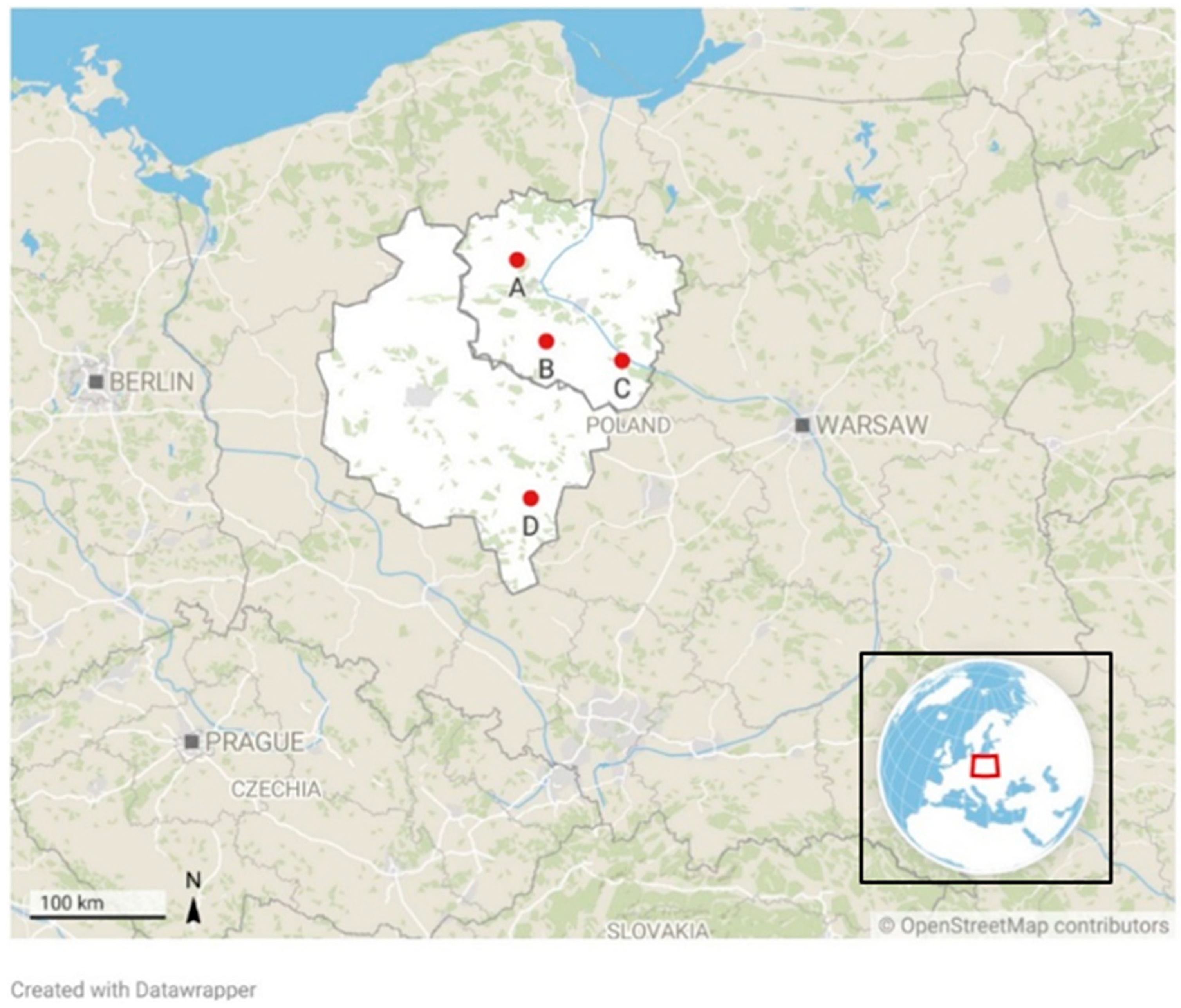
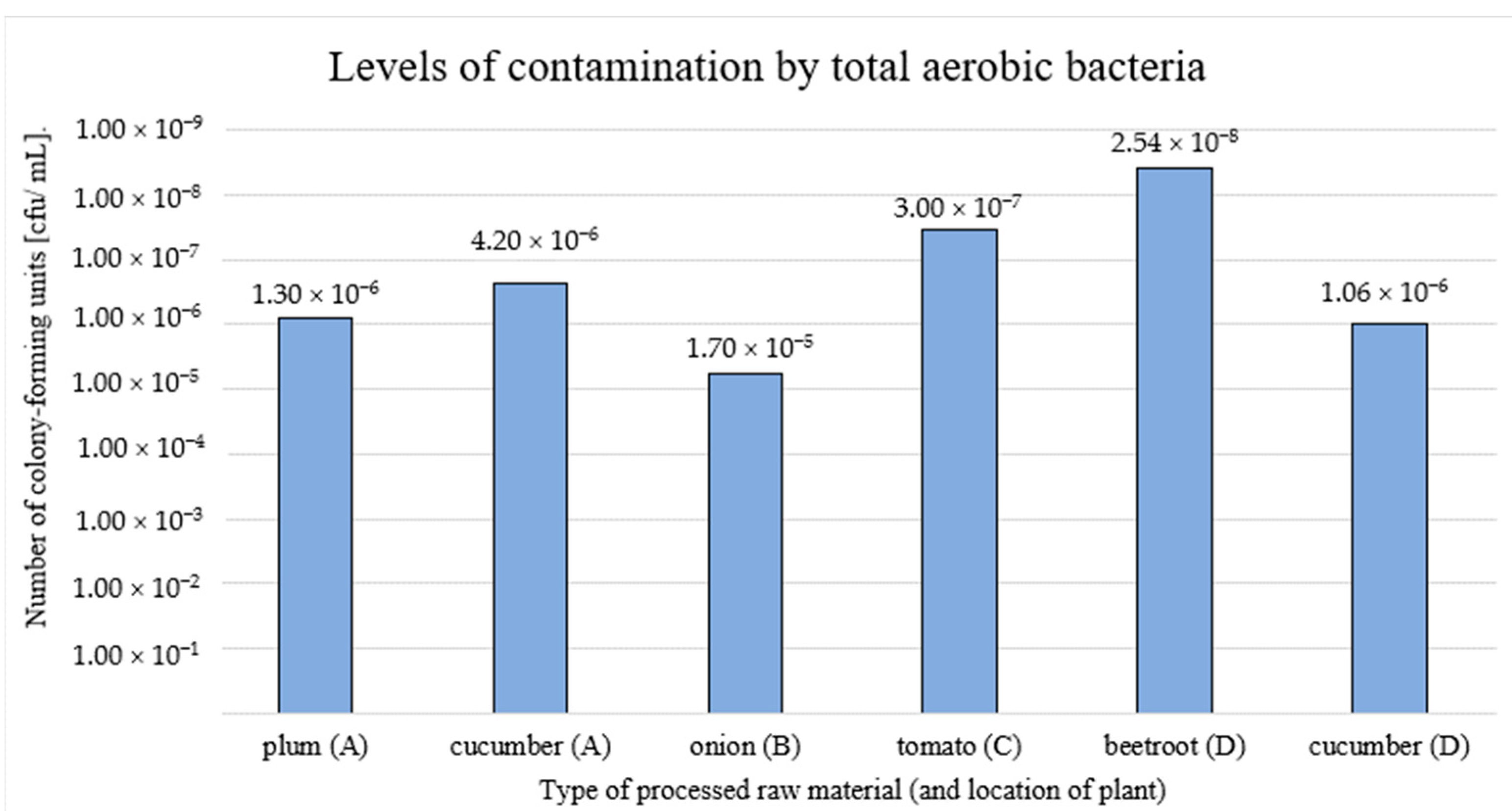
| Plant | Type of Processed Fruit/Vegetable | Type of Final Product |
|---|---|---|
| A | strawberries, raspberries, cherries, currants, rhubarb, plums, apples, tomatoes, cucumbers, leeks, broccoli, cauliflower | ready-to-eat fruits and vegetables, frozen products |
| B | onions | peeled onion, onion rings |
| C | tomatoes, apples | ready-to-eat fruits and vegetables, apple and tomato concentrate |
| D | beetroots, cucumbers, onions | ready-to-eat fruits and vegetables, pickled cucumbers, vegetable salads |
| No. | Location | Raw Material Type | Species |
|---|---|---|---|
| 1 | A | cucumber | Staphylococcus sciuri |
| 2 | A | cucumber | Micrococcus luteus |
| 3 | A | plum | Staphylococcus sciuri |
| 4 | A | cucumber | Lelliottia amnigena |
| 5 | A | cucumber | Enterococcus casseliflavus |
| 6 | A | cucumber | Comamonas testosteroni |
| 7 | B | onion | Enterobacter ludwigii |
| 8 | B | onion | Kerstersia gyiorum |
| 9 | B | onion | Citrobacter braakii |
| 10 | B | onion | Pseudomonas aeruginosa |
| 11 | B | onion | Enterococcus faecalis |
| 12 | B | onion | Klebsiella pneumoniae |
| 13 | C | tomato | Pediococcus pentosaceus |
| 14 | C | tomato | Enterobacter ludwigii |
| 15 | C | tomato | Micrococcus luteus |
| 16 | C | tomato | Klebsiella oxytoca |
| 17 | C | tomato | Pseudomonas protegens |
| 18 | C | tomato | Serratia marcescens |
| 19 | D | cucumber | Lelliottia amnigena |
| 20 | D | cucumber | Pseudomonas putida |
| 21 | D | beetroot | Proteus vulgaris |
| 22 | D | cucumber | Staphylococcus equorum |
| 23 | D | onion | Empedobacter falsenii |
| 24 | D | cucumber | Pseudomonas aeruginosa |
| 25 | D | cucumber | Providencia alcalifaciens |
| Type of Antibiotic | Antibiotic Susceptibility Profiles of Bacteria | ||||||
|---|---|---|---|---|---|---|---|
| Class | Antibiotic | K. oxytoca | K. pneumoniae | S. marcescens | P. vulgaris | P. aeruginosa | E. faecalis |
| Penicillins | piperacillin | S | S | S | S | I | n. a. |
| Penicillins | amoxicillin-clavulanic acid | S | S | n. a. | S | n. a. | n. a. |
| Penicillins | ticarcillin-clavulanic acid | S | S | S | S | I | S |
| Penicillins | ampicillin | n. a. | n. a. | n. a. | n. a. | n. a. | S |
| Cephalosporins | cefepime | R | S | S | S | I | n. a. |
| Cephalosporins | cefiderocol | S | S | S | S | S | n. a. |
| Cephalosporins | ceftazidime | S | S | S | S | I | n. a. |
| Carbapenems | meropenem | S | S | S | S | S | n. a. |
| Carbapenems | imipenem | I | I | I | I | I | I |
| Fluoroquinolones | ciprofloxacin | S | S | S | S | I | S |
| Fluoroquinolones | moxifloxacin | S | S | n. a. | S | n. a | * |
| Fluoroquinolones | levofloxacin | S | S | S | S | I | S |
| Aminoglycosides | gentamicin | S | S | S | S | n. a. | n. a. |
| Aminoglycosides | amikacin | S | S | S | S | S | n. a. |
| Aminoglycosides | tobramycin | S | S | I | S | S | n. a. |
| Chemotherapeutics | trimethoprim | S | S | S | S | n. a. | n. a. |
| Chemotherapeutics | trimethoprim | n. a. | n. a. | n. a. | n. a. | n. a. | S |
| Glycopeptides | vancomycin | n. a. | n. a. | n. a. | n. a. | n. a. | S |
| Tetracyclines | tigecycline | n. a. | n. a. | n. a. | n. a. | n. a. | S |
| Oxazolidinones | linezolid | n. a. | n. a. | n. a. | n. a. | n. a. | S |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanarek, P.; Breza-Boruta, B.; Bogiel, T. In the Depths of Wash Water: Isolation of Opportunistic Bacteria from Fresh-Cut Processing Plants. Pathogens 2024, 13, 768. https://doi.org/10.3390/pathogens13090768
Kanarek P, Breza-Boruta B, Bogiel T. In the Depths of Wash Water: Isolation of Opportunistic Bacteria from Fresh-Cut Processing Plants. Pathogens. 2024; 13(9):768. https://doi.org/10.3390/pathogens13090768
Chicago/Turabian StyleKanarek, Piotr, Barbara Breza-Boruta, and Tomasz Bogiel. 2024. "In the Depths of Wash Water: Isolation of Opportunistic Bacteria from Fresh-Cut Processing Plants" Pathogens 13, no. 9: 768. https://doi.org/10.3390/pathogens13090768
APA StyleKanarek, P., Breza-Boruta, B., & Bogiel, T. (2024). In the Depths of Wash Water: Isolation of Opportunistic Bacteria from Fresh-Cut Processing Plants. Pathogens, 13(9), 768. https://doi.org/10.3390/pathogens13090768






