Modeling of the HIV-1 Life Cycle in Productively Infected Cells to Predict Novel Therapeutic Targets
Abstract
1. Introduction
2. Results
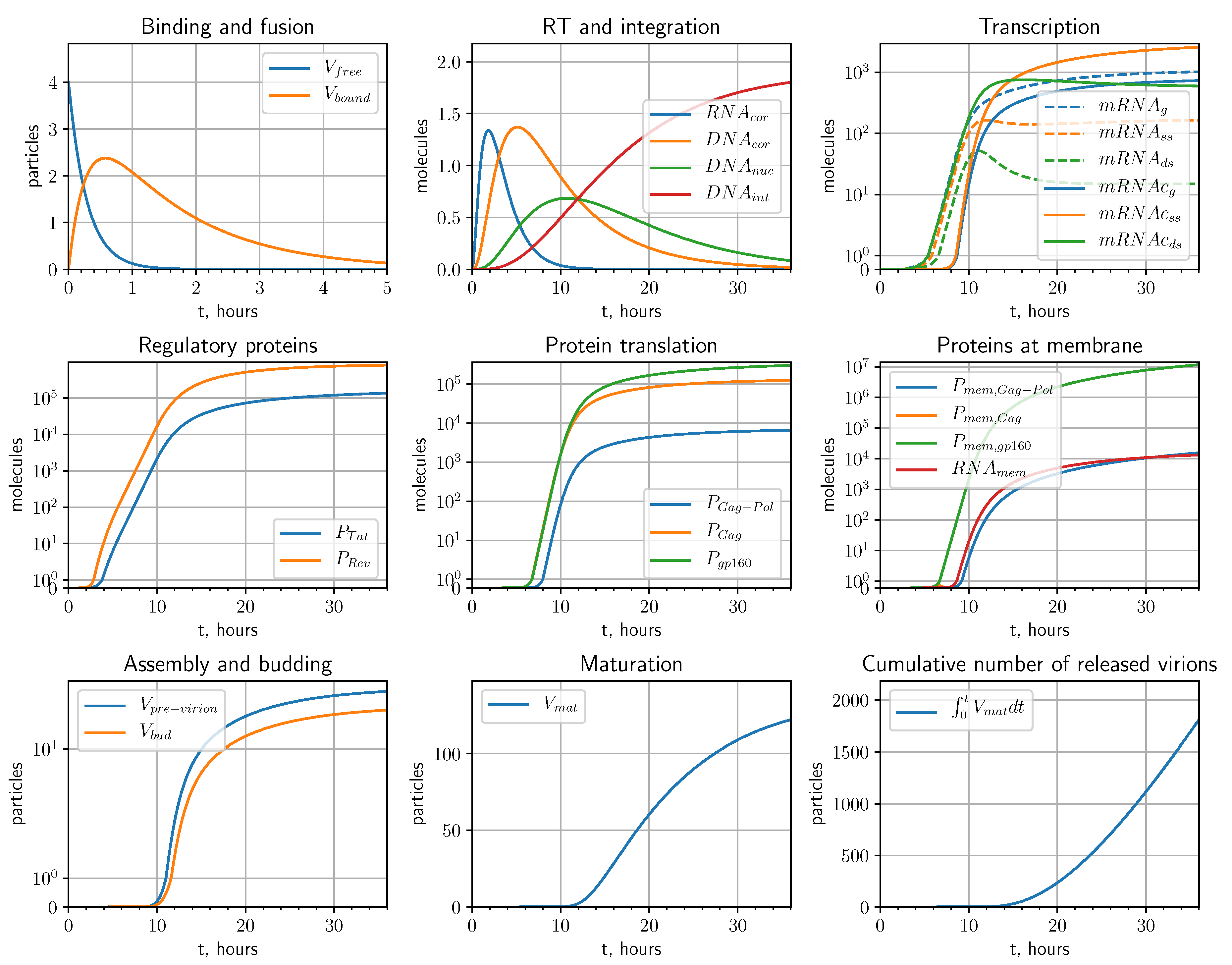
2.1. Mathematical Model
2.1.1. Entry
- binding of virion to CD4 receptors (glycoprotein gp120 binds to CD4 receptors on the T cell surface),
- binding to the co-receptor (CCR5 or CXCR4),
- fusion, i.e., the nucleocapsid is uncoated and the viral RNA is injected into the cell.
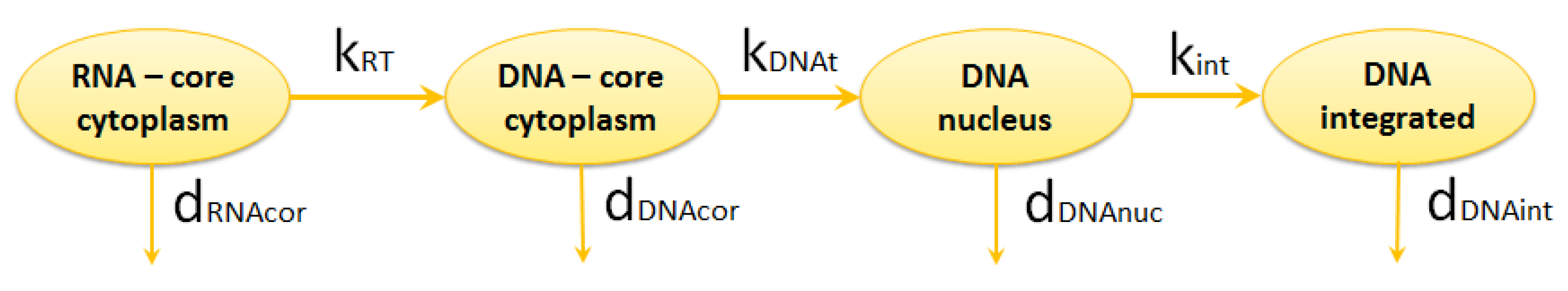
2.1.2. Reverse Transcription
- synthesis of minus-strand DNA from viral RNA,
- synthesis of plus-strand DNA,
- double-strand DNA formation.
2.1.3. Integration
2.1.4. Transcription
2.1.5. Translation
2.1.6. Assembly, Budding and Maturation
2.2. Model Parameters
3. Sensitivity Analysis
- maximal achievable level of transcription rate that can be induced by Tat;
- HIV-1 assembly, specifically sensitive to availability and translation rate of Gag molecules;
- full-length RNA transport to membrane and degradation;
- transport of pre-integration complex into nucleus and DNA integration;
- Rev-mediated regulation of splicing rates and export of full-length RNA;
- reverse transcription (rate of reverse transcription, RNA degradation, DNA degradation);
- kinetics of membrane-bound pre-virion complexes and virions;
- binding and fusion of free virions.
4. Discussion
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| HIV-1 | Human immunodeficiency virus type 1 |
| ART | antiretroviral therapy |
| Tat | trans-activator of transcription |
| Rev | regulator of expression of virion proteins |
References
- Zheltkova, V.; Argilaguet, J.; Peligero, C.; Bocharov, G.; Meyerhans, A. Prediction of PD-L1 inhibition effects for HIV-infected individuals. PLoS Comput. Biol. 2019, 15, e1007401. [Google Scholar] [CrossRef] [PubMed]
- Grebennikov, D.; Bouchnita, A.; Volpert, V.; Bessonov, N.; Meyerhans, A.; Bocharov, G. Spatial Lymphocyte Dynamics in Lymph Nodes Predicts the Cytotoxic T Cell Frequency Needed for HIV Infection Control. Front. Immunol. 2019, 10, 1213. [Google Scholar] [CrossRef] [PubMed]
- Chereshnev, V.A.; Bocharov, G.; Bazhan, S.; Bachmetyev, B.; Gainova, I.; Likhoshvai, V.; Argilaguet, J.M.; Martinez, J.P.; Rump, J.A.; Mothe, B.; et al. Pathogenesis and Treatment of HIV Infection: The Cellular, the Immune System and the Neuroendocrine Systems Perspective. Int. Rev. Immunol. 2013, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Reddy, B.; Yin, J. Quantitative intracellular kinetics of HIV type 1. AIDS Res. Hum. Retroviruses 1999, 15, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Yin, J. Robust growth of human immunodeficiency virus type 1 (HIV-1). Biophys. J. 2005, 89, 2210–2221. [Google Scholar] [CrossRef] [PubMed]
- Likhoshvai, V.; Khlebodarova, T.; Bazhan, S.; Gainova, I.; Chereshnev, V.; Bocharov, G. Mathematical model of the Tat-Rev regulation of HIV-1 replication in an activated cell predicts the existence of oscillatory dynamics in the synthesis of viral components. BMC Genom. 2014, 15 (Suppl. 12), S1. [Google Scholar] [CrossRef] [PubMed]
- Tameru, B.; Habtemariam, T.; Nganwa, D.; Ayanwale, L.; Beyene, G.; Robnett, V.; Wilson, W. Computational Modelling of Intracellular Viral Kinetics and CD4+ Cellular Population Dynamics of HIV/AIDS. Adv. Syst. Sci. Appl. 2008, 8, 40–45. [Google Scholar]
- Könnyű, B.; Sadiq, S.K.; Turányi, T.; Hírmondó, R.; Müller, B.; Kräusslich, H.G.; Coveney, P.V.; Müller, V. Gag-Pol Processing during HIV-1 Virion Maturation: A Systems Biology Approach. PLoS Comput. Biol. 2013, 9, e1003103. [Google Scholar] [CrossRef]
- Zarrabi, N.; Mancini, E.; Tay, J.; Shahand, S.; Sloot, P.M. Modeling HIV-1 intracellular replication: Two simulation approaches. Procedia Comput. Sci. 2010, 1, 555–564. [Google Scholar] [CrossRef]
- Mohammadi, P.; Desfarges, S.; Bartha, I.; Joos, B.; Zangger, N.; Muñoz, M.; Günthard, H.F.; Beerenwinkel, N.; Telenti, A.; Ciuffi, A. 24 Hours in the Life of HIV-1 in a T Cell Line. PLoS Pathog. 2013, 9, e1003161. [Google Scholar] [CrossRef]
- Hu, W.S.; Hughes, S.H. HIV-1 Reverse Transcription. Cold Spring Harb. Perspect. Med. 2012, 2, a006882. [Google Scholar] [CrossRef] [PubMed]
- Craigie, R.; Bushman, F.D. HIV DNA Integration. Cold Spring Harb. Perspect. Med. 2012, 2, a006890. [Google Scholar] [CrossRef] [PubMed]
- Freed, E.O. HIV-1 assembly, release and maturation. Nat. Rev. Microbiol. 2015, 13, 484–496. [Google Scholar] [CrossRef] [PubMed]
- Doranz, B.J.; Baik, S.S.W.; Doms, R.W. Use of a gp120 Binding Assay To Dissect the Requirements and Kinetics of Human Immunodeficiency Virus Fusion Events. J. Virol. 1999, 73, 10346–10358. [Google Scholar] [CrossRef] [PubMed]
- Raviv, Y.; Viard, M.; Bess, J., Jr.; Blumenthal, R. Quantitative Measurement of Fusion of HIV-1 and SIV with Cultured Cells Using Photosensitized Labeling. Virology 2002, 293, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.M.; McBride, K.; Boesecke, C.; Bailey, M.; Amin, J.; Suzuki, K.; Baker, D.; Zaunders, J.J.; Emery, S.; Cooper, D.A.; et al. Integrated HIV DNA accumulates prior to treatment while episomal HIV DNA records ongoing. transmission afterwards. AIDS 2012, 26, 543–550. [Google Scholar] [CrossRef]
- Loya, S.; Tal, R.; Hughes, S.H.; Hizi, A. The Effects of Cysteine Mutations on the Catalytic Activities of the Reverse Transcriptase of Human Immunodeficiency Virus Type-1. JBC 1992, 267, 13879–13883. [Google Scholar]
- HIV Sequence Database. Available online: https://www.hiv.lanl.gov/content/sequence/HIV/MAP/landmark.html (accessed on 28 February 2020).
- Sundquist, W.I.; Krausslich, H.G. HIV-1 Assembly, Budding, and Maturation. Cold Spring Harb. Perspect. Med. 2012, 2, a006924. [Google Scholar] [CrossRef]
- Ribeiro, R.M.; Qin, L.; Chavez, L.L.; Li, D.; Self, S.G.; Perelson, A.S. Estimation of the Initial Viral Growth Rate and Basic Reproductive Number during Acute HIV-1 Infection. J. Virol. 2010, 84, 6096–6102. [Google Scholar] [CrossRef]
- Swanson, C.M.; Malim, M.H. SnapShot: HIV-1 Proteins. Cell 2008, 133, 742–742.e1. [Google Scholar] [CrossRef]
- Schultz, A.; Sopper, S.; Sauermann, U.; Meyerhans, A.; Suspène, R. Stable multi-infection of splenocytes during SIV infection - the basis for continuous recombination. Retrovirology 2012, 9, 31. [Google Scholar] [CrossRef]
- Bocharov, G.; Chereshnev, V.; Gainova, I.; Bazhan, S.; Bachmetyev, B.; Argilaguet, J.; Martinez, J.; Meyerhans, A. Human Immunodeficiency Virus Infection: From Biological Observations to Mechanistic Mathematical Modelling. Math. Model. Nat. Phenom. 2012, 7, 78–104. [Google Scholar] [CrossRef]
- Doms, R.W.; Moore, J.P. HIV-1 Membrane Fusion. J. Cell Biol. 2000, 151, F9–F14. [Google Scholar] [CrossRef] [PubMed]
- Gallo, S.A.; Finnegan, C.M.; Viard, M.; Raviv, Y.; Dimitrov, A.; Rawat, S.S.; Puri, A.; Durell, S.; Blumenthal, R. The HIV Env-mediated fusion reaction. Biochim. Et Biophys. Acta (BBA) - Biomembr. 2003, 1614, 36–50. [Google Scholar] [CrossRef]
- Ramratnam, B.; Bonhoeffer, S.; Binley, J.; Hurley, A.; Zhang, L.; Mittler, J.E.; Markowitz, M.; Moore, J.P.; Perelson, A.S.; Ho, D.D. Rapid production and clearance of HIV-1 and hepatitis C virus assessed by large volume plasma apheresis. Lancet 1999, 354, 1782–1785. [Google Scholar] [CrossRef]
- Finzi, D.; Siliciano, R.F. Viral Dynamics in HIV-1 Infection. Cell 1998, 93, 665–671. [Google Scholar] [CrossRef]
- Heesters, B.A.; Lindqvist, M.; Vagefi, P.A.; Scully, E.P.; Schildberg, F.A.; Altfeld, M.; Walker, B.D.; Kaufmann, D.E.; Carroll, M.C. Follicular Dendritic Cells Retain Infectious HIV in Cycling Endosomes. PLoS Pathog. 2015, 11, 1–18. [Google Scholar] [CrossRef]
- Gallo, S.A.; Reeves, J.D.; Garg, H.; Foley, B.; Doms, R.W.; Blumenthal, R. Kinetic studies of HIV-1 and HIV-2 envelope glycoprotein-mediated fusion. Retrovirology 2006, 3, 90. [Google Scholar] [CrossRef]
- Brussel, A.; Sonigo, P. Evidence for Gene Expression by Unintegrated Human Immunodeficiency Virus Type 1 DNA Species. J. Virol. 2004, 78, 11263–11271. [Google Scholar] [CrossRef]
- Vandegraaff, N.; Kumar, R.; Burrell, C.J.; Li, P. Kinetics of Human Immunodeficiency Virus Type 1 (HIV) DNA Integration in Acutely Infected Cells as Determined Using a Novel Assay for Detection of Integrated HIV DNA. J. Virol. 2001, 75, 11253–11260. [Google Scholar] [CrossRef]
- Barbosa, P.; Charneau, P.; Dumey, N.; Clavel, F. Kinetic Analysis of HIV-1 Early Replicative Steps in a Coculture System. AIDS Res. Hum. Retroviruses 1994, 10, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Andreadis, S.T.; Palsson, B.O. Kinetics of Retrovirus Mediated Gene Transfer: The Importance of Intracellular Half-Life of Retroviruses. J. Theor. Biol. 1996, 182, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, M.G.; Li, G.; Potash, M.J.; Volsky, D.J. Contribution of multiple rounds of viral entry and reverse transcription to expression of human immunodeficiency virus type 1. A quantitative kinetic study. J. Biol. Chem. 1991, 266, 1783–1788. [Google Scholar] [PubMed]
- Butler, S.L.; Hansen, M.S.; Bushman, F.D. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 2001, 7, 631–634. [Google Scholar] [CrossRef] [PubMed]
- Kustikova, O.S.; Wahlers, A.; Kuhlcke, K.; Stahle, B.; Zander, A.R.; Baum, C.; Fehse, B. Dose finding with retroviral vectors: Correlation of retroviral vector copy numbers in single cells with gene transfer efficiency in a cell population. Blood 2003, 102, 3934–3937. [Google Scholar] [CrossRef]
- Siliciano, J.D.; Kajdas, J.; Finzi, D.; Quinn, T.C.; Chadwick, K.; Margolick, J.B.; Kovacs, C.; Gange, S.J.; Siliciano, R. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 2003, 9, 727–728. [Google Scholar] [CrossRef]
- Chen, J.; Grunwald, D.; Sardo, L.; Galli, A.; Plisov, S.; Nikolaitchik, O.A.; Chen, D.; Lockett, S.; Larson, D.R.; Pathak, V.K.; et al. Cytoplasmic HIV-1 RNA is mainly transported by diffusion in the presence or absence of Gag protein. Proc. Natl. Acad. Sci. USA 2014, 111, E5205–E5213. [Google Scholar] [CrossRef]
- Schwanhäusser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Correction: Corrigendum: Global quantification of mammalian gene expression control. Nature 2013, 495, 126–127. [Google Scholar] [CrossRef]
- Qu, N.; Ma, Z.; Zhang, M.; Rushdi, M.N.; Krueger, C.J.; Chen, A.K. Inhibition of retroviral Gag assembly by non-silencing miRNAs promotes autophagic viral degradation. Protein Cell 2018, 9, 640–651. [Google Scholar] [CrossRef]
- Hare, J.F.; Taylor, K. Mechanisms of plasma membrane protein degradation: Recycling proteins are degraded more rapidly than those confined to the cell surface. Proc. Natl. Acad. Sci. USA 1991, 88, 5902–5906. [Google Scholar] [CrossRef]
- Inamdar, K.; Floderer, C.; Favard, C.; Muriaux, D. Monitoring HIV-1 Assembly in Living Cells: Insights from Dynamic and Single Molecule Microscopy. Viruses 2019, 11, 72. [Google Scholar] [CrossRef] [PubMed]
- Ivanchenko, S.; Godinez, W.J.; Lampe, M.; Kräusslich, H.G.; Eils, R.; Rohr, K.; Bräuchle, C.; Müller, B.; Lamb, D.C. Dynamics of HIV-1 Assembly and Release. PLoS Pathog. 2009, 5, e1000652. [Google Scholar] [CrossRef] [PubMed]
- Chojnacki, J.; Staudt, T.; Glass, B.; Bingen, P.; Engelhardt, J.; Anders, M.; Schneider, J.; Muller, B.; Hell, S.W.; Krausslich, H.G. Maturation-Dependent HIV-1 Surface Protein Redistribution Revealed by Fluorescence Nanoscopy. Science 2012, 338, 524–528. [Google Scholar] [CrossRef] [PubMed]
- Marchuk, G.I.; Shutyaev, V.; Bocharov, G. Adjoint equations and analysis of complex systems: Application to virus infection modelling. J. Comput. Appl. Math. 2005, 184, 177–204. [Google Scholar] [CrossRef]
- Thenin-Houssier, S.; T. Valente, S. HIV-1 Capsid Inhibitors as Antiretroviral Agents. Curr. HIV Res. 2016, 14, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.; Kim, H.G.; Park, C.M.; Choi, M.S.; Kim, D.E.; Choi, B.S.; Kim, K.; Yoon, C.H. Identification of novel compounds against Tat-mediated human immunodeficiency virus-1 transcription by high-throughput functional screening assay. Biochem. Biophys. Res. Commun. 2020, 523, 368–374. [Google Scholar] [CrossRef]
- Bouchnita, A.; Bocharov, G.; Meyerhans, A.; Volpert, V. Towards a Multiscale Model of Acute HIV Infection. Computation 2017, 5, 6. [Google Scholar] [CrossRef]
- Grebennikov, D.S.; Bocharov, G.A. Spatially resolved modelling of immune responses following a multiscale approach: From computational implementation to quantitative predictions. Russ. J. Numer. Anal. Math. Model. 2019, 34, 253–260. [Google Scholar] [CrossRef]






| Parameter | Description | Value | Range, Relev. Refs. |
|---|---|---|---|
| rate of virion binding to CD4+ T cell membrane | [14,24,25] | ||
| d | clearance rate of free mature virions | [23,26] | |
| degradation rate of bound virions | |||
| [27,28] | |||
| rate of virion fusion with the cell | [15,25,29] | ||
| reverse transcription rate | [4,9,11] | ||
| degradation rate of RNA in cytoplasm | [30] | ||
| degradation rate of DNA in cytoplasm | [10,16,31,32] | ||
| transport rate of DNA from cytoplasm to nucleus | [4,32] | ||
| degradation rate of DNA in nucleus | [16] | ||
| integration rate | [4,33,34,35,36] | ||
| degradation rate of DNA integrated into chromosome | 0.00002 | [37] | |
| cell intrinsic rate of basal transcription | 15 | [4,5] | |
| level of transcription induced by Tat transactivation | 1500 | [4,5] | |
| threshold for half-maximal boosting of transcription by Tat | 1000 molec. | [4,5], calibrated by [10] | |
| threshold for half-maximal boosting on export of and by Rev | 77,000 molec. | , calibrated by [10] | |
| inhibitory effect of Rev on the splicing rates implying their 1 / ( 1 − β ) -fold reduction at saturation level of Rev | 0.9 | [4] | |
| transport rate of to cell membrane | 2.8 | [38] | |
| rate of export from nucleus, | [4,5,6] | ||
| rate of export from nucleus | [4,5,6] | ||
| rate of splicing for full-length virus RNA | [4,5,6] | ||
| rate of splicing for singly spliced virus RNA | [4,5,6] | ||
| degradation rate of , | [4,5,6] | ||
| degradation rate of protein gp160 | [4] | ||
| degradation rate of protein j, | [4,9] | ||
| degradation rate of Tat protein | [4,5,6] | ||
| degradation rate of Rev protein | [4,5,6] | ||
| fraction of coding | |||
| , | |||
| — | 0.05 | [4] | |
| — | 0.95 | [4] | |
| — | 0.64 | [18] | |
| — | 0.025 | [4] | |
| — | 0.2 | [4,5] | |
| rate of mRNA to proteins translation | 524 proteins/mRNA/h | [4,5,6,39] | |
| degradation rate for the membrane anchored protein Gag-Pol | [40] | ||
| degradation rate for the membrane anchored protein Gag | [40] | ||
| degradation rate for membrane associated gp160 (Env) | [41] | ||
| rate of protein transport to membrane, | [4,38,42] | ||
| incorporation rate of molecules into pre-virion complexes | 8 | [19,43] | |
| number of viral RNA transcripts in a new virion | 2 | [13,19] | |
| number of Gag molecules in a new virion | 5000 | [19,21] | |
| number of Gag-Pol molecules in a new virion | 250 | [19,21] | |
| number of gp160 molecules in a new virion | 24 | [21,44] | |
| degradation rate of assembled pre-virion complex | [43] | ||
| budding rate of new virions | [43] | ||
| degradation rate for budded immature viral like particle | [26] | ||
| (= clearance rate of mature virions) | |||
| maturation rate | [8] |
| Processes Having Negative Effect on J | p | Processes Having Positive Effect on J | p | ||
|---|---|---|---|---|---|
| Gag contribution to virion assembly | 1810 | Transcription induced by Tat | 1971 | ||
| Degradation of free and mature virions | d | 1721 | Translation of Gag molecules | 1811 | |
| Transport of genomic mRNA to membrane | 1697 | Assembly of pre-virion complexes | 1810 | ||
| Degradation of RNA during RT | 602 | Transport of proviral DNA to nucleus | 938 | ||
| Degradation of assembled complexes | 364 | Inhibitory effect of Rev on splicing rates | 895 | ||
| Degradation of DNA during RT | 268 | Reverse transcription | 781 | ||
| Splicing of full-length genomic RNA | 262 | Integration of proviral DNA | 712 | ||
| Degradation of budded immature particles | 242 | Export of full-length genomic RNA | 415 | ||
| Degradation of genomic mRNA | 161 | Budding of immature particles | 412 | ||
| Tolerance of mRNA export and | 118 | Maturation of budded particles | 285 | ||
| splicing to Rev-mediated regulation | Binding of virions to the cell membrane | 250 | |||
| Fusion of virions with the cell | 166 | ||||
| Translation of Rev molecules | 118 | ||||
| Splicing of singly spliced RNA | 104 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shcherbatova, O.; Grebennikov, D.; Sazonov, I.; Meyerhans, A.; Bocharov, G. Modeling of the HIV-1 Life Cycle in Productively Infected Cells to Predict Novel Therapeutic Targets. Pathogens 2020, 9, 255. https://doi.org/10.3390/pathogens9040255
Shcherbatova O, Grebennikov D, Sazonov I, Meyerhans A, Bocharov G. Modeling of the HIV-1 Life Cycle in Productively Infected Cells to Predict Novel Therapeutic Targets. Pathogens. 2020; 9(4):255. https://doi.org/10.3390/pathogens9040255
Chicago/Turabian StyleShcherbatova, Olga, Dmitry Grebennikov, Igor Sazonov, Andreas Meyerhans, and Gennady Bocharov. 2020. "Modeling of the HIV-1 Life Cycle in Productively Infected Cells to Predict Novel Therapeutic Targets" Pathogens 9, no. 4: 255. https://doi.org/10.3390/pathogens9040255
APA StyleShcherbatova, O., Grebennikov, D., Sazonov, I., Meyerhans, A., & Bocharov, G. (2020). Modeling of the HIV-1 Life Cycle in Productively Infected Cells to Predict Novel Therapeutic Targets. Pathogens, 9(4), 255. https://doi.org/10.3390/pathogens9040255






