Circulating Phylotypes of White Spot Syndrome Virus in Bangladesh and Their Virulence
Abstract
:1. Introduction
2. Materials and Methods
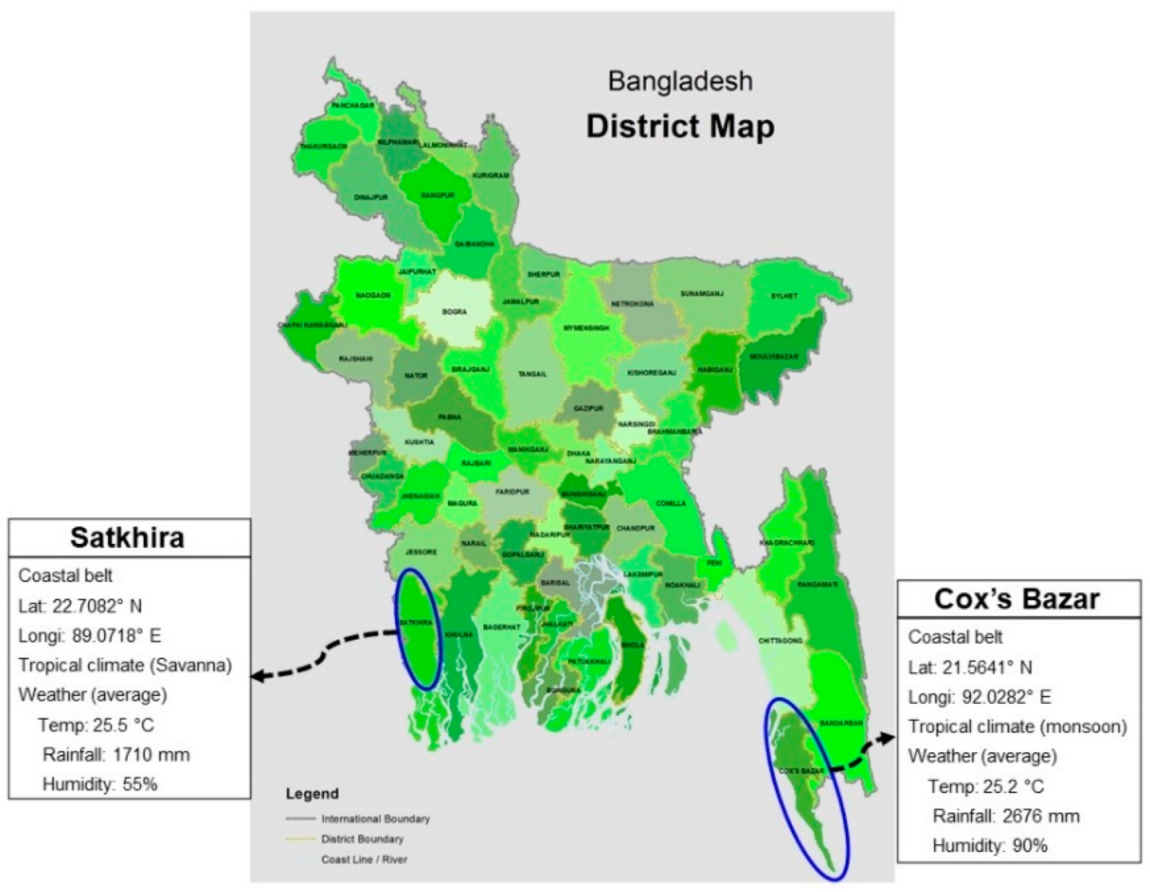
2.1. Sampling and Measurement of Physicochemical Parameters
2.2. DNA Extraction
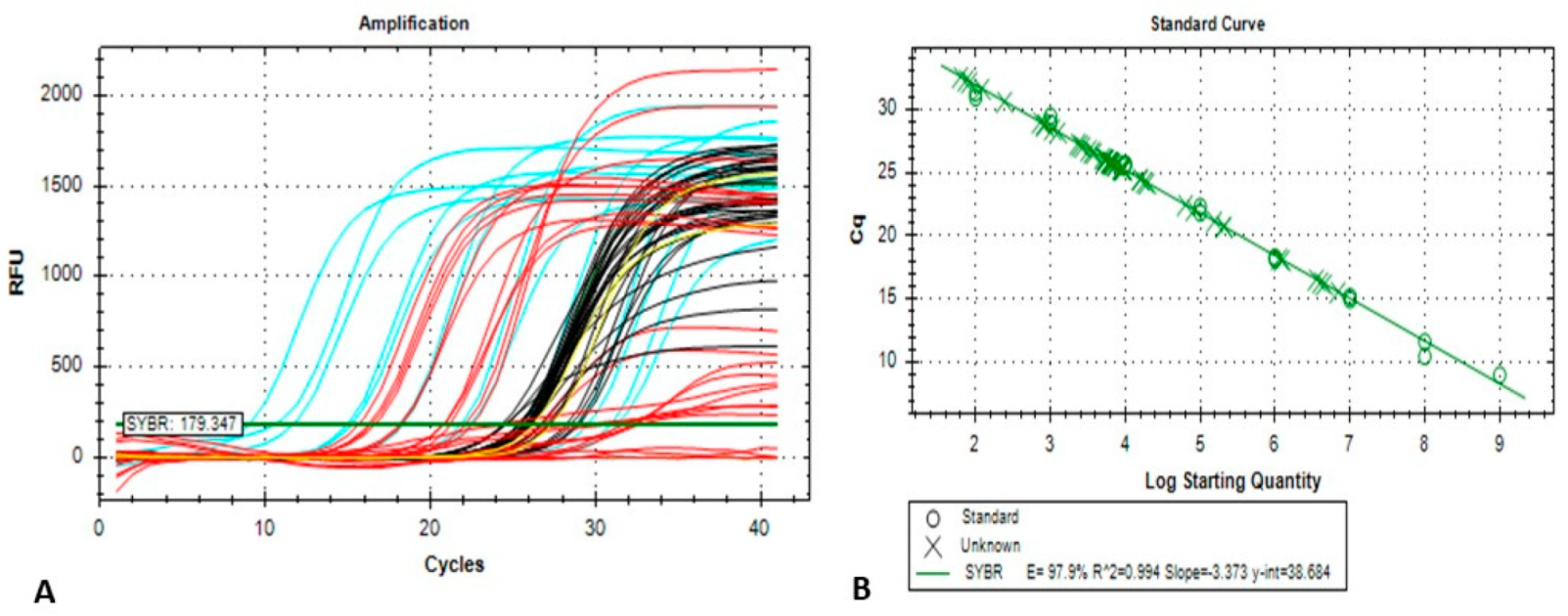
2.3. Conventional and Quantitative Real-Time PCR (qPCR) Assay
template DNA per reaction) × dilution factor] ± Standard Deviation (SD).
2.4. Sequencing of VP28 Protein, Phylogenetic and Mutation Analyses
2.5. Experimental Infection
2.6. RNA Extraction and Preparation of cDNA
2.7. Gene Expression Analysis
2.8. Observation of Binding Affinity of VP28 and Its Receptor Protein Rab7
2.9. Statistical Analysis
3. Results
3.1. Prevalence of WSSV and Physicochemical Parameters in the Study Ghers
3.2. Detection of WSSV in Scylla Olivacea
3.3. Viral Loads in Circulating Phylotypes of WSSV Differed in Crustacean Samples
3.4. Variations in Amino-Acid Mutations in VP28 of WSSV
3.5. Shrimp Post-Larvae Mortality Rates and Lethal Time Differed between Phylotypes
3.6. Quantitative Detection of WSSV in Challenged Shrimp PL
3.7. Crab Mortality Rates and Viral Load Counts Differed in Treated Crabs
3.8. Gene Expression Profiling of Immunity Genes in Both the Infected Groups
3.9. Rab7-VP28 Binding Affinity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nisar, U.; Zhang, H.; Navghan, M.; Zhu, Y.; Mu, Y. Comparative analysis of profitability and resource use efficiency between Penaeus monodon and Litopenaeus vannamei in India. PLoS ONE 2021, 16, e0250727. [Google Scholar] [CrossRef]
- Trang, T.T.; Hung, N.H.; Ninh, N.H.; Knibb, W.; Nguyen, N.H. Genetic variation in disease resistance against White Spot Syndrome Virus (WSSV) in Liptopenaeus vannamei. Front. Genet. 2019, 10, 264. [Google Scholar] [CrossRef]
- Rahman, M.; Hossain, M. Production and export of shrimp of Bangladesh: Problems and prospects. Progress. Agric. 2009, 20, 163–171. [Google Scholar] [CrossRef] [Green Version]
- DoF. National Fish Week Compendium 2013; Ministry of Fisheries and Livestock, People’s Republic of Bangladesh, DoF: Dhaka, Bangladesh, 2013. (In Bengali) [Google Scholar]
- FRSS. Fisheries Statistical Yearbook of Bangladesh; Fisheries Resources Survey System (FRSS), Department of Fisheries: Dhaka, Bangladesh, 2017. [Google Scholar]
- Tendencia, E.A.; Verreth, J.A. Temperature fluctuation, low salinity, water microflora: Risk factors for WSSV outbreaks in Penaeus monodon. Isr. J. Aquac.-Bamidgeh 2011, 63, 1–7. [Google Scholar]
- de la Luz Vazquez-Sauceda, M.; Sánchez-Martínez, J.G.; Pérez-Castañeda, R.; Rábago-Castro, J.L.; Aguirre-Guzmán, G.; Vargas-Cruz, D.Y. White Spot Syndrome Virus (WSSV) and Necrotizing Hepatopancreatitis (NHP) detection in wild shrimpof the San Andrés Lagoon, Mexico. Rev. Biol. Mar. Oceanogr. 2016, 51, 455–459. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zeng, X.; Sun, Y.; Wang, Y.; Zhang, Z. Enhanced Immune Protection of Mud Crab Scylla paramamosain in Response to the Secondary Challenge by Vibrio parahaemolyticus. Front. Immunol. 2020, 11, 565958. [Google Scholar] [CrossRef]
- Molla, M.; Islam, M.; Islam, S.; Salam, M. Socio-economic status of crab collectors and fatteners in the southwest region of Bangladesh. J. Bangladesh Agric. Univ. 2009, 7, 411–419. [Google Scholar] [CrossRef] [Green Version]
- Escobedo-Bonilla, C.M.; Alday-Sanz, V.; Wille, M.; Sorgeloos, P.; Pensaert, M.; Nauwynck, H. A review on the morphology, molecular characterization, morphogenesis and pathogenesis of white spot syndrome virus. J. Fish Dis. 2008, 31, 1–18. [Google Scholar] [CrossRef]
- Somboonna, N.; Mangkalanan, S.; Udompetcharaporn, A.; Krittanai, C.; Sritunyalucksana, K.; Flegel, T. Mud crab susceptibility to disease from white spot syndrome virus is species-dependent. BMC Res. Notes 2010, 3, 315. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Paz, A. White spot syndrome virus: An overview on an emergent concern. Vet. Res. 2010, 41, 43. [Google Scholar] [CrossRef] [Green Version]
- Gong, Y.; Kong, T.; Ren, X.; Chen, J.; Lin, S.; Zhang, Y.; Li, S. Exosome-mediated apoptosis pathway during WSSV infection in crustacean mud crab. PLoS Pathog. 2020, 16, e1008366. [Google Scholar] [CrossRef]
- Zhu, G.; Li, S.; Wu, J.; Li, F.; Zhao, X.-M. Identification of functional gene modules associated with STAT-mediated antiviral responses to white spot syndrome virus in shrimp. Front. Physiol. 2019, 10, 212. [Google Scholar] [CrossRef]
- Yu, Y.; Liu, J.; Li, F.; Zhang, X.; Zhang, C.; Xiang, J. Gene set based association analyses for the WSSV resistance of Pacific white shrimp Litopenaeus vannamei. Sci. Rep. 2017, 7, 40549. [Google Scholar] [CrossRef]
- Corbel, V.; Zuprizal, Z.; Shi, C.; Arcier, J.M.; Bonami, J.R. Experimental infection of European crustaceans with white spot syndrome virus (WSSV). J. Fish Dis. 2001, 24, 377–382. [Google Scholar] [CrossRef]
- Sun, Y.; Li, F.; Sun, Z.; Zhang, X.; Li, S.; Zhang, C.; Xiang, J. Transcriptome analysis of the initial stage of acute WSSV infection caused by temperature change. PLoS ONE 2014, 9, e90732. [Google Scholar]
- Xie, S.; Zhang, X.; Zhang, J.; Li, F.; Xiang, J. Envelope proteins of white spot syndrome virus (WSSV) interact with Litopenaeus vannamei peritrophin-like protein (LvPT). PLoS ONE 2015, 10, e0144922. [Google Scholar] [CrossRef]
- Kwang, J. Oral vaccination of baculovirus-expressed VP28 displays enhanced protection against white spot syndrome virus in Penaeus monodon. PLoS ONE 2011, 6, e26428. [Google Scholar]
- Hossain, A.; Nandi, S.; Siddique, M.; Sanyal, S.; Sultana, M.; Hossain, M. Prevalence and distribution of W hite S pot S yndrome V irus in cultured shrimp. Lett. Appl. Microbiol. 2015, 60, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.-M.; Wang, H.-C.; Leu, J.-H.; Hsiao, H.-H.; Wang, A.H.-J.; Kou, G.-H.; Lo, C.-F. Genomic and proteomic analysis of thirty-nine structural proteins of shrimp white spot syndrome virus. J. Virol. 2004, 78, 11360–11370. [Google Scholar] [CrossRef] [Green Version]
- Sarathi, M.; Simon, M.C.; Ahmed, V.I.; Kumar, S.R.; Hameed, A.S. Silencing VP28 gene of white spot syndrome virus of shrimp by bacterially expressed dsRNA. Mar. Biotechnol. 2008, 10, 198. [Google Scholar] [CrossRef]
- Musthaq, S.S.; Madhan, S.; Hameed, A.S.; Kwang, J. Localization of VP28 on the baculovirus envelope and its immunogenicity against white spot syndrome virus in Penaeus monodon. Virology 2009, 391, 315–324. [Google Scholar] [CrossRef] [Green Version]
- Verbruggen, B.; Bickley, L.K.; Van Aerle, R.; Bateman, K.S.; Stentiford, G.D.; Santos, E.M.; Tyler, C.R. Molecular mechanisms of white spot syndrome virus infection and perspectives on treatments. Viruses 2016, 8, 23. [Google Scholar] [CrossRef] [Green Version]
- Nilsen, P.; Karlsen, M.; Sritunyalucksana, K.; Thitamadee, S. White spot syndrome virus VP28 specific double-stranded RNA provides protection through a highly focused siRNA population. Sci. Rep. 2017, 7, 1028. [Google Scholar] [CrossRef]
- Zwart, M.P.; Dieu, B.T.M.; Hemerik, L.; Vlak, J.M. Evolutionary trajectory of white spot syndrome virus (WSSV) genome shrinkage during spread in Asia. PLoS ONE 2010, 5, e13400. [Google Scholar] [CrossRef] [Green Version]
- Parrilla-Taylor, D.P.; Vibanco-Pérez, N.; Durán-Avelar, M.D.J.; Gomez-Gil, B.; Llera-Herrera, R.; Vázquez-Juárez, R. Molecular variability and genetic structure of white spot syndrome virus strains from northwest Mexico based on the analysis of genomes. FEMS Microbiol. Lett. 2018, 365, fny216. [Google Scholar] [CrossRef] [Green Version]
- Joseph, T.C.; Rajan, L.A.; James, R.; Lalitha, K.; Surendran, P. Variations of structural protein sequences among geographical isolates of white spot syndrome virus. Int. Aquat. Res. 2015, 7, 85–91. [Google Scholar] [CrossRef] [Green Version]
- Siddique, M.A.; Haque, M.I.-M.; Sanyal, S.K.; Hossain, A.; Nandi, S.P.; Alam, A.R.U.; Sultana, M.; Hasan, M.; Hossain, M.A. Circulatory white spot syndrome virus in South-West region of Bangladesh from 2014 to 2017: Molecular characterization and genetic variation. AMB Express 2018, 8, 25. [Google Scholar] [CrossRef] [Green Version]
- Rout, N.; Kumar, S.; Jaganmohan, S.; Murugan, V. DNA vaccines encoding viral envelope proteins confer protective immunity against WSSV in black tiger shrimp. Vaccine 2007, 25, 2778–2786. [Google Scholar] [CrossRef]
- Mendoza-Cano, F.; Sánchez-Paz, A. Development and validation of a quantitative real-time polymerase chain assay for universal detection of the White Spot Syndrome Virus in marine crustaceans. Virol. J. 2013, 10, 186. [Google Scholar] [CrossRef] [Green Version]
- Rahman, M.S.; Islam, M.R.; Hoque, M.N.; Alam, A.S.M.R.U.; Akther, M.; Puspo, J.A.; Akter, S.; Anwar, A.; Sultana, M.; Hossain, M.A. Comprehensive annotations of the mutational spectra of SARS-CoV-2 spike protein: A fast and accurate pipeline. Transbound. Emerg. Dis. 2020, 68, 1625–1638. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2016, 45, D37–D42. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Saha, O.; Rakhi, N.N.; Hoque, M.N.; Sultana, M.; Hossain, M.A. Genome-wide genetic marker analysis and genotyping of Escherichia fergusonii strain OTSVEF-60. Braz. J. Microbiol. 2021, 52, 989–1004. [Google Scholar] [CrossRef]
- Hoque, M.N.; Istiaq, A.; Clement, R.A.; Gibson, K.M.; Saha, O.; Islam, O.K.; Abir, R.A.; Sultana, M.; Siddiki, A.; Crandall, K.A. Insights into the resistome of bovine clinical mastitis microbiome, a key factor in disease complication. Front. Microbiol. 2020, 11, 860. [Google Scholar] [CrossRef]
- Katoh, K.; Asimenos, G.; Toh, H. Multiple alignment of DNA sequences with MAFFT. In Bioinformatics for DNA Sequence Analysis; Springer: Berlin/Heidelberg, Germany, 2009; pp. 39–64. [Google Scholar]
- Rahman, M.S.; Islam, M.R.; Alam, A.R.U.; Islam, I.; Hoque, M.N.; Akter, S.; Rahaman, M.M.; Sultana, M.; Hossain, M.A. Evolutionary dynamics of SARS-CoV-2 nucleocapsid protein and its consequences. J. Med. Virol. 2021, 93, 2177–2195. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.S.; Hoque, M.N.; Islam, M.R.; Islam, I.; Mishu, I.D.; Rahaman, M.M.; Sultana, M.; Hossain, M.A. Mutational insights into the envelope protein of SARS-CoV-2. Gene Rep. 2021, 22, 100997. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-L.; Lo, C.-F.; Chiu, Y.-L.; Chang, C.-F.; Kou, G.-H. Natural and experimental infection of white spot syndrome virus (WSSV) in benthic larvae of mud crab Scylla serrata. Dis. Aquat. Org. 2000, 40, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Gunasekaran, T.; Gopalakrishnan, A.; Deivasigamani, B.; Seralathan, M.V.; Kathirkaman, P. Spontaneous white spot syndrome virus (WSSV) infection in mud crab (Scylla serrata Forskal 1775) fattening pens farm of south east coast of India. Comp. Clin. Pathol. 2018, 27, 413–419. [Google Scholar] [CrossRef]
- Jeena, K.; Prasad, K.P.; Pathan, M.K.; Babu, P.G. Expression profiling of WSSV ORF 199 and shrimp ubiquitin conjugating enzyme in WSSV Infected Penaeus monodon. Asian-Australas. J. Anim. Sci. 2012, 25, 1184. [Google Scholar] [CrossRef] [Green Version]
- Deris, Z.M.; Iehata, S.; Ikhwanuddina, M.; Sahimi, M.B.M.K.; Do, T.D.; Sorgeloos, P.; Sung, Y.Y.; Wong, L.L. Immune and bacterial toxin genes expression in different giant tiger prawn, penaeus monodon post-larvae stages following AHPND-causing strain of vibrio parahaemolyticus challenge. Aquac. Rep. 2020, 16, 100248. [Google Scholar] [CrossRef]
- Shekhar, M.S.; Gomathi, A.; Gopikrishna, G.; Ponniah, A.G. Gene expression profiling in gill tissues of White spot syndrome virus infected black tiger shrimp Penaeus monodon by DNA microarray. Virus Dis. 2015, 26, 9–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vangone, A.; Bonvin, A.M.J.J. Contact-based prediction of binding affinity in protein-protein complexes. eLife 2015, 4, e07454. [Google Scholar] [CrossRef]
- Xue, L.; Rodrigues, J.; Kastritis, P.; Bonvin, A.M.J.J.; Vangone, A. PRODIGY: A web-server for predicting the binding affinity in protein-protein complexes. Bioinformatics 2016, 32, 3676–3678. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.S.; Hoque, M.N.; Islam, M.R.; Akter, S.; Alam, A.R.U.; Siddique, M.A.; Saha, O.; Rahaman, M.M.; Sultana, M.; Crandall, K.A.; et al. Epitope-based chimeric peptide vaccine design against S, M and E proteins of SARS-CoV-2, the etiologic agent of COVID-19 pandemic: An in silico approach. PeerJ 2020, 8, e9572. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.H.M.; Pires, D.E.V.; Ascher, D.B. DynaMut: Predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018, 46, W350–W355. [Google Scholar] [CrossRef]
- Hoque, M.N.; Istiaq, A.; Clement, R.A.; Sultana, M.; Crandall, K.A.; Siddiki, A.Z.; Hossain, M.A. Metagenomic deep sequencing reveals association of microbiome signature with functional biases in bovine mastitis. Sci. Rep. 2019, 9, 13536. [Google Scholar] [CrossRef] [Green Version]
- Ayub, F.; Sarker, M.Y.; Alam, M.S. Prevalence of white spot syndrome virus infection detected by one-step and nested PCR in selected tiger shrimp (Penaeus monodon) hatcheries. Aquac. Int. 2008, 16, 405–415. [Google Scholar] [CrossRef] [Green Version]
- Rouf, M.; Shahriar, S.; Sarower, M.; Ahsan, M. Taxonomic clarification of mud crab species of genus Scylla (Brachyura: Portunidae) available in the Coastal Regions of Bangladesh. Asian Fish. Sci. 2016, 29, 124–136. [Google Scholar] [CrossRef]
- Tuyen, N.; Verreth, J.; Vlak, J.; De Jong, M. Horizontal transmission dynamics of White spot syndrome virus by cohabitation trials in juvenile Penaeus monodon and P. vannamei. Prev. Vet. Med. 2014, 117, 286–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaseeharan, B.; Jayakumar, R.; Ramasamy, P. PCR-based detection of white spot syndrome virus in cultured and captured crustaceans in India. Lett. Appl. Microbiol. 2003, 37, 443–447. [Google Scholar] [CrossRef] [Green Version]
- Zafar, M.; Haque, M.; Aziz, M.; Alam, M. Study on water and soil quality parameters of shrimp and prawn farming in the southwest region of Bangladesh. J. Bangladesh Agric. Univ. 2015, 13, 153–160. [Google Scholar] [CrossRef] [Green Version]
- Páez-Osuna, F.; Gracia, A.; Flores-Verdugo, F.; Lyle-Fritch, L.P.; Alonso-Rodrıguez, R.; Roque, A.; Ruiz-Fernández, A.C. Shrimp aquaculture development and the environment in the Gulf of California ecoregion. Mar. Pollut. Bull. 2003, 46, 806–815. [Google Scholar] [CrossRef]
- Le Moullac, G.; Haffner, P. Environmental factors affecting immune responses in Crustacea. Aquaculture 2000, 191, 121–131. [Google Scholar] [CrossRef]
- Soo, T.C.C.; Bhassu, S. Differential STAT gene expressions of Penaeus monodon and Macrobrachium rosenbergii in response to white spot syndrome virus (WSSV) and bacterial infections: Additional insight into genetic variations and transcriptomic highlights. PLoS ONE 2021, 16, e0258655. [Google Scholar] [CrossRef]
- Chang, Y.-S.; Liu, W.-J.; Lee, C.-C.; Chou, T.-L.; Lee, Y.-T.; Wu, T.-S.; Huang, J.-Y.; Huang, W.-T.; Lee, T.-L.; Kou, G.-H. A 3D model of the membrane protein complex formed by the white spot syndrome virus structural proteins. PLoS ONE 2010, 5, e10718. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.; Wu, J.; Sivaraman, J.; Hew, C.L. Crystal structures of major envelope proteins VP26 and VP28 from white spot syndrome virus shed light on their evolutionary relationship. J. Virol. 2007, 81, 6709–6717. [Google Scholar] [CrossRef] [Green Version]
- Verma, A.K.; Gupta, S.; Verma, S.; Mishra, A.; Nagpure, N.; Singh, S.P.; Pathak, A.K.; Sarkar, U.K.; Singh, S.P.; Singh, M. Interaction between shrimp and white spot syndrome virus through PmRab7-VP28 complex: An insight using simulation and docking studies. J. Mol. Model. 2013, 19, 1285–1294. [Google Scholar] [CrossRef]
- Sritunyalucksana, K.; Wannapapho, W.; Lo, C.F.; Flegel, T.W. PmRab7 is a VP28-binding protein involved in white spot syndrome virus infection in shrimp. J. Virol. 2006, 80, 10734–10742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandrika, S.K.; Puthiyedathu, S.T. Challenges and prospects of Viral Envelope protein VP28-based control strategies to combat white spot syndromevirus in penaeid shrimps: A review. Rev. Aquac. 2021, 13, 734–743. [Google Scholar] [CrossRef]
- Durand, S.; Lightner, D. Quantitative real time PCR for the measurement of white spot syndrome virus in shrimp. J. Fish Dis. 2002, 25, 381–389. [Google Scholar] [CrossRef]
- Rahman, M.; Corteel, M.; Escobedo-Bonilla, C.M.; Wille, M.; Alday-Sanz, V.; Pensaert, M.; Sorgeloos, P.; Nauwynck, H. Virulence of white spot syndrome virus (WSSV) isolates may be correlated with the degree of replication in gills of Penaeus vannamei juveniles. Dis. Aquat. Org. 2008, 79, 191–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hameed, A.S.; Balasubramanian, G.; Musthaq, S.S.; Yoganandhan, K. Experimental infection of twenty species of Indian marine crabs with white spot syndrome virus (WSSV). Dis. Aquat. Org. 2003, 57, 157–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escobedo-Bonilla, C.M.; Wille, M.; Sanz, V.A.; Sorgeloos, P.; Pensaert, M.; Nauwynck, H. In vivo titration of white spot syndrome virus (WSSV) in specific pathogen-free Litopenaeus vannamei by intramuscular and oral routes. Dis. Aquat. Org. 2005, 66, 163–170. [Google Scholar] [CrossRef]
- Woramongkolchai, N.; Supungul, P.; Tassanakajon, A. The possible role of penaeidin5 from the black tiger shrimp, Penaeus monodon, in protection against viral infection. Dev. Comp. Immunol. 2011, 35, 530–536. [Google Scholar] [CrossRef]
- Kawabata, S.I.; Nagayama, R.; Hirata, M.; Shigenaga, T.; Agarwala, K.L.; Saito, T.; Cho, J.; Nakajima, H.; Takagi, T.; Iwanaga, S. Tachycitin, a Small Granular Component in Horseshoe Crab Hemocytes is an Antimicrobial Protein with Chitin-Binding Activity. J. Biochem. 1996, 120, 1253–1260. [Google Scholar] [CrossRef] [PubMed]
- Bachere, E.; Destoumieux, D.; Bulet, P. Penaeidins, antimicrobial peptides of shrimp: A comparison with other effectors of innate immunity. Aquaculture 2000, 19, 71–88. [Google Scholar] [CrossRef]
- Munoz, M.; Vandenbulcke, F.; Saulnier, D.; Bachere, E. Expression and distribution of penaeidin antimicrobial peptides are regulated by haemocyte reactions in microbial challenged shrimp. Eur. J. Biochem. 2002, 269, 2678–2689. [Google Scholar] [CrossRef]
- Li, C.Y.; Yan, H.Y.; Son, Y.L. Tiger shrimp (Penaeus monodon) penaeidin possesses cytokine features to promote integrin-mediated granulocyte and semi-granulocyte adhesion. Fish Shellfish Immunol. 2010, 28, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.L.; Li, C.Y. Shrimp immune system-special focus on penaeidin. J. Mar. Sci. Technol. 2014, 22, 1–8. [Google Scholar]
- Sotelo-Mundo, R.R.; Islas-Osuna, M.A.; De-la-Re-Vega, E.; Hernández-López, J.; Vargas-Albores, F.; Yepiz-Plascencia, G. cDNA cloning of the lysozyme of the white shrimp Penaeus vannamei. Fish Shellfish Immunol. 2003, 15, 325–331. [Google Scholar] [CrossRef]
- Xing, Y.; Feng-Ying, G.; Qing-Mei, Z.; Jun-Jie, B.; Huan, W.; Hai-Hua, L.; Qing, J. Cloning and characterization of the tiger shrimp lysozyme. Mol. Biol. Rep. 2009, 36, 1239–1246. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.X.; Zhou, N.; Fu, R.; Cao, D.; Si, Y.; Li, A.; Zhao, H.; Zhang, Q.; Yu, H. The polymorphism of chicken-type lysozyme gene in Japanese flounder (Paralichthys olivaceus) and its association with resistance/susceptibility to List. Anguillarum. Fish Shellfish Immunol. 2017, 66, 43–49. [Google Scholar] [CrossRef]
- Mai, W.; Wang, W. Protection of blue shrimp (Litopenaeus stylirostris) against the White Spot Syndrome Virus (WSSV) when injected with shrimp lysozyme. Fish Shellfish Immunol. 2010, 28, 727–733. [Google Scholar] [CrossRef]









| Name of Gene | Primer Sequence (5′-3′) | Reference |
|---|---|---|
| Penaeidin | F: TGGTCTGCCTGGTCTTCCT R: AAGCACGAGCTTGTAAGGG | [44] |
| Lysozyme | F: TGGTGTGGCAGCGATTATG R: GATCGAGGTCGCGATTCTTAC | [44] |
| Beta-actin | F: CCCTGTTCCAGCCCTCATT R: GGATGTCCACGTCGCACTT | [45] |
| VP28 | F: GCGCGCGGATCCAATCATGGATCTTTCTTTCAC R: GCGCGCGAATTCTTACTCGGTCTCAGTGCC | [30] |
| qVP28 | F: TGTGACCAAGACCATCGAAA R: CTTGATTTTGCCCAAGGTGT | [29] |
| Gher ID | Salinity (ppt) | Dissolved Oxygen (ppm) | Temperature (°C) | pH | Shrimp Samples Selected | Crab Samples Selected |
|---|---|---|---|---|---|---|
| Cox1 | 14 | 3.8 | 33.9 | 8.4 | 18 | 6 |
| Cox2 | 21 | 3.75 | 33.7 | 8 | 18 | 6 |
| Cox3 | 21 | 3.65 | 33.7 | 7.8 | 18 | 6 |
| Cox4 | 20 | 3.90 | 33 | 8.1 | 18 | 6 |
| Cox5 | 20 | 3.70 | 32.9 | 8.2 | 18 | 6 |
| SS1 | 14 | 3.8 | 34 | 8.5 | 18 | 6 |
| SS2 | 15 | 3.6 | 33.7 | 8.6 | 18 | 6 |
| SS3 | 15 | 3.7 | 34 | 7.8 | 18 | 6 |
| D1 | 19 | 3.8 | 33.8 | 7.8 | 18 | 6 |
| D2 | 17.5 | 3.7 | 33.5 | 8 | 18 | 6 |
| D3 | 20 | 3.6 | 33 | 8.6 | 18 | 6 |
| A1 | 16.5 | 3.5 | 33.3 | 7.7 | 18 | 6 |
| A2 | 16 | 3.8 | 33.8 | 7.8 | 18 | 6 |
| A3 | 15 | 3.6 | 34 | 8 | 18 | 6 |
| K1 | 14 | 3.8 | 34.6 | 8.6 | 18 | 6 |
| K2 | 15 | 3.6 | 34.7 | 7.8 | 18 | 6 |
| K3 | 16 | 3.6 | 33.9 | 7.9 | 18 | 6 |
| S1 | 13 | 3.6 | 33.9 | 7.9 | 18 | 6 |
| S2 | 16 | 3.8 | 34 | 8.1 | 18 | 6 |
| S3 | 16 | 3.7 | 34 | 8 | 18 | 6 |
| Sample ID | CT | CT | CT Mean | CT SD | WSSV Copies | WSSV Copies | Mean WSSV Copies/Reaction | Mean WSSV Copies/mg Tissue |
|---|---|---|---|---|---|---|---|---|
| G1 | 27.09 | 27.21 | 27.15 | 0.085 | 2.74 × 103 | 2.52 × 103 | 2.63 × 103 | 2.59 × 104 |
| G2 | 25.61 | 25.9 | 25.755 | 0.205 | 7.53 × 103 | 6.21 × 103 | 6.87 × 103 | 8.41 × 104 |
| G3 | 28.80 | 29.02 | 28.91 | 0.156 | 8.50 × 102 | 7.35 × 102 | 7.93 × 102 | 1.03 × 104 |
| G4 | 25.83 | 25.77 | 25.80 | 0.042 | 6.45 × 103 | 6.74 × 103 | 6.60 × 103 | 7.76 × 104 |
| G5 | 25.29 | 25.38 | 25.34 | 0.064 | 9.37 × 103 | 8.78 × 103 | 9.08 × 103 | 5.56 × 104 |
| G6 | 26.05 | 25.77 | 25.91 | 0.198 | 5.55 × 103 | 6.72 × 103 | 6.14 × 103 | 6.94 × 104 |
| G7 | 25.30 | 25.35 | 25.325 | 0.035 | 9.31 × 103 | 8.98 × 103 | 9.15 × 103 | 1.28 × 105 |
| G8 | 26.10 | 26.67 | 26.385 | 0.403 | 5.37 × 103 | 3.64 × 103 | 4.51 × 103 | 4.16 × 104 |
| G9 | 28.21 | 28.47 | 28.34 | 0.184 | 1.27 × 103 | 1.07 × 103 | 1.17 × 103 | 1.30 × 104 |
| G10 | 25.89 | 26.05 | 25.97 | 0.113 | 6.20 × 103 | 5.58 × 103 | 5.89 × 103 | 6.93 × 104 |
| G11 | 25.74 | 24.49 | 25.12 | 0.884 | 6.87 × 103 | 1.61 × 104 | 1.15 × 103 | 6.89 × 104 |
| G12 | 25.85 | 26.09 | 25.97 | 0.17 | 6.36 × 103 | 5.40 × 103 | 5.88 × 103 | 7.20 × 104 |
| G13 | 25.64 | 25.86 | 25.75 | 0.156 | 7.36 × 103 | 6.34 × 103 | 6.85 × 103 | 7.34 × 104 |
| G14 | 25.32 | 25.36 | 25.34 | 0.028 | 9.16 × 103 | 8.89 × 103 | 9.03 × 103 | 6.85 × 104 |
| G15 | 24.30 | 24.52 | 24.41 | 0.156 | 1.83 × 104 | 1.58 × 104 | 1.71 × 104 | 2.44 × 105 |
| G16 | 25.70 | 25.46 | 25.58 | 0.170 | 7.07 × 103 | 8.33 × 103 | 7.67 × 103 | 7.55 × 104 |
| G17 | 24.52 | 24.70 | 24.61 | 0.127 | 1.58 × 104 | 1.40 × 104 | 1.49 × 104 | 1.82 × 105 |
| G18 | 27.10 | 26.92 | 27.01 | 0.127 | 2.72 × 103 | 3.07 × 103 | 2.89 × 103 | 3.77 × 104 |
| G19 | 27.40 | 27.04 | 27.22 | 0.255 | 2.22 × 103 | 2.83 × 103 | 2.5 × 103 | 2.95 × 104 |
| G20 | 24.90 | 24.68 | 24.79 | 0.156 | 1.22 × 104 | 1.42 × 104 | 1.32 × 104 | 8.06 × 104 |
| G21 | 24.27 | 24.37 | 24.32 | 0.071 | 1.88 × 104 | 1.75 × 104 | 1.81 × 104 | 2.05 × 105 |
| G22 | 23.98 | 23.48 | 23.73 | 0.354 | 2.29 × 104 | 3.22 × 104 | 2.71 × 104 | 3.79 × 105 |
| G23 | 24.00 | 24.26 | 24.13 | 0.184 | 2.26 × 104 | 1.89 × 104 | 2.06 × 104 | 1.91 × 105 |
| G24 | 26.53 | 27.03 | 26.78 | 0.354 | 4.01 × 103 | 2.85 × 103 | 3.38 × 103 | 3.76 × 104 |
| G25 | 23.85 | 24.35 | 24.1 | 0.354 | 2.50 × 104 | 1.78 × 104 | 2.10 × 104 | 2.48 × 105 |
| G26 | 20.95 | 21.23 | 21.09 | 0.198 | 1.81 × 105 | 1.49 × 105 | 1.64 × 105 | 9.87 × 105 |
| G27 | 24.63 | 25.09 | 24.86 | 0.325 | 1.47 × 104 | 1.07 × 104 | 1.25 × 104 | 1.54 × 105 |
| G28 | 23.50 | 23.88 | 23.69 | 0.269 | 3.17 × 104 | 2.45 × 104 | 2.79 × 104 | 2.99 × 105 |
| G29 | 23.60 | 24.08 | 23.84 | 0.339 | 2.96 × 104 | 2.14 × 104 | 2.52 × 104 | 1.91 × 105 |
| G30 | 22.22 | 22.62 | 22.42 | 0.283 | 7.61 × 104 | 5.79 × 104 | 6.63 × 104 | 9.48 × 105 |
| E1 | 33.48 | 23.65 | 28.565 | 6.951 | 3.48 × 101 | 4.24 × 104 | 2.12 × 104 | 9.50 × 104 |
| E2 | 32.19 | 32.35 | 32.27 | 0.113 | 8.44 × 101 | 7.54 × 101 | 7.99 × 101 | 8.88 × 102 |
| E3 | 31.15 | 29.51 | 30.33 | 1.16 | 1.71 × 102 | 5.24 × 102 | 3.48 × 102 | 1.86 × 103 |
| E4 | 12.37 | 7.65 | 10.01 | 3.338 | 6.0 × 107 | 1.59 × 109 | 8.25 × 108 | 1.10 × 1010 |
| E5 | 18.23 | 18.11 | 18.17 | 0.085 | 1.16 × 106 | 1.25 × 106 | 1.21 × 106 | 9.04 × 106 |
| E6 | 30.63 | 24.20 | 27.415 | 4.547 | 2.44 × 102 | 1.97 × 104 | 9.97 × 103 | 1.03 × 105 |
| E7 | 7.19 | 28.84 | 18.015 | 15.31 | 2.17 × 109 | 8.29 × 102 | 1.09 × 109 | 7.15 × 109 |
| E8 | 16.38 | 16.58 | 16.48 | 0.141 | 4.10 × 106 | 3.56 × 106 | 3.83 × 106 | 2.74 × 107 |
| E9 | 26.82 | 26.59 | 26.705 | 0.163 | 3.30 × 103 | 3.86 × 103 | 3.58 × 103 | 3.07 × 103 |
| E10 | 23.36 | 23.65 | 23.505 | 0.205 | 3.48 × 104 | 2.85 × 104 | 3.17 × 104 | 1.62 × 105 |
| E11 | 20.76 | 20.74 | 20.73 | 0.014 | 2.06 × 105 | 2.08 × 105 | 2.07 × 105 | 5.18 × 105 |
| E12 | 22.48 | 22.08 | 22.28 | 0.283 | 6.37 × 104 | 8.33 × 104 | 7.35 × 104 | 1.91 × 105 |
| E13 | 16.22 | 15.60 | 15.91 | 0.438 | 4.57 × 106 | 6.99 × 106 | 5.78 × 106 | 4.08 × 107 |
| E14 | 16.92 | 21.17 | 19.05 | 3.005 | 3.38 × 107 | 1.56 × 105 | 1.70 × 107 | 7.78 × 107 |
| E15 | 32.60 | 31.65 | 32.125 | 0.672 | 6.35 × 101 | 1.21 × 102 | 9.23 × 101 | 3.31 × 102 |
| E16 | 23.96 | 23.34 | 23.65 | 0.438 | 2.32 × 104 | 3.54 × 104 | 2.87 × 104 | 1.28 × 105 |
| E17 | 12.21 | 12.53 | 12.27 | 0.226 | 7.06 × 107 | 5.67 × 107 | 6.78 × 107 | 7.53 × 108 |
| E18 | 23.23 | 23.73 | 23.48 | 0.354 | 3.82 × 104 | 2.71 × 104 | 3.22 × 104 | 1.72 × 105 |
| E19 | 12.05 | 12.69 | 12.37 | 0.453 | 7.87 × 107 | 5.09 × 107 | 6.33 × 107 | 8.44 × 108 |
| E20 | 15.44 | 15.20 | 15.32 | 0.170 | 7.78 × 106 | 9.17 × 106 | 8.45 × 106 | 6.34 × 107 |
| E21 | 12.00 | 11.26 | 11.63 | 0.523 | 8.15 × 107 | 1.35 × 108 | 1.05 × 108 | 1.09 × 109 |
| E22 | 19.88 | 19.38 | 19.63 | 0.354 | 3.75 × 105 | 5.29 × 105 | 4.46 × 105 | 2.94 × 106 |
| E23 | 12.02 | 12.42 | 12.22 | 0.283 | 8.04 × 107 | 6.12 × 107 | 7.01 × 107 | 5.01 × 108 |
| E24 | 21.95 | 22.39 | 22.17 | 0.311 | 9.14 × 104 | 6.77 × 104 | 7.87 × 104 | 6.75 × 104 |
| E25 | 26.40 | 26.82 | 26.61 | 0.297 | 4.38 × 103 | 3.29 × 103 | 3.80 × 103 | 1.95 × 104 |
| E26 | 16.02 | 16.86 | 16.44 | 0.594 | 5.24 × 106 | 2.95 × 106 | 3.93 × 106 | 9.83 × 106 |
| E27 | 12.25 | 11.79 | 12.02 | 0.325 | 6.87 × 107 | 9.40 × 107 | 8.04 × 107 | 2.09 × 108 |
| E28 | 18.40 | 16.44 | 17.42 | 1.386 | 1.03 × 106 | 3.93 × 106 | 2.01 × 106 | 1.42 × 107 |
| E29 | 17.10 | 16.74 | 16.92 | 0.255 | 2.51 × 106 | 3.20 × 106 | 2.83 × 106 | 1.30 × 107 |
| E30 | 16.70 | 16.24 | 16.47 | 0.325 | 3.29 × 106 | 4.51 × 106 | 3.85 × 106 | 1.38 × 107 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hasan, M.M.; Hoque, M.N.; Ahmed, F.; Haque, M.I.-M.; Sultana, M.; Hossain, M.A. Circulating Phylotypes of White Spot Syndrome Virus in Bangladesh and Their Virulence. Microorganisms 2022, 10, 191. https://doi.org/10.3390/microorganisms10010191
Hasan MM, Hoque MN, Ahmed F, Haque MI-M, Sultana M, Hossain MA. Circulating Phylotypes of White Spot Syndrome Virus in Bangladesh and Their Virulence. Microorganisms. 2022; 10(1):191. https://doi.org/10.3390/microorganisms10010191
Chicago/Turabian StyleHasan, Mehedi Mahmudul, M. Nazmul Hoque, Firoz Ahmed, Md. Inja-Mamun Haque, Munawar Sultana, and M. Anwar Hossain. 2022. "Circulating Phylotypes of White Spot Syndrome Virus in Bangladesh and Their Virulence" Microorganisms 10, no. 1: 191. https://doi.org/10.3390/microorganisms10010191






