What Flips the Switch? Signals and Stress Regulating Extraintestinal Pathogenic Escherichia coli Type 1 Fimbriae (Pili)
Abstract
:1. Introduction
Role of Type 1 Fimbriae in Pathogenesis
2. Type 1 Fimbriae
2.1. Type 1 Fimbriae Biogenesis
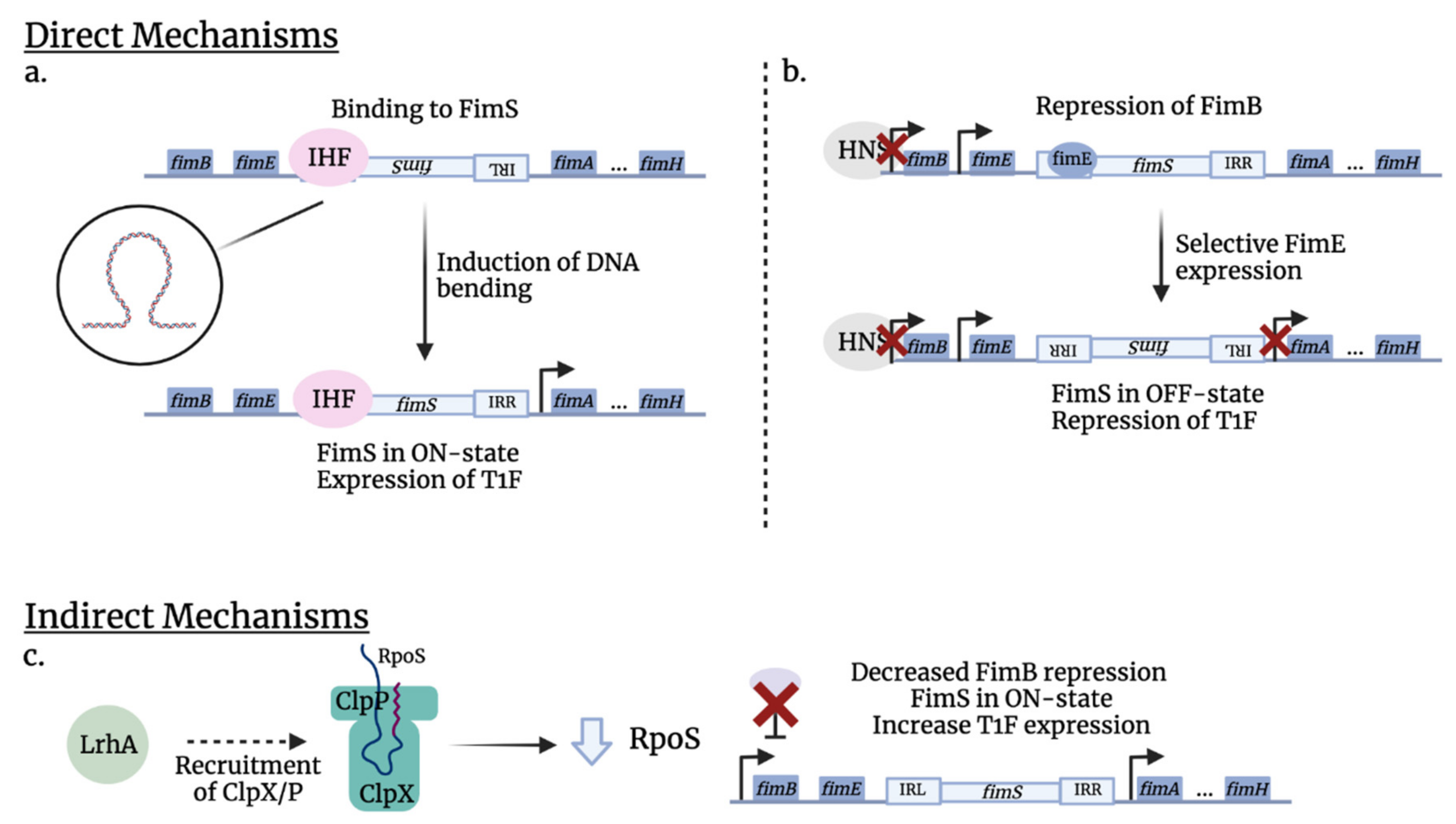
2.2. Genetic Organization of Fimbrial Gene Clusters and Transcriptional Regulation
3. Regulators of Stress Responses and Type 1 Fimbriae in ExPEC
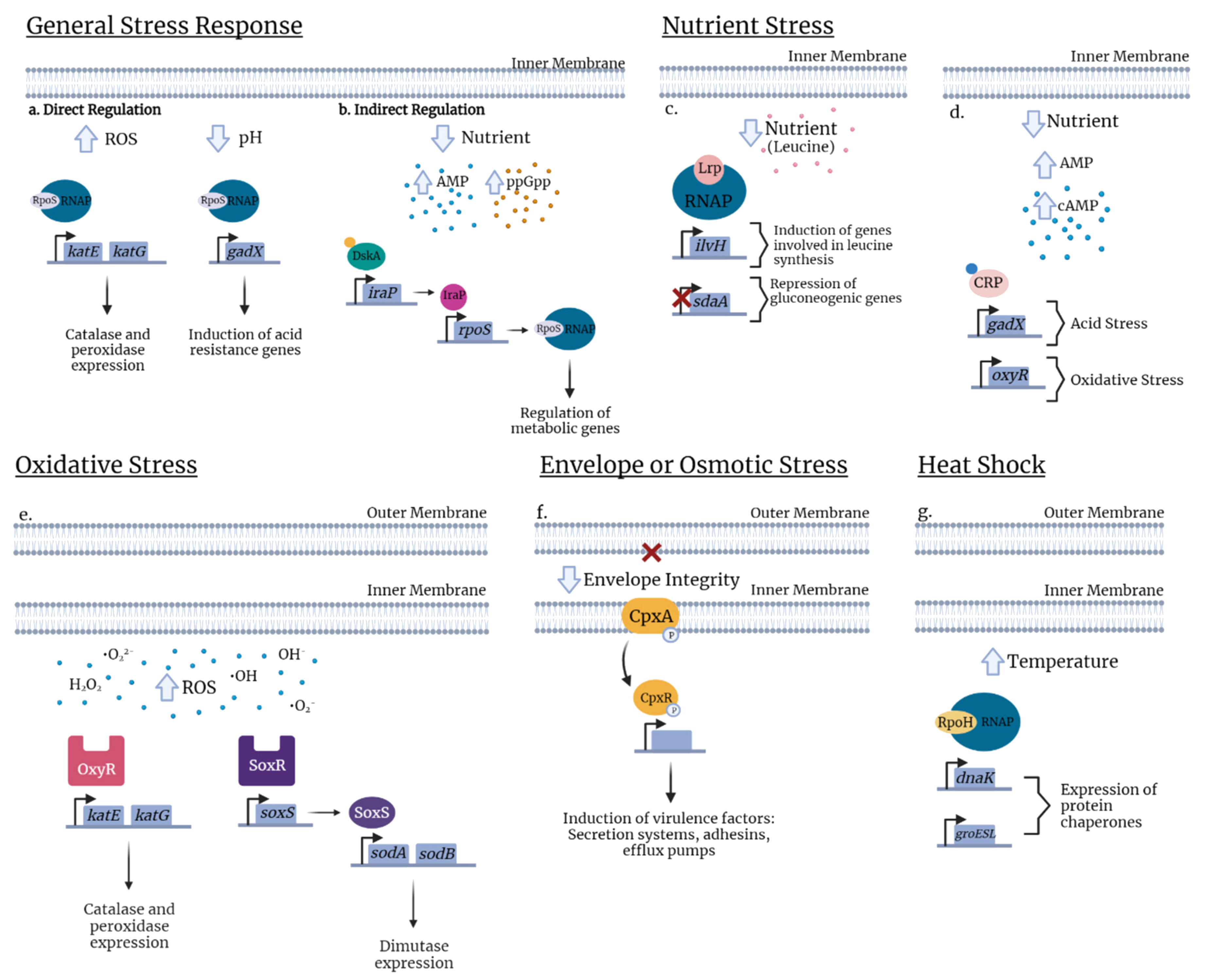
3.1. General Stress
3.2. Envelope Stress
3.3. Osmotic and Oxidative Stress
3.4. Nitrosative Stress
3.5. Nutritional Stress and Metabolism
3.6. Biofilm Formation
3.7. Physical Cues and Regulation of Type 1 Fimbriae
3.8. Shear Stress
4. Link between Stress and Type 1 Fimbriae in Non-Pathogenic E. coli
5. Conclusions
Perspectives and Outstanding Questions
6. Glossary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Gottesman, S. Trouble is coming: Signaling pathways that regulate general stress responses in bacteria. J. Biol. Chem. 2019, 294, 11685–11700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moriel, D.G.; Rosini, R.; Seib, K.; Serino, L.; Pizza, M.; Rappuoli, R. Escherichia coli: Great Diversity around a Common Core. mBio 2012, 3, e00118-12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manges, A.R.; Geum, H.M.; Guo, A.; Edens, T.J.; Fibke, C.D.; Pitout, J.D.D. Global Extraintestinal Pathogenic Escherichia coli (ExPEC) Lineages. Clin. Microbiol. Rev. 2019, 32, e00135-18. [Google Scholar] [CrossRef]
- Kaper, J.B.; Nataro, J.P.; Mobley, H. Pathogenic Escherichia coli. Nat. Rev. Genet. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Alteri, C.J.; Mobley, H. Escherichia coli physiology and metabolism dictates adaptation to diverse host microenvironments. Curr. Opin. Microbiol. 2012, 15, 3–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- La Ragione, R.; Woodward, M. Virulence factors of Escherichia coli serotypes associated with avian colisepticaemia. Res. Veter- Sci. 2002, 73, 27–35. [Google Scholar] [CrossRef]
- Johnson, J.R.; Russo, T.A. Extraintestinal pathogenic Escherichia coli: “The other bad E. coli”. J. Lab. Clin. Med. 2002, 139, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.L.; Fratamico, P.M.; Gunther, N.W. Extraintestinal Pathogenic Escherichia coli. Foodborne Pathog. Dis. 2007, 4, 134–163. [Google Scholar] [CrossRef] [Green Version]
- Lindstedt, B.-A.; Finton, M.D.; Porcellato, D.; Brandal, L.T. High frequency of hybrid Escherichia coli strains with combined Intestinal Pathogenic Escherichia coli (IPEC) and Extraintestinal Pathogenic Escherichia coli (ExPEC) virulence factors isolated from human faecal samples. BMC Infect. Dis. 2018, 18, 544. [Google Scholar] [CrossRef]
- Denamur, E.; Clermont, O.; Bonacorsi, S.; Gordon, D. The population genetics of pathogenic Escherichia coli. Nat. Rev. Genet. 2020, 19, 37–54. [Google Scholar] [CrossRef]
- Sarowska, J.; Futoma-Koloch, B.; Jama-Kmiecik, A.; Frej-Madrzak, M.; Ksiazczyk, M.; Bugla-Ploskonska, G.; Choroszy-Krol, I. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: Recent reports. Gut Pathog. 2019, 11, 10. [Google Scholar] [CrossRef] [Green Version]
- Welch, R.A.; Burland, V.; Plunkett, G.; Redford, P.; Roesch, P.; Rasko, D.; Buckles, E.L.; Liou, S.R.; Boutin, A.; Hackett, J.; et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 2002, 99, 17020–17024. [Google Scholar] [CrossRef] [Green Version]
- Dobrindt, U.; Chowdary, M.G.; Krumbholz, G.; Hacker, J. Genome dynamics and its impact on evolution of Escherichia coli. Med. Microbiol. Immunol. 2010, 199, 145–154. [Google Scholar] [CrossRef]
- Klemm, P.; Hancock, V.; Schembri, M.A. Fimbrial adhesins from extraintestinal Escherichia coli. Environ. Microbiol. Rep. 2010, 2, 628–640. [Google Scholar] [CrossRef]
- Spurbeck, R.R.; Stapleton, A.; Johnson, J.R.; Walk, S.T.; Hooton, T.M.; Mobley, H.L.T. Fimbrial Profiles Predict Virulence of Uropathogenic Escherichia coli Strains: Contribution of Ygi and Yad Fimbriae. Infect. Immun. 2011, 79, 4753–4763. [Google Scholar] [CrossRef] [Green Version]
- Nowicki, B.; Rhen, M.; Väisänen-Rhen, V.; Pere, A.; Korhonen, T.K. Immunofluorescence study of fimbrial phase variation in Escherichia coli KS71. J. Bacteriol. 1984, 160, 691–695. [Google Scholar] [CrossRef] [Green Version]
- Snyder, J.A.; Haugen, B.J.; Lockatell, C.V.; Maroncle, N.; Hagan, E.C.; Johnson, D.E.; Welch, R.A.; Mobley, H.L.T. Coordinate Expression of Fimbriae in Uropathogenic Escherichia coli. Infect. Immun. 2005, 73, 7588–7596. [Google Scholar] [CrossRef] [Green Version]
- Epler Barbercheck, C.R.; Bullitt, E.; Andersson, M. Bacterial adhesion pili. In Membrane Protein Complexes: Structure and Function; Harris, J.R., Boekema, E.J., Eds.; Springer: Singapore, 2018; Volume 87, pp. 1–18. [Google Scholar]
- Ottow, J.C.G. Ecology, physiology, and genetics of fimbriae and pili. Annu. Rev. Microbiol. 1975, 29, 79–108. [Google Scholar] [CrossRef]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.G.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef]
- Teng, C.-H.; Cai, M.; Shin, S.; Xie, Y.; Kim, K.-J.; Khan, N.A.; Di Cello, F.; Kim, K.S. Escherichia coli K1 RS218 Interacts with Human Brain Microvascular Endothelial Cells via Type 1 Fimbria Bacteria in the Fimbriated State. Infect. Immun. 2005, 73, 2923–2931. [Google Scholar] [CrossRef] [Green Version]
- Pourbakhsh, S.A.; Boulianne, M.; Martineau-Doize, B.; Dozois, C.M.; Desautels, C.; Fairbrother, J.M. Dynamics of Escherichia coli Infection in Experimentally Inoculated Chickens. Avian Dis. 1997, 41, 221. [Google Scholar] [CrossRef]
- Dozois, C.M.; Chanteloup, N.; Dho-Moulin, M.; Bree, A.; Desautels, C.; Fairbrother, J.M. Bacterial Colonization and in vivo Expression of F1 (Type 1) Fimbrial Antigens in Chickens Experimentally Infected with Pathogenic Escherichia coli. Avian Dis. 1994, 38, 231. [Google Scholar] [CrossRef]
- Fronzes, R.; Remaut, H.; Waksman, G. Architectures and biogenesis of non-flagellar protein appendages in Gram-negative bacteria. EMBO J. 2008, 27, 2271–2280. [Google Scholar] [CrossRef] [Green Version]
- Soto, G.E.; Hultgren, S.J. Bacterial Adhesins: Common Themes and Variations in Architecture and Assembly. J. Bacteriol. 1999, 181, 1059–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busch, A.; Waksman, G. Chaperone–usher pathways: Diversity and pilus assembly mechanism. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 1112–1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, C.H.; Danese, P.N.; Pinkner, J.S.; Silhavy, T.; Hultgren, S.J. The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J. 1997, 16, 6394–6406. [Google Scholar] [CrossRef] [PubMed]
- Chahales, P.; Thanassi, D.G. Structure, Function, and Assembly of Adhesive Organelles by Uropathogenic Bacteria. Microbiol. Spectr. 2015, 3, 3–5. [Google Scholar] [CrossRef] [Green Version]
- Pan, K.-L.; Hsiao, H.-C.; Weng, C.-L.; Wu, M.-S.; Chou, C.P. Roles of DegP in Prevention of Protein Misfolding in the Periplasm upon Overexpression of Penicillin Acylase in Escherichia coli. J. Bacteriol. 2003, 185, 3020–3030. [Google Scholar] [CrossRef] [Green Version]
- Klemm, P.; Jørgensen, B.J.; Van Die, I.; De Ree, H.; Bergmans, H. The fim genes responsible for synthesis of type 1 fimbriae in Escherichia coli, cloning and genetic organization. Mol. Genet. Genom. 1985, 199, 410–414. [Google Scholar] [CrossRef]
- Klemm, P. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J. 1986, 5, 1389–1393. [Google Scholar] [CrossRef]
- Orndorff, P.E.; Falkow, S. Organization and expression of genes responsible for type 1 piliation in Escherichia coli. J. Bacteriol. 1984, 159, 736–744. [Google Scholar] [CrossRef] [Green Version]
- Jones, C.H.; Pinkner, J.; Roth, R.; Heuser, J.; Nicholes, A.V.; Abraham, S.N.; Hultgren, S.J. FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc. Natl. Acad. Sci. USA 1995, 92, 2081–2085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abraham, J.M.; Freitag, C.S.; Clements, J.R.; Eisenstein, B.I. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc. Natl. Acad. Sci. USA 1985, 82, 5724–5727. [Google Scholar] [CrossRef] [Green Version]
- Klemm, P.; Christiansen, G. Three fim genes required for the regulation of length and mediation of adhesion of Escherichia coli type 1 fimbriae. Mol. Genet. Genom. 1987, 208, 439–445. [Google Scholar] [CrossRef]
- Abraham, S.N.; Goguen, J.D.; Sun, D.; Klemm, P.; Beachey, E.H. Identification of two ancillary subunits of Escherichia coli type 1 fimbriae by using antibodies against synthetic oligopeptides of fim gene products. J. Bacteriol. 1987, 169, 5530–5536. [Google Scholar] [CrossRef] [Green Version]
- Valenski, M.L.; Harris, S.L.; Spears, P.A.; Horton, J.R.; Orndorff, P.E. The Product of the fimI Gene Is Necessary for Escherichia coli Type 1 Pilus Biosynthesis. J. Bacteriol. 2003, 185, 5007–5011. [Google Scholar] [CrossRef] [Green Version]
- Gally, D.L.; Leathart, J.; Blomfield, I.C. Interaction of FimB and FimE with thefimswitch that controls the phase variation of type 1 fimbriae in Escherichia coli K-12. Mol. Microbiol. 1996, 21, 725–738. [Google Scholar] [CrossRef]
- Giampapa, C.S.; Abraham, S.N.; Chiang, T.M.; Beachey, E.H. Isolation and characterization of a receptor for type 1 fimbriae of Escherichia coli from guinea pig erythrocytes. J. Biol. Chem. 1988, 263, 5362–5367. [Google Scholar] [CrossRef]
- Gottesman, S. Stress Reduction, Bacterial Style. J. Bacteriol. 2017, 199, e00433-17. [Google Scholar] [CrossRef] [Green Version]
- Hengge-Aronis, R. Back to log phase: σ S as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol. Microbiol. 1996, 21, 887–893. [Google Scholar] [CrossRef]
- Soutourina, O.; Kolb, A.; Krin, E.; Laurent-Winter, C.; Rimsky, S.; Danchin, A.; Bertin, P. Multiple Control of Flagellum Biosynthesis in Escherichia coli: Role of H-NS Protein and the Cyclic AMP-Catabolite Activator Protein Complex in Transcription of the flhDC Master Operon. J. Bacteriol. 1999, 181, 7500–7508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roesch, P.L.; Blomfield, I.C. Leucine alters the interaction of the leucine-responsive regulatory protein (Lrp) with thefimswitch to stimulate site-specific recombination in Escherichia coli. Mol. Microbiol. 1998, 27, 751–761. [Google Scholar] [CrossRef] [PubMed]
- Dalebroux, Z.D.; Svensson, S.L.; Gaynor, E.C.; Swanson, M.S. ppGpp Conjures Bacterial Virulence. Microbiol. Mol. Biol. Rev. 2010, 74, 171–199. [Google Scholar] [CrossRef] [Green Version]
- Lemke, J.J.; Durfee, T.; Gourse, R.L. DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol. Microbiol. 2009, 74, 1368–1379. [Google Scholar] [CrossRef] [Green Version]
- Foster, J.W. Escherichia coli acid resistance: Tales of an amateur acidophile. Nat. Rev. Genet. 2004, 2, 898–907. [Google Scholar] [CrossRef]
- Johnson, J.R.; Clabots, C.; Rosen, H. Effect of Inactivation of the Global Oxidative Stress Regulator oxyR on the Colonization Ability of Escherichia coli O1:K1:H7 in a Mouse Model of Ascending Urinary Tract Infection. Infect. Immun. 2006, 74, 461–468. [Google Scholar] [CrossRef] [Green Version]
- Hung, D.L.; Raivio, T.; Jones, C.; Silhavy, T.; Hultgren, S.J. Cpx signaling pathway monitors biogenesis and affects assembly and expression of P pili. EMBO J. 2001, 20, 1508–1518. [Google Scholar] [CrossRef]
- Andersen, J.; Forst, S.A.; Zhao, K.; Inouye, M.; Delihas, N. The function of micF RNA. J. Biol. Chem. 1989, 264, 17961–17970. [Google Scholar] [CrossRef]
- Porcheron, G.; Habib, R.; Houle, S.; Caza, M.; Lépine, F.; Daigle, F.; Massé, E.; Dozois, C.M. The Small RNA RyhB Contributes to Siderophore Production and Virulence of Uropathogenic Escherichia coli. Infect. Immun. 2014, 82, 5056–5068. [Google Scholar] [CrossRef] [Green Version]
- Opdyke, J.A.; Kang, J.-G.; Storz, G. GadY, a Small-RNA Regulator of Acid Response Genes in Escherichia coli. J. Bacteriol. 2004, 186, 6698–6705. [Google Scholar] [CrossRef] [Green Version]
- Bessaiah, H.; Pokharel, P.; Loucif, H.; Kulbay, M.; Sasseville, C.; Habouria, H.; Houle, S.; Bernier, J.; Massé, E.; Van Grevenynghe, J.; et al. The RyfA small RNA regulates oxidative and osmotic stress responses and virulence in uropathogenic Escherichia coli. PLoS Pathog. 2021, 17, e1009617. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, G.; Blankschien, M.; Herman, C.; Gross, C.A.; Rhodius, V.A. Regulon and promoter analysis of the E. coli heat-shock factor, σ32, reveals a multifaceted cellular response to heat stress. Genes Dev. 2006, 20, 1776–1789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Biase, D.; Tramonti, A.; Bossa, F.; Visca, P. The response to stationary-phase stress conditions in Escherichia coli: Role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 1999, 32, 1198–1211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolter, R.; Siegele, D.A.; Tormo, A. The stationary phase of the bacterial life cycle. Annu. Rev. Microbiol. 1993, 47, 855–874. [Google Scholar] [CrossRef]
- Gruber, T.M.; Gross, C.A. Multiple Sigma Subunits and the Partitioning of Bacterial Transcription Space. Annu. Rev. Microbiol. 2003, 57, 441–466. [Google Scholar] [CrossRef]
- Brown, L.; Gentry, D.; Elliott, T.; Cashel, M. DksA Affects ppGpp Induction of RpoS at a Translational Level. J. Bacteriol. 2002, 184, 4455–4465. [Google Scholar] [CrossRef] [Green Version]
- Kvint, K.; Farewell, A.; Nyström, T. RpoS-dependent Promoters Require Guanosine Tetraphosphate for Induction Even in the Presence of High Levels of ςs. J. Biol. Chem. 2000, 275, 14795–14798. [Google Scholar] [CrossRef] [Green Version]
- Girard, M.E.; Gopalkrishnan, S.; Grace, E.D.; Halliday, J.A.; Gourse, R.L.; Herman, C. DksA and ppGpp Regulate the σ S Stress Response by Activating Promoters for the Small RNA DsrA and the Anti-Adapter Protein IraP. J. Bacteriol. 2018, 200. [Google Scholar] [CrossRef] [Green Version]
- Schellhorn, H.E.; Hassan, H.M. Transcriptional regulation of katE in Escherichia coli K-12. J. Bacteriol. 1988, 170, 4286–4292. [Google Scholar] [CrossRef] [Green Version]
- Hryckowian, A.J.; Welch, R.A. RpoS Contributes to Phagocyte Oxidase-Mediated Stress Resistance during Urinary Tract Infection by Escherichia coli CFT073. mBio 2013, 4, e00023-13. [Google Scholar] [CrossRef] [Green Version]
- Tramonti, A.; Visca, P.; De Canio, M.; Falconi, M.; De Biase, D. Functional Characterization and Regulation of gadX, a Gene Encoding an AraC/XylS-Like Transcriptional Activator of the Escherichia coli Glutamic Acid Decarboxylase System. J. Bacteriol. 2002, 184, 2603–2613. [Google Scholar] [CrossRef] [Green Version]
- Sommerfeldt, N.; Possling, A.; Becker, G.; Pesavento, C.; Tschowri, N.; Hengge, R. Gene expression patterns and differential input into curli fimbriae regulation of all GGDEF/EAL domain proteins in Escherichia coli. Microbiology 2009, 155, 1318–1331. [Google Scholar] [CrossRef] [Green Version]
- Kulesus, R.R.; Diaz-Perez, K.; Slechta, E.S.; Eto, D.S.; Mulvey, M.A. Impact of the RNA Chaperone Hfq on the Fitness and Virulence Potential of Uropathogenic Escherichia coli. Infect. Immun. 2008, 76, 3019–3026. [Google Scholar] [CrossRef] [Green Version]
- Dong, T.; Schellhorn, H.E. Role of RpoS in Virulence of Pathogens. Infect. Immun. 2010, 78, 887–897. [Google Scholar] [CrossRef] [Green Version]
- Dove, S.L.; Smith, S.; Dorman, C. Control of Escherichia coli type 1 fimbrial gene expression in stationary phase: A negative role for RpoS. Mol. Genet. Genom. 1997, 254, 13–20. [Google Scholar] [CrossRef]
- Aberg, A.; Shingler, V.; Balsalobre, C. (p)ppGpp regulates type 1 fimbriation of Escherichia coli by modulating the expression of the site-specific recombinase FimB. Mol. Microbiol. 2006, 60, 1520–1533. [Google Scholar] [CrossRef]
- Müller, C.M.; Åberg, A.; Straseviçiene, J.; Emődy, L.; Uhlin, B.E.; Balsalobre, C. Type 1 Fimbriae, a Colonization Factor of Uropathogenic Escherichia coli, Are Controlled by the Metabolic Sensor CRP-cAMP. PLoS Pathog. 2009, 5, e1000303. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Gottesman, S. Modes of Regulation of RpoS by H-NS. J. Bacteriol. 2006, 188, 7022–7025. [Google Scholar] [CrossRef] [Green Version]
- Blomfield, I.C.; Kulasekara, D.H.; Eisenstein, B.I. Integration host factor stimulates both FimB- and FimE-mediated site-specific DNA inversion that controls phase variation of type 1 fimbriae expression in Escherichia coli. Mol. Microbiol. 1997, 23, 705–707. [Google Scholar] [CrossRef]
- Gally, D.; Bogan, J.A.; Eisenstein, B.I.; Blomfield, I.C. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: Effects of temperature and media. J. Bacteriol. 1993, 175, 6186–6193. [Google Scholar] [CrossRef] [Green Version]
- Olsen, P.B.; Schembri, M.A.; Gally, D.L.; Klemm, P. Differential temperature modulation by H-NS of thefimBandfimErecombinase genes which control the orientation of the type 1 fimbrial phase switch. FEMS Microbiol. Lett. 1998, 162, 17–23. [Google Scholar] [CrossRef] [Green Version]
- Blumer, C.; Kleefeld, A.; Lehnen, D.; Heintz, M.; Dobrindt, U.; Nagy, G.; Michaelis, K.; Emödy, L.; Polen, T.; Rachel, R.; et al. Regulation of type 1 fimbriae synthesis and biofilm formation by the transcriptional regulator LrhA of Escherichia coli. Microbiology 2005, 151, 3287–3298. [Google Scholar] [CrossRef] [Green Version]
- Matter, L.B.; Ares, M.; Abundes-Gallegos, J.; Cedillo, M.L.; Yáñez, J.A.; Martínez-Laguna, Y.; De la Cruz, M.A.; Girón, J.A. The CpxRA stress response system regulates virulence features of avian pathogenic Escherichia coli. Environ. Microbiol. 2018, 20, 3363–3377. [Google Scholar] [CrossRef]
- Herren, C.D.; Mitra, A.; Palaniyandi, S.K.; Coleman, A.; Elankumaran, S.; Mukhopadhyay, S. The BarA-UvrY Two-Component System Regulates Virulence in Avian Pathogenic Escherichia coli O78:K80:H9. Infect. Immun. 2006, 74, 4900–4909. [Google Scholar] [CrossRef] [Green Version]
- Pavanelo, D.; Houle, S.; Matter, L.B.; Dozois, C.M.; Horn, F. The Periplasmic Trehalase Affects Type 1 Fimbria Production and Virulence of Extraintestinal Pathogenic Escherichia coli Strain MT78. Infect. Immun. 2018, 86, e00241-18. [Google Scholar] [CrossRef] [Green Version]
- Conover, M.S.; Hadjifrangiskou, M.; Palermo, J.J.; Hibbing, M.E.; Dodson, K.W.; Hultgren, S.J. Metabolic Requirements of Escherichia coli in Intracellular Bacterial Communities during Urinary Tract Infection Pathogenesis. mBio 2016, 7, e00104-16. [Google Scholar] [CrossRef] [Green Version]
- Cortes, M.A.M.; Gibon, J.; Chanteloup, N.K.; Moulin-Schouleur, M.; Gilot, P.; Germon, P. Inactivation of ibeA and ibeT Results in Decreased Expression of Type 1 Fimbriae in Extraintestinal Pathogenic Escherichia coli Strain BEN2908. Infect. Immun. 2008, 76, 4129–4136. [Google Scholar] [CrossRef] [Green Version]
- Bessaiah, H.; Pokharel, P.; Habouria, H.; Houle, S.; Dozois, C.M. yqhG Contributes to Oxidative Stress Resistance and Virulence of Uropathogenic Escherichia coli and Identification of Other Genes Altering Expression of Type 1 Fimbriae. Front. Cell. Infect. Microbiol. 2019, 9, 312. [Google Scholar] [CrossRef]
- Bateman, S.L.; Seed, P.C. Epigenetic regulation of the nitrosative stress response and intracellular macrophage survival by extraintestinal pathogenic Escherichia coli. Mol. Microbiol. 2012, 83, 908–925. [Google Scholar] [CrossRef] [Green Version]
- Crépin, S.; Houle, S.; Charbonneau, M.; Mourez, M.; Harel, J.; Dozois, C.M. Decreased Expression of Type 1 Fimbriae by apstMutant of Uropathogenic Escherichia coli Reduces Urinary Tract Infection. Infect. Immun. 2012, 80, 2802–2815. [Google Scholar] [CrossRef] [Green Version]
- Rouquet, G.; Porcheron, G.; Barra, C.; Reépeérant, M.; Chanteloup, N.K.; Schouler, C.; Gilot, P. A Metabolic Operon in Extraintestinal Pathogenic Escherichia coli Promotes Fitness under Stressful Conditions and Invasion of Eukaryotic Cells. J. Bacteriol. 2009, 191, 4427–4440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurabayashi, K.; Agata, T.; Asano, H.; Tomita, H.; Hirakawa, H. Fur Represses Adhesion to, Invasion of, and Intracellular Bacterial Community Formation within Bladder Epithelial Cells and Motility in Uropathogenic Escherichia coli. Infect. Immun. 2016, 84, 3220–3231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eberly, A.R.; Floyd, K.; Beebout, C.; Colling, S.J.; Fitzgerald, M.J.; Stratton, C.W.; Schmitz, J.E.; Hadjifrangiskou, M. Biofilm Formation by Uropathogenic Escherichia coli Is Favored under Oxygen Conditions That Mimic the Bladder Environment. Int. J. Mol. Sci. 2017, 18, 2077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vila, J.; Soto, S.M. Salicylate increases the expression of marA and reduces in vitro biofilm formation in uropathogenic Escherichia coli by decreasing type 1 fimbriae expression. Virulence 2012, 3, 280–285. [Google Scholar] [CrossRef] [Green Version]
- Kostakioti, M.; Hadjifrangiskou, M.; Pinkner, J.S.; Hultgren, S.J. QseC-mediated dephosphorylation of QseB is required for expression of genes associated with virulence in uropathogenic Escherichia coli. Mol. Microbiol. 2009, 73, 1020–1031. [Google Scholar] [CrossRef] [Green Version]
- Gibson, K.E.; Silhavy, T.J. The LysR Homolog LrhA Promotes RpoS Degradation by Modulating Activity of the Response Regulator SprE. J. Bacteriol. 1999, 181, 563–571. [Google Scholar] [CrossRef] [Green Version]
- Brandi, A.; Giangrossi, M.; Fabbretti, A.; Falconi, M. The hns Gene of Escherichia coli Is Transcriptionally Down-Regulated by (p)ppGpp. Microorganisms 2020, 8, 1558. [Google Scholar] [CrossRef]
- Dorman, C.J. H-NS, the genome sentinel. Nat. Rev. Genet. 2006, 5, 157–161. [Google Scholar] [CrossRef]
- Atlung, T.; Ingmer, H. H-NS: A modulator of environmentally regulated gene expression. Mol. Microbiol. 1997, 24, 7–17. [Google Scholar] [CrossRef]
- Williams, R.M.; Rimsky, S. Molecular aspects of the E. coli nucleoid protein, H-NS: A central controller of gene regulatory networks. FEMS Microbiol. Lett. 1997, 156, 175–185. [Google Scholar] [CrossRef]
- Bertin, P.; Hommais, F.; Krin, E.; Soutourina, O.; Tendeng, C.; Derzelle, S.; Danchin, A. H-NS and H-NS-like proteins in Gram-negative bacteria andtheir multiple role in the regulation of bacterial metabolism. Biochimie 2001, 83, 235–241. [Google Scholar] [CrossRef]
- Krin, E.; Danchin, A.; Soutourina, O. Decrypting the H-NS-dependent regulatory cascade of acid stress resistance in Escherichia coli. BMC Microbiol. 2010, 10, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Hommais, F.; Krin, E.; Laurent-Winter, C.; Soutourina, O.; Malpertuy, A.; Le Caer, J.-P.; Danchin, A.; Bertin, P. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 2001, 40, 20–36. [Google Scholar] [CrossRef]
- Johansson, J.; Uhlin, B.E. Differential protease-mediated turnover of H-NS and StpA revealed by a mutation altering protein stability and stationary-phase survival of Escherichia coli. Proc. Natl. Acad. Sci. USA 1999, 96, 10776–10781. [Google Scholar] [CrossRef] [Green Version]
- White-Ziegler, C.A.; Davis, T.R. Genome-Wide Identification of H-NS-Controlled, Temperature-Regulated Genes in Escherichia coli K-12. J. Bacteriol. 2009, 191, 1106–1110. [Google Scholar] [CrossRef] [Green Version]
- Muüller, C.M.; Dobrindt, U.; Nagy, G.; Emoödy, L.; Uhlin, B.E.; Hacker, J. Role of Histone-Like Proteins H-NS and StpA in Expression of Virulence Determinants of Uropathogenic Escherichia coli. J. Bacteriol. 2006, 188, 5428–5438. [Google Scholar] [CrossRef] [Green Version]
- White-Ziegler, C.A.; Hill, M.L.A.; Braaten, B.A.; van der Woude, M.W.; Low, D.A. Thermoregulation of Escherichia coli pap transcription: H-NS is a temperature-dependent DNA methylation blocking factor. Mol. Microbiol. 1998, 28, 1121–1137. [Google Scholar] [CrossRef] [Green Version]
- Göransson, M.; Sondén, B.; Nilsson, P.; Dagberg, B.; Foreman, K.; Emanuelsson, K.; Uhlin, B.E. Transcriptional silencing and thermoregulation of gene expression in Escherichia coli. Nature 1990, 344, 682–685. [Google Scholar] [CrossRef]
- Brambilla, E.; Sclavi, B. Gene Regulation by H-NS as a Function of Growth Conditions Depends on Chromosomal Position in Escherichia coli. G3 Genes Genomes Genet. 2015, 5, 605–614. [Google Scholar] [CrossRef] [Green Version]
- Oshima, T.; Ito, K.; Kabayama, H.; Nakamura, Y. Regulation of Irp gene expression by H-NS and Lrp proteins in Escherichia coli: Dominant negative mutations in Irp. Mol. Genet. Genom. 1995, 247, 521–528. [Google Scholar] [CrossRef]
- Calvo, J.M.; Matthews, R.G. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 1994, 58, 466–490. [Google Scholar] [CrossRef]
- Ernsting, B.R.; Atkinson, M.R.; Ninfa, A.J.; Matthews, R.G. Characterization of the regulon controlled by the leucine-responsive regulatory protein in Escherichia coli. J. Bacteriol. 1992, 174, 1109–1118. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Wu, J.; Friedberg, D.; Plakto, J.; Calvo, J.M. Regulation of the Escherichia coli lrp gene. J. Bacteriol. 1994, 176, 1831–1839. [Google Scholar] [CrossRef] [Green Version]
- Blomfield, I.C.; Calie, P.J.; Eberhardt, K.J.; McClain, M.; Eisenstein, B.I. Lrp stimulates phase variation of type 1 fimbriation in Escherichia coli K-12. J. Bacteriol. 1993, 175, 27–36. [Google Scholar] [CrossRef] [Green Version]
- Cho, B.-K.; Barrett, C.L.; Knight, E.M.; Park, Y.S.; Palsson, B. Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli. Proc. Natl. Acad. Sci. USA 2008, 105, 19462–19467. [Google Scholar] [CrossRef] [Green Version]
- Kroner, G.M.; Wolfe, M.B.; Freddolino, P.L. Escherichia coli Lrp Regulates One-Third of the Genome via Direct, Cooperative, and Indirect Routes. J. Bacteriol. 2019, 201, e00411-18. [Google Scholar] [CrossRef] [Green Version]
- Newman, E.B.; Lin, R. Leucine-responsive regulatory protein: A Global Regulator of Gene Expression in E. Coli. Annu. Rev. Microbiol. 1995, 49, 747–775. [Google Scholar] [CrossRef]
- Hart, B.R.; Blumenthal, R.M. Unexpected Coregulator Range for the Global Regulator Lrp of Escherichia coli and Proteus mirabilis. J. Bacteriol. 2011, 193, 1054–1064. [Google Scholar] [CrossRef] [Green Version]
- Landgraf, J.R.; Wu, J.; Calvo, J.M. Effects of nutrition and growth rate on Lrp levels in Escherichia coli. J. Bacteriol. 1996, 178, 6930–6936. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.Y.; Lim, C.J.; Droge, P.; Yan, J. Regulation of Bacterial DNA Packaging in Early Stationary Phase by Competitive DNA Binding of Dps and IHF. Sci. Rep. 2015, 5, 18146. [Google Scholar] [CrossRef] [Green Version]
- Neidhardt, F.C.; Curtiss, R. Escherichia coli and Salmonella: Cellular and Molecular Biology, ed., 2nd ed.; American Society for Micro-Biology: Washington, DC, USA, 1996. [Google Scholar]
- Gourse, R.L.; Chen, A.Y.; Gopalkrishnan, S.; Sanchez-Vazquez, P.; Myers, A.; Ross, W. Transcriptional Responses to ppGpp and DksA. Annu. Rev. Microbiol. 2018, 72, 163–184. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Vazquez, P.; Dewey, C.N.; Kitten, N.; Ross, W.; Gourse, R.L. Genome-wide effects on Escherichia coli transcription from ppGpp binding to its two sites on RNA polymerase. Proc. Natl. Acad. Sci. USA 2019, 116, 8310–8319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abranches, J.; Martinez, A.R.; Kajfasz, J.K.; Chaévez, V.; Garsin, D.A.; Lemos, J.A. The Molecular Alarmone (p)ppGpp Mediates Stress Responses, Vancomycin Tolerance, and Virulence in Enterococcus faecalis. J. Bacteriol. 2009, 191, 2248–2256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wells, D.H.; Gaynor, E.C. Helicobacter pylori Initiates the Stringent Response upon Nutrient and pH Downshift. J. Bacteriol. 2006, 188, 3726–3729. [Google Scholar] [CrossRef] [Green Version]
- Gentry, D.R.; Hernandez, V.J.; Nguyen, L.H.; Jensen, D.B.; Cashel, M. Synthesis of the stationary-phase sigma factor sigma s is positively regulated by ppGpp. J. Bacteriol. 1993, 175, 7982–7989. [Google Scholar] [CrossRef] [Green Version]
- Cashel, M.; Gallant, J. Two Compounds implicated in the Function of the RC Gene of Escherichia coli. Nature 1969, 221, 838–841. [Google Scholar] [CrossRef]
- Gallant, J.A. Stringent control in E. coli. Annu. Rev. Genet. 1979, 13, 393–415. [Google Scholar] [CrossRef]
- Godfrey, H. The role of the stringent response in the pathogenesis of bacterial infections. Trends Microbiol. 2002, 10, 349–351. [Google Scholar] [CrossRef]
- Magnusson, L.U.; Farewell, A.; Nyström, T. ppGpp: A global regulator in Escherichia coli. Trends Microbiol. 2005, 13, 236–242. [Google Scholar] [CrossRef]
- Germain, E.; Guiraud, P.; Byrne, D.; Douzi, B.; Djendli, M.; Maisonneuve, E. YtfK activates the stringent response by triggering the alarmone synthetase SpoT in Escherichia coli. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Traxler, M.F.; Summers, S.M.; Nguyen, H.-T.; Zacharia, V.M.; Hightower, G.A.; Smith, J.T.; Conway, T. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 2008, 68, 1128–1148. [Google Scholar] [CrossRef] [Green Version]
- Durfee, T.; Hansen, A.-M.; Zhi, H.; Blattner, F.R.; Jin, D.J. Transcription Profiling of the Stringent Response in Escherichia coli. J. Bacteriol. 2008, 190, 1084–1096. [Google Scholar] [CrossRef] [Green Version]
- Botsford, J.L.; Harman, J.G. Cyclic AMP in prokaryotes. Microbiol. Rev. 1992, 56, 100–122. [Google Scholar] [CrossRef]
- McDonough, K.A.; Rodriguez, A. The myriad roles of cyclic AMP in microbial pathogens: From signal to sword. Nat. Rev. Genet. 2011, 10, 27–38. [Google Scholar] [CrossRef] [Green Version]
- Kalia, D.; Merey, G.; Nakayama, S.; Zheng, Y.; Zhou, J.; Luo, Y.; Guo, M.; Roembke, B.T.; Sintim, H.O. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 2012, 42, 305–341. [Google Scholar] [CrossRef]
- Kolb, A.; Busby, S.; Buc, H.; Garges, S.; Adhya, S. Transcriptional regulation by camp and its receptor protein. Annu. Rev. Biochem. 1993, 62, 749–797. [Google Scholar] [CrossRef]
- Görke, B.; Stülke, J. Carbon catabolite repression in bacteria: Many ways to make the most out of nutrients. Nat. Rev. Genet. 2008, 6, 613–624. [Google Scholar] [CrossRef]
- Brã¼Ckner, R.; Titgemeyer, F. Carbon catabolite repression in bacteria: Choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 2002, 209, 141–148. [Google Scholar] [CrossRef] [Green Version]
- Fong, J.C.N.; Yildiz, F.H. Interplay between Cyclic AMP-Cyclic AMP Receptor Protein and Cyclic di-GMP Signaling in Vibrio cholerae Biofilm Formation. J. Bacteriol. 2008, 190, 6646–6659. [Google Scholar] [CrossRef] [Green Version]
- Gosset, G.; Zhang, Z.; Nayyar, S.; Cuevas, W.A.; Saier, M.H. Transcriptome Analysis of Crp-Dependent Catabolite Control of Gene Expression in Escherichia coli. J. Bacteriol. 2004, 186, 3516–3524. [Google Scholar] [CrossRef] [Green Version]
- Castanie-Cornet, M.-P.; Penfound, T.A.; Smith, D.; Elliott, J.F.; Foster, J.W. Control of Acid Resistance in Escherichia coli. J. Bacteriol. 1999, 181, 3525–3535. [Google Scholar] [CrossRef] [Green Version]
- Barth, E.; Gora, K.V.; Gebendorfer, K.M.; Settele, F.; Jakob, U.; Winter, J. Interplay of cellular cAMP levels, σ S activity and oxidative stress resistance in Escherichia coli. Microbiology 2009, 155, 1680–1689. [Google Scholar] [CrossRef]
- Molina-Quiroz, R.C.; Silva, C.A.; Brewster, J.; Castro-Nallar, E.; Levy, S.B.; Camilli, A. Cyclic AMP Regulates Bacterial Persistence through Repression of the Oxidative Stress Response and SOS-Dependent DNA Repair in Uropathogenic Escherichia coli. mBio 2018, 9, e02144-17. [Google Scholar] [CrossRef] [Green Version]
- Balsalobre, C.; Johansson, J.; Uhlin, B.E. Cyclic AMP-Dependent Osmoregulation of crp Gene Expression in Escherichia coli. J. Bacteriol. 2006, 188, 5935–5944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishizuka, H.; Hanamura, A.; Kunimura, T.; Aiba, H. A lowered concentration of cAMP receptor protein caused by glucose is an important determinant for catabolite repression in Escherichia coli. Mol. Microbiol. 1993, 10, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Hengge-Aronis, R. Signal Transduction and Regulatory Mechanisms Involved in Control of the σ S (RpoS) Subunit of RNA Polymerase. Microbiol. Mol. Biol. Rev. 2002, 66, 373–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donovan, G.T.; Norton, J.P.; Bower, J.M.; Mulvey, M.A. Adenylate Cyclase and the Cyclic AMP Receptor Protein Modulate Stress Resistance and Virulence Capacity of Uropathogenic Escherichia coli. Infect. Immun. 2012, 81, 249–258. [Google Scholar] [CrossRef] [Green Version]
- Holmqvist, E.; Wagner, E.G.H. Impact of bacterial sRNAs in stress responses. Biochem. Soc. Trans. 2017, 45, 1203–1212. [Google Scholar] [CrossRef] [Green Version]
- Hoe, C.-H.; Raabe, C.A.; Rozhdestvensky, T.S.; Tang, T.-H. Bacterial sRNAs: Regulation in stress. Int. J. Med. Microbiol. 2013, 303, 217–229. [Google Scholar] [CrossRef]
- Richards, G.R.; Vanderpool, C.K. Molecular call and response: The physiology of bacterial small RNAs. Biochim. Biophys. Acta (BBA) Bioenerg. 2011, 1809, 525–531. [Google Scholar] [CrossRef] [Green Version]
- Fröhlich, K.S.; Gottesman, S. Small Regulatory RNAs in the Enterobacterial Response to Envelope Damage and Oxidative Stress. Microbiol. Spectr. 2018, 6, 211–228. [Google Scholar] [CrossRef]
- Raghavan, R.; Groisman, E.A.; Ochman, H. Genome-wide detection of novel regulatory RNAs in E. coli. Genome Res. 2011, 21, 1487–1497. [Google Scholar] [CrossRef] [Green Version]
- Masseé, E.; Vanderpool, C.K.; Gottesman, S. Effect of RyhB Small RNA on Global Iron Use in Escherichia coli. J. Bacteriol. 2005, 187, 6962–6971. [Google Scholar] [CrossRef] [Green Version]
- Yuzawa, H.; Nagai, H.; Mori, H.; Yura, T. Heat induction of θ32synthesis mediated by mRNA secondary structure: A primary step of the heat shock response in Escherichia coli. Nucleic Acids Res. 1993, 21, 5449–5455. [Google Scholar] [CrossRef]
- Guisbert, E.; Yura, T.; Rhodius, V.A.; Gross, C.A. Convergence of Molecular, Modeling, and Systems Approaches for an Understanding of the Escherichia coli Heat Shock Response. Microbiol. Mol. Biol. Rev. 2008, 72, 545–554. [Google Scholar] [CrossRef] [Green Version]
- Ritossa, F. Discovery of the heat shock response. Cell Stress Chaperones 1996, 1, 97–98. [Google Scholar] [CrossRef] [Green Version]
- Bukau, B.; Horwich, A.L. The Hsp70 and Hsp60 Chaperone Machines. Cell 1998, 92, 351–366. [Google Scholar] [CrossRef] [Green Version]
- Morimoto, R.I. Regulation of the heat shock transcriptional response: Cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998, 12, 3788–3796. [Google Scholar] [CrossRef] [Green Version]
- Guisbert, E.; Herman, C.; Lu, C.Z.; Gross, C.A. A chaperone network controls the heat shock response in E. coli. Genes Dev. 2004, 18, 2812–2821. [Google Scholar] [CrossRef] [Green Version]
- Arsène, F.; Tomoyasu, T.; Bukau, B. The heat shock response of Escherichia coli. Int. J. Food Microbiol. 2000, 55, 3–9. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Tomoyasu, T.; Takaya, A.; Morioka, M.; Yamamoto, T. Effects of disruption of heat shock genes on susceptibility of Escherichia coli to fluoroquinolones. BMC Microbiol. 2003, 3, 16. [Google Scholar] [CrossRef] [Green Version]
- Silhavy, T.J.; Kahne, D.; Walker, S. The Bacterial Cell Envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Fang, F.C.; Frawley, E.R.; Tapscott, T.; Vázquez-Torres, A. Bacterial Stress Responses during Host Infection. Cell Host Microbe 2016, 20, 133–143. [Google Scholar] [CrossRef] [Green Version]
- Raivio, T.L. MicroReview: Envelope stress responses and Gram-negative bacterial pathogenesis. Mol. Microbiol. 2005, 56, 1119–1128. [Google Scholar] [CrossRef]
- Delhaye, A.; Collet, J.-F.; Laloux, G. A Fly on the Wall: How Stress Response Systems Can Sense and Respond to Damage to Peptidoglycan. Front. Cell. Infect. Microbiol. 2019, 9, 380. [Google Scholar] [CrossRef]
- Hunke, S.; Keller, R.; Müller, V.S. Signal integration by the Cpx-envelope stress system. FEMS Microbiol. Lett. 2011, 326, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Raivio, T.L.; Leblanc, S.K.D.; Price, N.L. The Escherichia coli Cpx Envelope Stress Response Regulates Genes of Diverse Function That Impact Antibiotic Resistance and Membrane Integrity. J. Bacteriol. 2013, 195, 2755–2767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landini, P. Cross-talk mechanisms in biofilm formation and responses to environmental and physiological stress in Escherichia coli. Res. Microbiol. 2009, 160, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Debnath, I.; Norton, J.P.; Barber, A.E.; Ott, E.M.; Dhakal, B.K.; Kulesus, R.R.; Mulvey, M.A. The Cpx Stress Response System Potentiates the Fitness and Virulence of Uropathogenic Escherichia coli. Infect. Immun. 2013, 81, 1450–1459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taschner, N.P.; Yagil, E.; Spira, B. A differential effect of σ S on the expression of the PHO regulon genes of Escherichia coli. Microbiology 2004, 150, 2985–2992. [Google Scholar] [CrossRef] [Green Version]
- Teng, C.-H.; Xie, Y.; Shin, S.; Di Cello, F.; Paul-Satyaseela, M.; Cai, M.; Kim, K.S. Effects of ompA Deletion on Expression of Type 1 Fimbriae in Escherichia coli K1 Strain RS218 and on the Association of E. coli with Human Brain Microvascular Endothelial Cells. Infect. Immun. 2006, 74, 5609–5616. [Google Scholar] [CrossRef] [Green Version]
- Miller, R.A.; Britigan, B.E. Role of oxidants in microbial pathophysiology. Clin. Microbiol. Rev. 1997, 10, 1–18. [Google Scholar] [CrossRef]
- Fang, F.C. Antimicrobial reactive oxygen and nitrogen species: Concepts and controversies. Nat. Rev. Genet. 2004, 2, 820–832. [Google Scholar] [CrossRef]
- Schwan, W.R.; Lee, J.L.; Lenard, F.A.; Matthews, B.T.; Beck, M.T. Osmolarity and pH Growth Conditions Regulate fim Gene Transcription and Type 1 Pilus Expression in Uropathogenic Escherichia coli. Infect. Immun. 2002, 70, 1391–1402. [Google Scholar] [CrossRef] [Green Version]
- Snyder, J.A.; Haugen, B.J.; Buckles, E.L.; Lockatell, C.V.; Johnson, D.E.; Donnenberg, M.S.; Welch, R.A.; Mobley, H.L.T. Transcriptome of Uropathogenic Escherichia coli during Urinary Tract Infection. Infect. Immun. 2004, 72, 6373–6381. [Google Scholar] [CrossRef] [Green Version]
- Withman, B.; Gunasekera, T.S.; Beesetty, P.; Agans, R.; Paliy, O. Transcriptional Responses of Uropathogenic Escherichia coli to Increased Environmental Osmolality Caused by Salt or Urea. Infect. Immun. 2012, 81, 80–89. [Google Scholar] [CrossRef] [Green Version]
- Greene, S.E.; Hibbing, M.E.; Janetka, J.; Chen, S.; Hultgren, S.J. Human Urine Decreases Function and Expression of Type 1 Pili in Uropathogenic Escherichia coli. mBio 2015, 6, e00820-15. [Google Scholar] [CrossRef] [Green Version]
- Greenberg, J.; Monach, P.; Chou, J.; Josephy, P.D.; Demple, B. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc. Natl. Acad. Sci. USA 1990, 87, 6181–6185. [Google Scholar] [CrossRef] [Green Version]
- Demple, B. Regulation of bacterial oxidative stress genes. Annu. Rev. Genet. 1991, 25, 315–337. [Google Scholar] [CrossRef]
- Imlay, J.A. Cellular Defenses against Superoxide and Hydrogen Peroxide. Annu. Rev. Biochem. 2008, 77, 755–776. [Google Scholar] [CrossRef] [Green Version]
- Zheng, M.; Storz, G. Redox sensing by prokaryotic transcription factors. Biochem. Pharmacol. 1999, 59, 1–6. [Google Scholar] [CrossRef]
- Farr, S.B.; Kogoma, T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol. Rev. 1991, 55, 561–585. [Google Scholar] [CrossRef]
- Nunoshiba, T.; Hidalgo, E.; Cuevas, C.F.A.; Demple, B. Two-stage control of an oxidative stress regulon: The Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J. Bacteriol. 1992, 174, 6054–6060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, M.; Wang, X.; Templeton, L.J.; Smulski, D.R.; LaRossa, R.A.; Storz, G. DNA Microarray-Mediated Transcriptional Profiling of the Escherichia coli Response to Hydrogen Peroxide. J. Bacteriol. 2001, 183, 4562–4570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, S.M.; Schellhorn, H.E. Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Arch. Biochem. Biophys. 2012, 525, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Lee, K.-L.; Yeo, W.-S.; Park, S.-J.; Roe, J.-H. SoxRS-Mediated Lipopolysaccharide Modification Enhances Resistance against Multiple Drugs in Escherichia coli. J. Bacteriol. 2009, 191, 4441–4450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, J.R.; Russo, T.A.; Drawz, S.M.; Clabots, C.; Olson, R.; Kuskowski, M.A.; Rosen, H. OxyR contributes to the virulence of a Clonal Group A Escherichia coli strain (O17:K+:H18) in animal models of urinary tract infection, subcutaneous infection, and systemic sepsis. Microb. Pathog. 2013, 64, 1–5. [Google Scholar] [CrossRef]
- Fléchard, M.; Cortes, M.A.M.; Répérant, M.; Germon, P. New Role for the ibeA Gene in H2O2 Stress Resistance of Escherichia coli. J. Bacteriol. 2012, 194, 4550–4560. [Google Scholar] [CrossRef] [Green Version]
- Hannan, T.J.; Mysorekar, I.U.; Chen, S.L.; Walker, J.N.; Jones, J.M.; Pinkner, J.S.; Hultgren, S.J.; Seed, P.C. LeuX tRNA-dependent and -independent mechanisms of Escherichia coli pathogenesis in acute cystitis. Mol. Microbiol. 2007, 67, 116–128. [Google Scholar] [CrossRef] [Green Version]
- Bertrand, N.; Houle, S.; LeBihan, G.; Poirier, E.; Dozois, C.M.; Harel, J. Increased Pho Regulon Activation Correlates with Decreased Virulence of an Avian Pathogenic Escherichia coli O78 Strain. Infect. Immun. 2010, 78, 5324–5331. [Google Scholar] [CrossRef] [Green Version]
- Lamarche, M.G.; Wanner, B.L.; Crépin, S.; Harel, J. The phosphate regulon and bacterial virulence: A regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol. Rev. 2008, 32, 461–473. [Google Scholar] [CrossRef] [Green Version]
- Crépin, S.; Lamarche, M.G.; Garneau, P.; Séguin, J.; Proulx, J.; Dozois, C.M.; Harel, J. Genome-wide transcriptional response of an avian pathogenic Escherichia coli (APEC) pst mutant. BMC Genom. 2008, 9, 568. [Google Scholar] [CrossRef] [Green Version]
- Becker, K.; Skaar, E.P. Metal limitation and toxicity at the interface between host and pathogen. FEMS Microbiol. Rev. 2014, 38, 1235–1249. [Google Scholar] [CrossRef] [Green Version]
- Klemm, P.; Schembri, M. Type 1 Fimbriae, Curli, and Antigen 43: Adhesion, Colonization, and Biofilm Formation. EcoSal Plus 2004, 1. [Google Scholar] [CrossRef]
- Rasko, D.A.; Moreira, C.G.; Li, D.R.; Reading, N.C.; Ritchie, J.M.; Waldor, M.K.; Williams, N.; Taussig, R.; Wei, S.; Roth, M.; et al. Targeting QseC Signaling and Virulence for Antibiotic Development. Science 2008, 321, 1078–1080. [Google Scholar] [CrossRef] [Green Version]
- Schwan, W.R.; Beck, M.T.; Hung, C.S.; Hultgren, S.J. Differential Regulation of Escherichia coli fimGenes following Binding to Mannose Receptors. J. Pathog. 2018, 2018, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Rentschler, A.E.; Lovrich, S.D.; Fitton, R.; Enos-Berlage, J.; Schwan, W.R. OmpR regulation of the uropathogenic Escherichia coli fimB gene in an acidic/high osmolality environment. Microbiology 2013, 159, 316–327. [Google Scholar] [CrossRef] [Green Version]
- Tchesnokova, V.; Aprikian, P.; Kisiela, D.; Gowey, S.; Korotkova, N.; Thomas, W.; Sokurenko, E. Type 1 Fimbrial Adhesin FimH Elicits an Immune Response That Enhances Cell Adhesion of Escherichia coli. Infect. Immun. 2011, 79, 3895–3904. [Google Scholar] [CrossRef] [Green Version]
- Ashkar, A.A.; Mossman, K.L.; Coombes, B.K.; Gyles, C.L.; Mackenzie, R. FimH Adhesin of Type 1 Fimbriae Is a Potent Inducer of Innate Antimicrobial Responses Which Requires TLR4 and Type 1 Interferon Signalling. PLoS Pathog. 2008, 4, e1000233. [Google Scholar] [CrossRef]
- Mian, M.F.; Lauzon, N.M.; Andrews, D.; Lichty, B.D.; Ashkar, A.A. FimH Can Directly Activate Human and Murine Natural Killer Cells via TLR4. Mol. Ther. 2010, 18, 1379–1388. [Google Scholar] [CrossRef]
- Wang, L.; Keatch, R.; Zhao, Q.; Wright, J.A.; Bryant, C.E.; Redmann, A.L.; Terentjev, E.M. Influence of Type I Fimbriae and Fluid Shear Stress on Bacterial Behavior and Multicellular Architecture of Early Escherichia coli Biofilms at Single-Cell Resolution. Appl. Environ. Microbiol. 2018, 84, e02343-17. [Google Scholar] [CrossRef] [Green Version]
- Thomas, W.E.; Nilsson, L.M.; Forero, M.; Sokurenko, E.V.; Vogel, V. Shear-dependent ‘stick-and-roll’ adhesion of type 1 fimbriated Escherichia coli. Mol. Microbiol. 2004, 53, 1545–1557. [Google Scholar] [CrossRef]
- Eisenstein, B.I. Phase Variation of Type 1 Fimbriae in Escherichia coli Is Under Transcriptional Control. Science 1981, 214, 337–339. [Google Scholar] [CrossRef]
- Sohanpal, B.K.; El-Labany, S.; Lahooti, M.; Plumbridge, J.A.; Blomfield, I.C. Integrated regulatory responses of fimB to N-acetylneuraminic (sialic) acid and GlcNAc in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 2004, 101, 16322–16327. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Outten, F.W. IscR Controls Iron-Dependent Biofilm Formation in Escherichia coli by Regulating Type I Fimbria Expression. J. Bacteriol. 2009, 191, 1248–1257. [Google Scholar] [CrossRef] [Green Version]
- McVicker, G.; Sun, L.; Sohanpal, B.K.; Gashi, K.; Williamson, R.A.; Plumbridge, J.; Blomfield, I.C. SlyA Protein Activates fimB Gene Expression and Type 1 Fimbriation in Escherichia coli K-12. J. Biol. Chem. 2011, 286, 32026–32035. [Google Scholar] [CrossRef] [Green Version]
- Sohanpal, B.K.; Friar, S.; Roobol, J.; Plumbridge, J.A.; Blomfield, I.C. Multiple co-regulatory elements and IHF are necessary for the control of fimB expression in response to sialic acid and N-acetylglucosamine in Escherichia coli K-12. Mol. Microbiol. 2007, 63, 1223–1236. [Google Scholar] [CrossRef]
- Liu, Y.; Han, R.; Wang, J.; Yang, P.; Wang, F.; Yang, B. Magnesium Sensing Regulates Intestinal Colonization of Enterohemorrhagic Escherichia coli O157:H7. mBio 2020, 11, e02470-20. [Google Scholar] [CrossRef]
- Nakashima, N.; Goh, S.; Good, L.; Tamura, T. Multiple-Gene Silencing Using Antisense RNAs in Escherichia coli; Springer: New York, NY, USA, 2011; pp. 307–319. [Google Scholar] [CrossRef]
- Nakashima, N.; Tamura, T.; Good, L. Paired termini stabilize antisense RNAs and enhance conditional gene silencing in Escherichia coli. Nucleic Acids Res. 2006, 34, e138. [Google Scholar] [CrossRef]
- Magistro, G.; Magistro, C.; Stief, C.G.; Schubert, S. A simple and highly efficient method for gene silencing in Escherichia coli. J. Microbiol. Methods 2018, 154, 25–32. [Google Scholar] [CrossRef]

| Regulator | Stress Response | Role in Virulence | Reference |
|---|---|---|---|
| RpoS | Nutrient deprivation | Master regulator of stress | [41] |
| H-NS | Temperature | Regulates flagellar gene expression and fim and pap operon and many other genes | [42] |
| Lrp | Nutrient deprivation | Required for fim and pap fimbriae | [43] |
| ppGpp | Stringent response | Involved in biofilm formation and production of flagella | [44,45] |
| cAMP | Nutrient deprivation | Required for acid stress response, regulation of multiple virulence factors | [46] |
| SoxS/R and OxyR | Oxidative stress | Required for virulence in UPEC | [47] |
| CpxRA | Membrane damage | Required for type 1 and P fimbriae expression in UPEC | [48] |
| sRNA | Diverse | MicF regulates gene expression for the outer membrane | [49] |
| RyhB is required for nutrient stress/iron homeostasis | [50] | ||
| GadY is required for acid stress resistance | [51] | ||
| RyfA is required for survival in human macrophages, resistance to multiple stresses | [52] | ||
| RpoH | Heat shock | Regulates gene expression in heat shock | [53] |
| Regulator | Switch | FimE | FimB | Effect on Fim Expression | Reference |
|---|---|---|---|---|---|
| General and specific stress regulators | |||||
| IHF | Switching on fimS | Positive or negative 1 | [70] | ||
| Lrp | +/− | +/− | Positive or negative 1 | [71] | |
| H-NS | − | <37 °C: − >37 °C: + | <37 °C: Negative >37 °C: Positive | [72] | |
| RpoS | − | Negative | [66] | ||
| LrhA | + | Negative | [73] | ||
| ppGpp | − | Negative | [67] | ||
| cAMP | − | Negative | [68] | ||
| Envelope stress | |||||
| CpxR-P | Regulates the inversion | - | Negative | [74] | |
| BarA/UvrY | Reduction of fimA | Unknown | Unknown 1 | [75] | |
| Oxidative and osmotic stress | |||||
| TreA | Unknown | Unknown | Positive | [76] | |
| YeaR | Unknown | Unknown | Positive? 1 | [77] | |
| IbeA | + ? | + ? | Positive? 1 | [78] | |
| YqhG | Unknown | Unknown | Positive? 1 | [79] | |
| RyfA | Unknown | Unknown | Positive? 1 | [52] | |
| Nitrosative stress | |||||
| FimX | Unknown | Unknown | Positive? | [80] | |
| Nutrient limitation and oxygenation | |||||
| Pst and Pho regulon | + | − | Negative | [81] | |
| Frz | Unknown | Unknown | Positive | [82] | |
| Fur | Increased fimA | Unknown | Positive | [83] | |
| Oxygenation | Unknown | Unknown | Positive | [84] | |
| Biofilm and quorum sensing | |||||
| Effect of salicylate on marA | − | Negative | [85] | ||
| QseC/B | Unknown | Unknown | Positive | [86] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bessaiah, H.; Anamalé, C.; Sung, J.; Dozois, C.M. What Flips the Switch? Signals and Stress Regulating Extraintestinal Pathogenic Escherichia coli Type 1 Fimbriae (Pili). Microorganisms 2022, 10, 5. https://doi.org/10.3390/microorganisms10010005
Bessaiah H, Anamalé C, Sung J, Dozois CM. What Flips the Switch? Signals and Stress Regulating Extraintestinal Pathogenic Escherichia coli Type 1 Fimbriae (Pili). Microorganisms. 2022; 10(1):5. https://doi.org/10.3390/microorganisms10010005
Chicago/Turabian StyleBessaiah, Hicham, Carole Anamalé, Jacqueline Sung, and Charles M. Dozois. 2022. "What Flips the Switch? Signals and Stress Regulating Extraintestinal Pathogenic Escherichia coli Type 1 Fimbriae (Pili)" Microorganisms 10, no. 1: 5. https://doi.org/10.3390/microorganisms10010005








