Hastening Progress in Cyclospora Requires Studying Eimeria Surrogates
Abstract
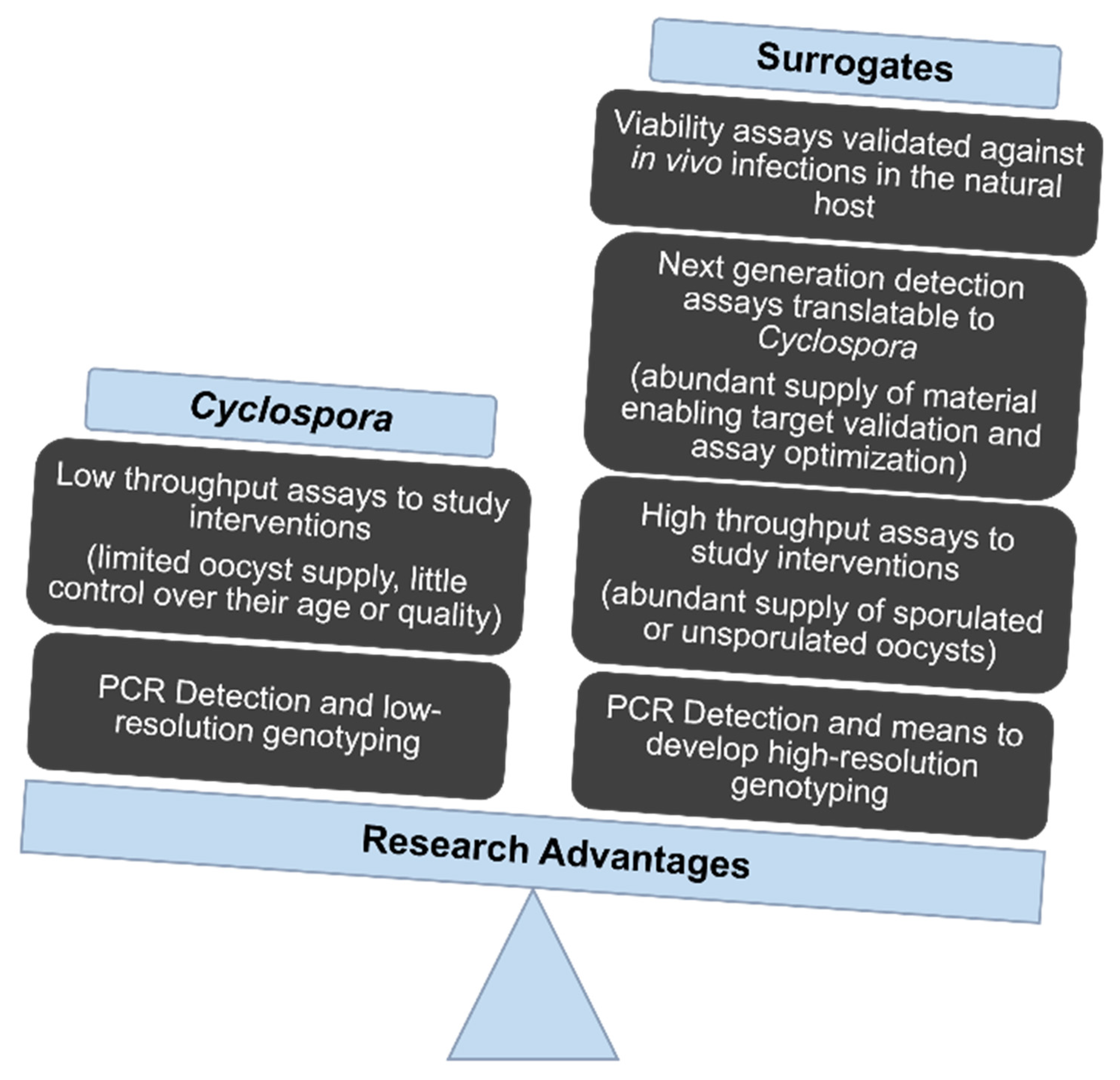
:1. Cyclospora: Simple to Understand, but Difficult to Study
- Specificity.
- Speed.
- Cost effectiveness.
- Means to assess parasite viability.
2. The Scarcity of Oocysts Slows Research Progress
- Filtration systems to remove oocysts from irrigation or water in processing plants.
- Detection.
- Sanitizing systems, including chemical and physical treatments, for fresh produce and food processing equipment.
- Cheaper, faster surveillance tools.
- More specific and sensitive diagnostic tools.
- Viability assays, to better determine food safety risk.
3. Suitable Animal Models Hasten Progress
3.1. Researchers Presently Lack a Species of Cyclospora That Cycles in an Established Laboratory animal Model
3.2. Eimeria That Infect Poultry Provide Especially Useful Surrogates for C. cayetanensis
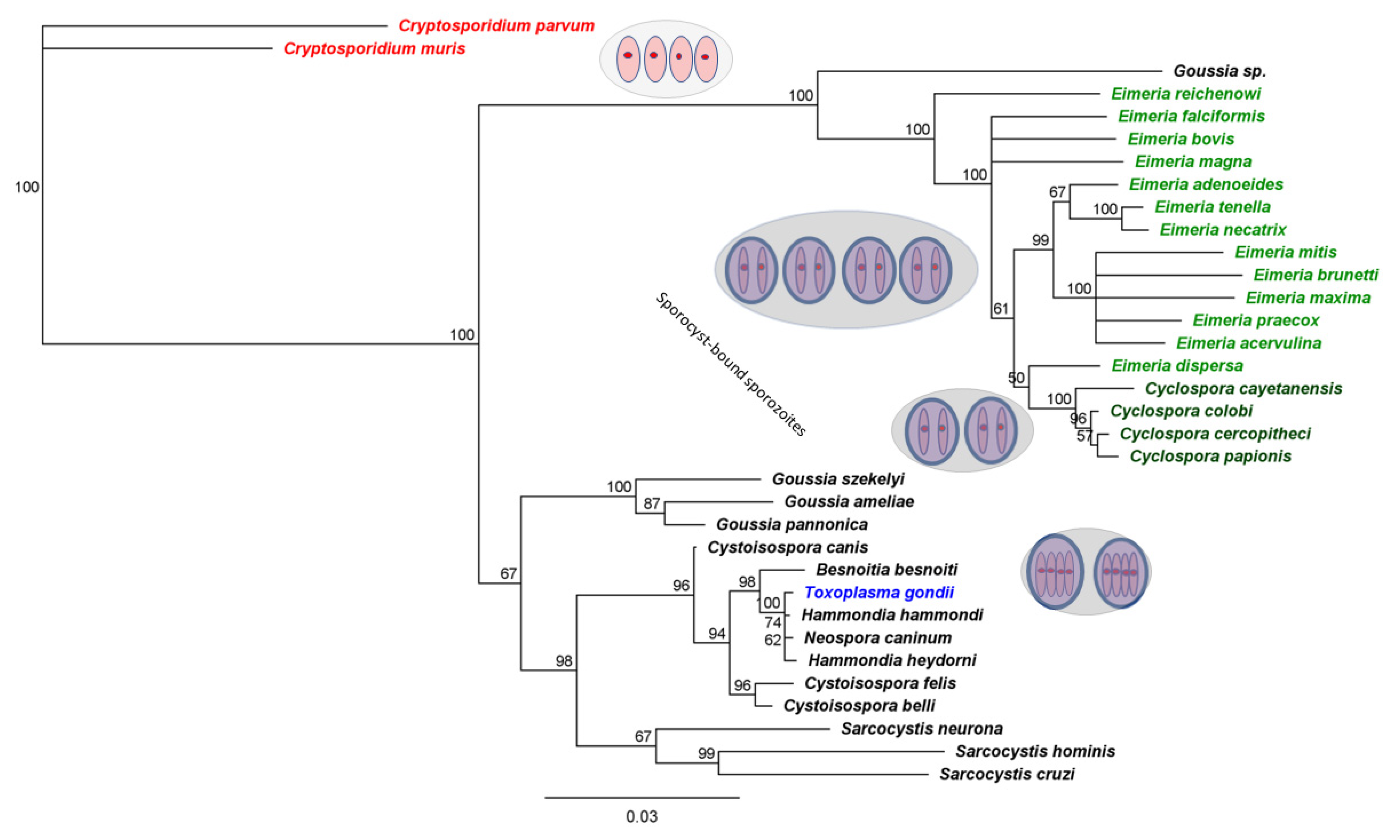
4. What Can We Learn from Such Surrogates?
4.1. Detection
- Provide an abundant and safe source of biological material necessary for establishing “proof of principle” and for optimizing assay conditions. For example, Eimeria oocysts may be well-suited to evaluating developmental pipelines for diagnostic aptamers [75,76] for inclusion in cheap, field-deployable ‘dip sticks.’
- Merit evaluation for their ability to trigger false positive test results. Cross-reactivity poses real risks for molecular diagnostic assays; because of their close evolutionary relationship and genetic similarity, Eimeria and Cyclospora can be easily mistaken for one another.
4.2. Maturation and Viability
4.3. Sanitation and Inactivation Treatments
5. A Path to Future Progress
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Dubey, J.P.; Khan, A.; Rosenthal, B.M. Life Cycle and Transmission of Cyclospora cayetanensis: Knowns and Unknowns. Microorganisms 2022, 10, 118. [Google Scholar] [CrossRef] [PubMed]
- Alfano-Sobsey, E.M.; Eberhard, M.L.; Seed, J.R.; Weber, D.J.; Won, K.Y.; Nace, E.K.; Moe, C.L. Human challenge pilot study with Cyclospora cayetanensis. Emerg. Infect. Dis. 2004, 10, 726–728. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Guo, Y.; Zhang, L.; Rowe, L.A.; Roellig, D.M.; Frace, M.A.; Li, N.; Liu, S.; Feng, Y.; Xiao, L. Genetic similarities between Cyclospora cayetanensis and cecum-infecting avian Eimeria spp. in apicoplast and mitochondrial genomes. Parasit Vectors 2015, 8, 358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qvarnstrom, Y.; Wei-Pridgeon, Y.; Li, W.; Nascimento, F.S.; Bishop, H.S.; Herwaldt, B.L.; Moss, D.M.; Nayak, V.; Srinivasamoorthy, G.; Sheth, M.; et al. Draft Genome Sequences from Cyclospora cayetanensis Oocysts Purified from a Human Stool Sample. Genome Announc. 2015, 3, e01324-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cinar, H.N.; Gopinath, G.; Jarvis, K.; Murphy, H.R. The Complete Mitochondrial Genome of the Foodborne Parasitic Pathogen Cyclospora cayetanensis. PLoS ONE 2015, 10, e0128645. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Wang, L.; Zheng, H.; Xu, Z.; Roellig, D.M.; Li, N.; Frace, M.A.; Tang, K.; Arrowood, M.J.; Moss, D.M.; et al. Comparative genomics reveals Cyclospora cayetanensis possesses coccidia-like metabolism and invasion components but unique surface antigens. BMC Genom. 2016, 17, 316. [Google Scholar] [CrossRef] [Green Version]
- Gopinath, G.R.; Cinar, H.N.; Murphy, H.R.; Durigan, M.; Almeria, M.; Tall, B.D.; DaSilva, A.J. A hybrid reference-guided de novo assembly approach for generating Cyclospora mitochondrion genomes. Gut Pathog. 2018, 10, 15. [Google Scholar] [CrossRef] [Green Version]
- Qvarnstrom, Y.; Wei-Pridgeon, Y.; Van Roey, E.; Park, S.; Srinivasamoorthy, G.; Nascimento, F.S.; Moss, D.M.; Talundzic, E.; Arrowood, M.J. Purification of Cyclospora cayetanensis oocysts obtained from human stool specimens for whole genome sequencing. Gut Pathog. 2018, 10, 45. [Google Scholar] [CrossRef]
- Yanta, C.A.; Pollo, S.M.J.; Barta, J.R.; Reiling, S.J.; Wasmuth, J.D.; Dixon, B.R.; Guy, R.A. Draft Hybrid Genome Assembly of a Canadian Cyclospora cayetanensis Isolate. Microbiol. Resour. Announc. 2022, 11, e0107221. [Google Scholar] [CrossRef]
- Cinar, H.N.; Qvarnstrom, Y.; Wei-Pridgeon, Y.; Li, W.; Nascimento, F.S.; Arrowood, M.J.; Murphy, H.R.; Jang, A.; Kim, E.; Kim, R.; et al. Comparative sequence analysis of Cyclospora cayetanensis apicoplast genomes originating from diverse geographical regions. Parasit Vectors 2016, 9, 611. [Google Scholar] [CrossRef]
- Guo, Y.; Roellig, D.M.; Li, N.; Tang, K.; Frace, M.; Ortega, Y.; Arrowood, M.J.; Feng, Y.; Qvarnstrom, Y.; Wang, L.; et al. Multilocus Sequence Typing Tool for Cyclospora cayetanensis. Emerg. Infect. Dis. 2016, 22, 1464–1467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nascimento, F.S.; Barta, J.R.; Whale, J.; Hofstetter, J.N.; Casillas, S.; Barratt, J.; Talundzic, E.; Arrowood, M.J.; Qvarnstrom, Y. Mitochondrial Junction Region as Genotyping Marker for Cyclospora cayetanensis. Emerg. Infect. Dis. 2019, 25, 1314–1319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barratt, J.L.N.; Park, S.; Nascimento, F.S.; Hofstetter, J.; Plucinski, M.; Casillas, S.; Bradbury, R.S.; Arrowood, M.J.; Qvarnstrom, Y.; Talundzic, E. Genotyping genetically heterogeneous Cyclospora cayetanensis infections to complement epidemiological case linkage. Parasitology 2019, 146, 1275–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofstetter, J.N.; Nascimento, F.S.; Park, S.; Casillas, S.; Herwaldt, B.L.; Arrowood, M.J.; Qvarnstrom, Y. Evaluation of Multilocus Sequence Typing of Cyclospora cayetanensis based on microsatellite markers. Parasite 2019, 26, 3. [Google Scholar] [CrossRef] [Green Version]
- Cinar, H.N.; Gopinath, G.; Murphy, H.R.; Almeria, S.; Durigan, M.; Choi, D.; Jang, A.; Kim, E.; Kim, R.; Choi, S.; et al. Molecular typing of Cyclospora cayetanensis in produce and clinical samples using targeted enrichment of complete mitochondrial genomes and next-generation sequencing. Parasit Vectors 2020, 13, 122. [Google Scholar] [CrossRef] [Green Version]
- Houghton, K.A.; Lomsadze, A.; Park, S.; Nascimento, F.S.; Barratt, J.; Arrowood, M.J.; VanRoey, E.; Talundzic, E.; Borodovsky, M.; Qvarnstrom, Y. Development of a workflow for identification of nuclear genotyping markers for Cyclospora cayetanensis. Parasite 2020, 27, 24. [Google Scholar] [CrossRef] [Green Version]
- Nascimento, F.S.; Barratt, J.; Houghton, K.; Plucinski, M.; Kelley, J.; Casillas, S.; Bennett, C.C.; Snider, C.; Tuladhar, R.; Zhang, J.; et al. Evaluation of an ensemble-based distance statistic for clustering MLST datasets using epidemiologically defined clusters of cyclosporiasis. Epidemiol. Infect. 2020, 148, e172. [Google Scholar] [CrossRef] [PubMed]
- Barratt, J.; Houghton, K.; Richins, T.; Straily, A.; Threlkel, R.; Bera, B.; Kenneally, J.; Clemons, B.; Madison-Antenucci, S.; Cebelinski, E.; et al. Investigation of US Cyclospora cayetanensis outbreaks in 2019 and evaluation of an improved Cyclospora genotyping system against 2019 cyclosporiasis outbreak clusters. Epidemiol. Infect. 2021, 149, e214. [Google Scholar] [CrossRef]
- Barratt, J.; Ahart, L.; Rice, M.; Houghton, K.; Richins, T.; Cama, V.; Arrowood, M.; Qvarnstrom, Y.; Straily, A. Genotyping Cyclospora cayetanensis From Multiple Outbreak Clusters With An Emphasis on a Cluster Linked to Bagged Salad Mix-United States, 2020. J. Infect. Dis. 2022, 225, 2176–2180. [Google Scholar] [CrossRef] [PubMed]
- Yanta, C.A.; Barta, J.R.; Corbeil, A.; Menan, H.; Thivierge, K.; Needle, R.; Morshed, M.; Dixon, B.R.; Wasmuth, J.D.; Guy, R.A. Genotyping Canadian Cyclospora cayetanensis Isolates to Supplement Cyclosporiasis Outbreak Investigations. Microorganisms 2022, 10, 447. [Google Scholar] [CrossRef]
- Li, J.; Chang, Y.; Shi, K.E.; Wang, R.; Fu, K.; Li, S.; Xu, J.; Jia, L.; Guo, Z.; Zhang, L. Multilocus sequence typing and clonal population genetic structure of Cyclospora cayetanensis in humans. Parasitology 2017, 144, 1890–1897. [Google Scholar] [CrossRef] [PubMed]
- Saleh, J.; Mercier, B.; Xi, W. Bioengineering methods for organoid systems. Biol. Cell 2021, 113, 475–491. [Google Scholar] [CrossRef] [PubMed]
- El Marjou, F.; Jouhanneau, C.; Krndija, D. Targeted Transgenic Mice Using CRISPR /Cas9 Technology. Methods Mol. Biol. 2021, 2214, 125–141. [Google Scholar] [CrossRef] [PubMed]
- Heo, I.; Dutta, D.; Schaefer, D.A.; Iakobachvili, N.; Artegiani, B.; Sachs, N.; Boonekamp, K.E.; Bowden, G.; Hendrickx, A.P.A.; Willems, R.J.L.; et al. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol. 2018, 3, 814–823. [Google Scholar] [CrossRef]
- Wilke, G.; Funkhouser-Jones, L.J.; Wang, Y.; Ravindran, S.; Wang, Q.; Beatty, W.L.; Baldridge, M.T.; VanDussen, K.L.; Shen, B.; Kuhlenschmidt, M.S.; et al. A Stem-Cell-Derived Platform Enables Complete Cryptosporidium Development In Vitro and Genetic Tractability. Cell Host Microbe 2019, 26, 123–134.e8. [Google Scholar] [CrossRef] [Green Version]
- Hassan, M.A.; Olijnik, A.A.; Frickel, E.M.; Saeij, J.P. Clonal and atypical Toxoplasma strain differences in virulence vary with mouse sub-species. Int. J. Parasitol. 2019, 49, 63–70. [Google Scholar] [CrossRef]
- Holthaus, D.; Delgado-Betancourt, E.; Aebischer, T.; Seeber, F.; Klotz, C. Harmonization of Protocols for Multi-Species Organoid Platforms to Study the Intestinal Biology of Toxoplasma gondii and Other Protozoan Infections. Front. Cell Infect. Microbiol. 2020, 10, 610368. [Google Scholar] [CrossRef]
- Seo, H.H.; Han, H.W.; Lee, S.E.; Hong, S.H.; Cho, S.H.; Kim, S.C.; Koo, S.K.; Kim, J.H. Modelling Toxoplasma gondii infection in human cerebral organoids. Emerg. Microbes Infect. 2020, 9, 1943–1954. [Google Scholar] [CrossRef]
- Minkah, N.K.; Schafer, C.; Kappe, S.H.I. Humanized Mouse Models for the Study of Human Malaria Parasite Biology, Pathogenesis, and Immunity. Front. Immunol. 2018, 9, 807. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.Y.; Zhang, Y.; Wu, X.D.; Zhang, K.; Lin, P.; Bian, H.J.; Qin, M.M.; Huang, W.; Wei, D.; Zhang, Z.; et al. Disrupting CD147-RAP2 interaction abrogates erythrocyte invasion by Plasmodium falciparum. Blood 2018, 131, 1111–1121. [Google Scholar] [CrossRef]
- Harbuzariu, A.; Pitts, S.; Cespedes, J.C.; Harp, K.O.; Nti, A.; Shaw, A.P.; Liu, M.; Stiles, J.K. Modelling heme-mediated brain injury associated with cerebral malaria in human brain cortical organoids. Sci. Rep. 2019, 9, 19162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, H.V.; Paton, C.A.; Girdwood, R.W.; Mtambo, M.M. Cyclospora in non-human primates in Gombe, Tanzania. Vet. Rec. 1996, 138, 528. [Google Scholar] [PubMed]
- Eberhard, M.L.; da Silva, A.J.; Lilley, B.G.; Pieniazek, N.J. Morphologic and molecular characterization of new Cyclospora species from Ethiopian monkeys: C. cercopitheci sp.n., C. colobi sp.n., and C. papionis sp.n. Emerg. Infect. Dis. 1999, 5, 651–658. [Google Scholar] [CrossRef]
- Lopez, F.A.; Manglicmot, J.; Schmidt, T.M.; Yeh, C.; Smith, H.V.; Relman, D.A. Molecular characterization of Cyclospora-like organisms from baboons. J. Infect. Dis. 1999, 179, 670–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eberhard, M.L.; Njenga, M.N.; DaSilva, A.J.; Owino, D.; Nace, E.K.; Won, K.Y.; Mwenda, J.M. A survey for Cyclospora spp. in Kenyan primates, with some notes on its biology. J. Parasitol. 2001, 87, 1394–1397. [Google Scholar] [CrossRef]
- Zhao, G.-H.; Cong, M.-M.; Bian, Q.-Q.; Cheng, W.-Y.; Wang, R.-J.; Qi, M.; Zhang, L.-X.; Lin, Q.; Zhu, X.-Q. Molecular characterization of Cyclospora -like organisms from golden snub-nosed monkeys in Qinling Mountain in Shaanxi province, northwestern China. PLoS ONE 2013, 8, e58216. [Google Scholar] [CrossRef] [Green Version]
- Eberhard, M.L.; Owens, J.R.; Bishop, H.S.; de Almeida, M.E.; da Silva, A.J. Cyclospora spp. in Drills, Bioko Island, Equatorial Guinea. Emerg. Infect. Dis. 2014, 20, 510–511. [Google Scholar] [CrossRef]
- Li, N.; Ye, J.; Arrowood, M.J.; Ma, J.; Wang, L.; Xu, H.; Feng, Y.; Xiao, L. Identification and morphologic and molecular characterization of Cyclospora macacae n. sp. from rhesus monkeys in China. Parasitol. Res. 2015, 114, 1811–1816. [Google Scholar] [CrossRef]
- Marangi, M.; Koehler, A.V.; Zanzani, S.A.; Manfredi, M.T.; Brianti, E.; Giangaspero, A.; Gasser, R.B. Detection of Cyclospora in captive chimpanzees and macaques by a quantitative PCR-based mutation scanning approach. Parasit Vectors 2015, 8, 274. [Google Scholar] [CrossRef] [Green Version]
- Eberhard, M.L.; Ortega, Y.R.; Hanes, D.E.; Nace, E.K.; Do, R.Q.; Robl, M.G.; Won, K.Y.; Gavidia, C.; Sass, N.L.; Mansfield, K.; et al. Attempts to establish experimental Cyclospora cayetanensis infection in laboratory animals. J. Parasitol. 2000, 86, 577–582. [Google Scholar] [CrossRef]
- Zerpa, R.; Uchima, N.; Huicho, L. Cyclospora cayetanensis associated with watery diarrhoea in Peruvian patients. J. Trop. Med. Hyg. 1995, 98, 325–329. [Google Scholar] [PubMed]
- García-López, H.L.; Rodríguez-Tovar, L.E.; Medina-De la Garza, C.E. Identification of Cyclospora in poultry. Emerg. Infect. Dis. 1996, 2, 356–357. [Google Scholar] [CrossRef] [PubMed]
- Sherchand, J.B.; Cross, J.H. Emerging pathogen Cyclospora cayetanensis infection in Nepal. Southeast Asian J. Trop. Med. Public Health 2001, 32 (Suppl. 2), 143–150. [Google Scholar] [PubMed]
- Pérez-Cordón, G.; Hitos Prados, A.; Romero, D.; Sánchez Moreno, M.; Pontes, A.; Osuna, A.; Rosales, M.J. Intestinal parasitism in the animals of the zoological garden “Peña Escrita” (Almuñecar, Spain). Vet. Parasitol. 2008, 156, 302–309. [Google Scholar] [CrossRef]
- Pérez-Cordón, G.; Prados, A.H.; Romero, D.; Moreno, M.S.; Pontes, A.; Osuna, A.; Rosales, M.J. Intestinal and haematic parasitism in the birds of the Almuñecar (Granada, Spain) ornithological garden. Vet. Parasitol. 2009, 165, 361–366. [Google Scholar] [CrossRef]
- Cama, V.A.; Ortega, Y.R. Cyclospora cayetanensis. In Foodborne Parasites, 2nd ed.; Ortega, Y.R., Sterling, C.R., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2018; pp. 41–56. [Google Scholar]
- Ortega, Y.R.; Sherchand, J.B. Cyclospora cayetanensis. In Biology of Foodborne Parasites, 1st ed.; Xiao, L., Ryan, U., Feng, Y., Eds.; CRC Press: Boca Raton, FL, USA, 2015; pp. 97–110. [Google Scholar]
- Li, G.; Xiao, S.; Zhou, R.; Li, W.; Wadeh, H. Molecular characterization of Cyclospora-like organism from dairy cattle. Parasitol. Res. 2007, 100, 955–961. [Google Scholar] [CrossRef]
- Chu, D.-M.T.; Sherchand, J.B.; Cross, J.H.; Orlandi, P.A. Detection of Cyclospora cayetanensis in animal fecal isolates from Nepal using an FTA filter-base polymerase chain reaction method. Am. J. Trop. Med. Hyg. 2004, 71, 373–379. [Google Scholar] [CrossRef] [Green Version]
- Ortega, Y.R.; Sanchez, R. Update on Cyclospora cayetanensis, a food-borne and waterborne parasite. Clin. Microbiol. Rev. 2010, 23, 218–234. [Google Scholar] [CrossRef] [Green Version]
- Ortega, Y.R.; Gilman, R.H.; Sterling, C.R. A new coccidian parasite (Apicomplexa: Eimeriidae) from humans. J. Parasitol. 1994, 80, 625–629. [Google Scholar] [CrossRef]
- Ford, P.L.; Duszynski, D.W.; McAllister, C.T. Coccidia (Apicomplexa) from heteromyid rodents in the southwestern United States, Baja California, and northern Mexico with three new species from Chaetodipus hispidus. J. Parasitol. 1990, 76, 325–331. [Google Scholar] [CrossRef]
- Rosenthal, B.M.; Dunams-Morel, D.; Ostoros, G.; Molnár, K. Coccidian parasites of fish encompass profound phylogenetic diversity and gave rise to each of the major parasitic groups in terrestrial vertebrates. Infect. Genet. Evol. 2016, 40, 219–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Relman, D.A.; Schmidt, T.M.; Gajadhar, A.; Sogin, M.; Cross, J.; Yoder, K.; Sethabutr, O.; Echeverria, P. Molecular phylogenetic analysis of Cyclospora, the human intestinal pathogen, suggests that it is closely related to Eimeria species. J. Infect. Dis. 1996, 173, 440–445. [Google Scholar] [CrossRef] [PubMed]
- Pieniazek, N.J.; Herwaldt, B.L. Reevaluating the molecular taxonomy: Is human-associated Cyclospora a mammalian Eimeria species? Emerg. Infect. Dis. 1997, 3, 381–383. [Google Scholar] [CrossRef] [PubMed]
- Jordan, B.; Albanese, G.; Tensa, L. Coccidiosis in Chickens (Gallus gallus). In Coccidiosis in Livestock, Poultry, Companion Animals, and Humans, 1st ed.; Dubey, J.P., Ed.; CRC Press: Boca Raton, FL, USA, 2019; pp. 169–174. [Google Scholar]
- Ogedengbe, M.E.; Qvarnstrom, Y.; da Silva, A.J.; Arrowood, M.J.; Barta, J.R. A linear mitochondrial genome of Cyclospora cayetanensis (Eimeriidae, Eucoccidiorida, Coccidiasina, Apicomplexa) suggests the ancestral start position within mitochondrial genomes of eimeriid coccidia. Int. J. Parasitol. 2015, 45, 361–365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogedengbe, M.E.; El-Sherry, S.; Ogedengbe, J.D.; Chapman, H.D.; Barta, J.R. Phylogenies based on combined mitochondrial and nuclear sequences conflict with morphologically defined genera in the eimeriid coccidia (Apicomplexa). Int. J. Parasitol. 2018, 48, 59–69. [Google Scholar] [CrossRef]
- Blake, D.P.; Knox, J.; Dehaeck, B.; Huntington, B.; Rathinam, T.; Ravipati, V.; Ayoade, S.; Gilbert, W.; Adebambo, A.O.; Jatau, I.D.; et al. Re-calculating the cost of coccidiosis in chickens. Vet. Res. 2020, 51, 115. [Google Scholar] [CrossRef]
- Lee, M.B.; Lee, E.H. Coccidial contamination of raspberries: Mock contamination with Eimeria acervulina as a model for decontamination treatment studies. J. Food Prot. 2001, 64, 1854–1857. [Google Scholar] [CrossRef]
- Kniel, K.E.; Shearer, A.E.H.; Cascarino, J.L.; Wilkins, G.C.; Jenkins, M.C. High hydrostatic pressure and UV light treatment of produce contaminated with Eimeria acervulina as a Cyclospora cayetanensis surrogate. J. Food Prot. 2007, 70, 2837–2842. [Google Scholar] [CrossRef] [Green Version]
- Shearer, A.E.H.; Wilkins, G.C.; Jenkins, M.C.; Kniel, K.E. Effects of high hydrostatic pressure on Eimeria acervulina pathogenicity, immunogenicity and structural integrity. Innov. Food Sci. Emerg. Technol. 2007, 8, 259–268. [Google Scholar] [CrossRef]
- Verweij, J.J.; Laeijendecker, D.; Brienen, E.A.T.; van Lieshout, L.; Polderman, A.M. Detection of Cyclospora cayetanensis in travellers returning from the tropics and subtropics using microscopy and real-time PCR. Int. J. Med. Microbiol. 2003, 293, 199–202. [Google Scholar] [CrossRef]
- Varma, M.; Hester, J.D.; Schaefer, F.W.; Ware, M.W.; Lindquist, H.D.A. Detection of Cyclospora cayetanensis using a quantitative real-time PCR assay. J. Microbiol. Methods 2003, 53, 27–36. [Google Scholar] [CrossRef]
- Murphy, H.R.; Lee, S.; da Silva, A.J. Evaluation of an Improved U.S. Food and Drug Administration Method for the Detection of Cyclospora cayetanensis in Produce Using Real-Time PCR. J. Food Prot. 2017, 80, 1133–1144. [Google Scholar] [CrossRef] [PubMed]
- Almeria, S.; da Silva, A.J.; Blessington, T.; Cloyd, T.C.; Cinar, H.N.; Durigan, M.; Murphy, H.R. Evaluation of the U.S. Food and Drug Administration validated method for detection of Cyclospora cayetanensis in high-risk fresh produce matrices and a method modification for a prepared dish. Food Microbiol. 2018, 76, 497–503. [Google Scholar] [CrossRef]
- Murphy, H.R.; Cinar, H.N.; Gopinath, G.; Noe, K.E.; Chatman, L.D.; Miranda, N.E.; Wetherington, J.H.; Neal-McKinney, J.; Pires, G.S.; Sachs, E.; et al. Interlaboratory validation of an improved method for detection of Cyclospora cayetanensis in produce using a real-time PCR assay. Food Microbiol. 2018, 69, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Qvarnstrom, Y.; Benedict, T.; Marcet, P.L.; Wiegand, R.E.; Herwaldt, B.L.; da Silva, A.J. Molecular Detection of Cyclospora cayetanensis in Human Stool Specimens using UNEX-based DNA extraction and real-time PCR. Parasitology 2018, 145, 865–870. [Google Scholar] [CrossRef] [PubMed]
- Murphy, H.R.; Almeria, S.; da Silva, A.J. BAM 19b: Molecular Detection of Cyclospora cayetanensis in Fresh Produce Using Real-Time PCR. Bacteriological Analytical Manual 2022. Available online: https://www.fda.gov/food/laboratory-methods-food-safety/bam-19b-molecular-detection-Cyclospora-cayetanensis-fresh-produce-using-real-time-pcr (accessed on 16 September 2022).
- Assurian, A.; Murphy, H.; Ewing, L.; Cinar, H.N.; da Silva, A.; Almeria, S. Evaluation of the U.S. Food and Drug Administration validated molecular method for detection of Cyclospora cayetanensis oocysts on fresh and frozen berries. Food Microbiol. 2020, 87, 103397. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.D.T.; Jacomasso, T.; Mattos, E.C.; Farias, A.B.; Rampazzo, R.C.P.; Pinto, R.S.; Tassi, W.; Marciano, M.A.M.; Pereira-Chioccola, V.L.; Murphy, H.R.; et al. Ready-to-use qPCR for detection of Cyclospora cayetanensis or Trypanosoma cruzi in food matrices. Food Waterborne Parasitol. 2021, 22, e00111. [Google Scholar] [CrossRef]
- Durigan, M.; Murphy, H.; Deng, K.; Kmet, M.; Lindemann, S.; Newkirk, R.; Patel, V.Y.; Ulaszek, J.; Warren, J.; Ewing, L.; et al. BAM 19c: Dead-end Ultrafiltration for the Detection of Cyclospora cayetanensis from Agricultural Water. Bacteriological Analytical Manual 2020. Available online: https://www.fda.gov/media/140309/download (accessed on 16 September 2022).
- Kahler, A.M.; Mattioli, M.C.; da Silva, A.J.; Hill, V. Detection of Cyclospora cayetanensis in produce irrigation and wash water using large-volume sampling techniques. Food Waterborne Parasitol. 2021, 22, e00110. [Google Scholar] [CrossRef]
- Durigan, M.; Patregnani, E.; Gopinath, G.R.; Ewing-Peeples, L.; Lee, C.; Murphy, H.R.; Almeria, S.; Cinar, H.N.; Negrete, F.; da Silva, A.J. Development of a Molecular Marker Based on the Mitochondrial Genome for Detection of Cyclospora cayetanensis in Food and Water Samples. Microorganisms 2022, 10, 1762. [Google Scholar] [CrossRef]
- Stanciu, L.A.; Wei, Q.; Barui, A.K.; Mohammad, N. Recent Advances in Aptamer-Based Biosensors for Global Health Applications. Annu. Rev. Biomed. Eng. 2021, 23, 433–459. [Google Scholar] [CrossRef]
- Vishwakarma, A.; Lal, R.; Ramya, M. Aptamer-based approaches for the detection of waterborne pathogens. Int. Microbiol. 2021, 24, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Dubey, J.P.; Lindsay, D.S.; Jenkins, M.C.; Bauer, C. Biology of Intestinal Coccidia. In Coccidiosis in Livestock, Poultry, Companion Animals, and Humans, 1st ed.; Dubey, J.P., Ed.; CRC Press: Boca Raton, FL, USA, 2019; pp. 1–36. [Google Scholar]
- Ortega, Y.R.; Sterling, C.R.; Gilman, R.H.; Cama, V.A.; Díaz, F. Cyclospora species--a new protozoan pathogen of humans. N. Engl. J. Med. 1993, 328, 1308–1312. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.V.; Paton, C.A.; Mitambo, M.M.; Girdwood, R.W. Sporulation of Cyclospora sp. oocysts. Appl. Environ. Microbiol. 1997, 63, 1631–1632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sathyanarayanan, L.; Ortega, Y. Effects of temperature and different food matrices on Cyclospora cayetanensis oocyst sporulation. J. Parasitol. 2006, 92, 218–222. [Google Scholar] [CrossRef]
- Tucker, M.S.; O’Brien, C.N.; Jenkins, M.C.; Rosenthal, B.M. Dynamically expressed genes provide candidate viability biomarkers in a model coccidian. PLoS ONE 2021, 16, e0258157. [Google Scholar] [CrossRef]
- Speer, C.A.; Clark, S.; Dubey, J.P. Ultrastructure of the oocysts, sporocysts, and sporozoites of Toxoplasma gondii. J. Parasitol. 1998, 84, 505–512. [Google Scholar] [CrossRef]
- Mai, K.; Sharman, P.A.; Walker, R.A.; Katrib, M.; De Souza, D.; McConville, M.J.; Wallach, M.G.; Belli, S.I.; Ferguson, D.J.P.; Smith, N.C. Oocyst wall formation and composition in coccidian parasites. Mem. Inst. Oswaldo Cruz 2009, 104, 281–289. [Google Scholar] [CrossRef] [Green Version]
- Bushkin, G.G.; Motari, E.; Carpentieri, A.; Dubey, J.P.; Costello, C.E.; Robbins, P.W.; Samuelson, J. Evidence for a structural role for acid-fast lipids in oocyst walls of Cryptosporidium, Toxoplasma, and Eimeria. mBio 2013, 4, e00387-13. [Google Scholar] [CrossRef] [Green Version]
- Freppel, W.; Ferguson, D.J.P.; Shapiro, K.; Dubey, J.P.; Puech, P.H.; Dumetre, A. Structure, composition, and roles of the Toxoplasma gondii oocyst and sporocyst walls. Cell Surf. 2019, 5, 100016. [Google Scholar] [CrossRef]
- Dumetre, A.; Dubey, J.P.; Ferguson, D.J.P. Effect of household bleach on the structure of the sporocyst wall of Toxoplasma gondii. Parasite 2021, 28, 68. [Google Scholar] [CrossRef]
- Fatica, M.K.; Schneider, K.R. The use of chlorination and alternative sanitizers in the produce industry. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 2009, 4, 94052. [Google Scholar] [CrossRef]
- Gil, M.I.; Selma, M.V.; López-Gálvez, F.; Allende, A. Fresh-cut product sanitation and wash water disinfection: Problems and solutions. Int. J. Food Microbiol. 2009, 134, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Finch, G.R.; Black, E.K.; Gyürék, L.; Belosevic, M. Ozone inactivation of Cryptosporidium parvum in demand-free phosphate buffer determined by in vitro excystation and animal infectivity. Appl. Environ. Microbiol. 1993, 59, 4203–4210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bukhari, Z.; Marshall, M.M.; Korich, D.G.; Fricker, C.R.; Smith, H.V.; Rosen, J.; Clancy, J.L. Comparison of Cryptosporidium parvum viability and infectivity assays following ozone treatment of oocysts. Appl. Environ. Microbiol. 2000, 66, 2972–2980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chauret, C.; Nolan, K.; Chen, P.; Springthorpe, S.; Sattar, S. Aging of Cryptosporidium parvum oocysts in river water and their susceptibility to disinfection by chlorine and monochloramine. Can. J. Microbiol. 1998, 44, 1154–1160. [Google Scholar] [CrossRef] [PubMed]
- Ruffell, K.M.; Rennecker, J.L.; Mariñas, B.J. Inactivation of Cryptosporidium parvum oocysts with chlorine dioxide. Water Res. 2000, 34, 868–876. [Google Scholar] [CrossRef]
- Chauret, C.P.; Radziminski, C.Z.; Lepuil, M.; Creason, R.; Andrews, R.C. Chlorine dioxide inactivation of Cryptosporidium parvum oocysts and bacterial spore indicators. Appl. Environ. Microbiol. 2001, 67, 2993–3001. [Google Scholar] [CrossRef] [Green Version]
- Driedger, A.M.; Rennecker, J.L.; Mariñas, B.J. Sequential inactivation of Cryptosporidium parvum oocysts with ozone and free chlorine. Water Res. 2000, 34, 3591–3597. [Google Scholar] [CrossRef]
- Driedger, A.M.; Rennecker, J.L.; Mariñas, B.J. Inactivation of Cryptosporidium parvum oocysts with ozone and monochloramine at low temperature. Water Res. 2001, 35, 41–48. [Google Scholar] [CrossRef]
- Rennecker, J.L.; Kim, J.H.; Corona-Vasquez, B.; Mariñas, B.J. Role of disinfectant concentration and pH in the inactivation kinetics of Cryptosporidium parvum oocysts with ozone and monochloramine. Environ. Sci. Technol. 2001, 35, 2752–2757. [Google Scholar] [CrossRef]
- Mirza Alizadeh, A.; Jazaeri, S.; Shemshadi, B.; Hashempour-Baltork, F.; Sarlak, Z.; Pilevar, Z.; Hosseini, H. A review on inactivation methods of Toxoplasma gondii in foods. Pathog. Glob. Health 2018, 112, 306–319. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, A.M.; El Temsahy, M.M.; Abou El Naga, I.F. Effect of ozone on the viability of some protozoa in drinking water. J. Egypt. Soc. Parasitol. 2001, 31, 603–616. [Google Scholar] [PubMed]
- Liou, C.-T.; Wang, J.-S.; Ooi, H.-K. Effect of ozone treatment on Eimeria colchici oocysts. J. Parasitol. 2002, 88, 159–162. [Google Scholar] [CrossRef]
- El Zawawy, L.A.; El-Said, D.; Ali, S.M.; Fathy, F.M. Disinfection efficacy of sodium dichloroisocyanurate (NADCC) against common food-borne intestinal protozoa. J. Egypt. Soc. Parasitol. 2010, 40, 165–185. [Google Scholar] [PubMed]
- Wainwright, K.E.; Miller, M.A.; Barr, B.C.; Gardner, I.A.; Melli, A.C.; Essert, T.; Packham, A.E.; Truong, T.; Lagunas-Solar, M.; Conrad, P.A. Chemical inactivation of Toxoplasma gondii oocysts in water. J. Parasitol. 2007, 93, 925–931. [Google Scholar] [CrossRef] [PubMed]
- Ortega, Y.R.; Mann, A.; Torres, M.P.; Cama, V. Efficacy of gaseous chlorine dioxide as a sanitizer against Cryptosporidium parvum, Cyclospora cayetanensis, and Encephalitozoon intestinalis on produce. J. Food Prot. 2008, 71, 2410–2414. [Google Scholar] [CrossRef]
- Lee, M.B.; Lee, E.H. The effectiveness of hydrogen peroxide liquid or gas plasma on protozoan oocysts. Can. J. Infect. Dis. Med. Microbiol. 2007, 18, 265. [Google Scholar] [CrossRef] [Green Version]
- Vassal, S.; Favennec, L.; Ballet, J.J.; Brasseur, P. Hydrogen peroxide gas plasma sterilization is effective against Cryptosporidium parvum oocysts. Am. J. Infect. Control. 1998, 26, 136–138. [Google Scholar] [CrossRef]
- Quilez, J.; Sanchez-Acedo, C.; Avendaño, C.; del Cacho, E.; Lopez-Bernad, F. Efficacy of Two Peroxygen-Based Disinfectants for Inactivation of Cryptosporidium parvum Oocysts. Appl. Environ. Microbiol. 2005, 71, 2479–2483. [Google Scholar] [CrossRef] [Green Version]
- Cha, J.O.; Zhao, J.; Yang, M.S.; Kim, W.I.; Cho, H.S.; Lim, C.W.; Kim, B. Oocyst-Shedding Patterns of Three Eimeria Species in Chickens and Shedding Pattern Variation Depending on the Storage Period of Eimeria tenella Oocysts. J. Parasitol. 2018, 104, 18–22. [Google Scholar] [CrossRef]
- Jenkins, M.C.; Tucker, M.; Parker, C.; O’Brien, C.; Miska, K. Cloning and expression of a cDNA coding for Eimeria acervulina 25 kDa protein associated with oocyst and sporocyst walls. Vet. Parasitol. 2022, 309, 109762. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Keeley, A. Comparison of propidium monoazide-quantitative PCR and reverse transcription quantitative PCR for viability detection of fresh Cryptosporidium oocysts following disinfection and after long-term storage in water samples. Water Res. 2012, 46, 5941–5953. [Google Scholar] [CrossRef] [PubMed]
- Alonso, J.L.; Amoros, I.; Guy, R.A. Quantification of viable Giardia cysts and Cryptosporidium oocysts in wastewater using propidium monoazide quantitative real-time PCR. Parasitol. Res. 2014, 113, 2671–2678. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Feng, Y.; Hu, Y.; Villegas, E.N.; Xiao, L. Human infective potential of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in urban wastewater treatment plant effluents. J. Water Health 2016, 14, 411–423. [Google Scholar] [CrossRef]
- Rousseau, A.; Escotte-Binet, S.; La Carbona, S.; Dumètre, A.; Chagneau, S.; Favennec, L.; Kubina, S.; Dubey, J.P.; Majou, D.; Bigot-Clivot, A.; et al. Toxoplasma gondii Oocyst Infectivity Assessed Using a Sporocyst-Based Cell Culture Assay Combined with Quantitative PCR for Environmental Applications. Appl. Environ. Microbiol. 2019, 85, e01189-19. [Google Scholar] [CrossRef] [Green Version]
- Kent, B.N.; Salichos, L.; Gibbons, J.G.; Rokas, A.; Newton, I.L.; Clark, M.E.; Bordenstein, S.R. Complete bacteriophage transfer in a bacterial endosymbiont (Wolbachia) determined by targeted genome capture. Genome Biol. Evol. 2011, 3, 209–218. [Google Scholar] [CrossRef] [Green Version]
- Schuenemann, V.J.; Bos, K.; DeWitte, S.; Schmedes, S.; Jamieson, J.; Mittnik, A.; Forrest, S.; Coombes, B.K.; Wood, J.W.; Earn, D.J.; et al. Targeted enrichment of ancient pathogens yielding the pPCP1 plasmid of Yersinia pestis from victims of the Black Death. Proc. Natl. Acad. Sci. USA 2011, 108, E746–E752. [Google Scholar] [CrossRef] [Green Version]
- Domagalska, M.A.; Imamura, H.; Sanders, M.; Van den Broeck, F.; Bhattarai, N.R.; Vanaerschot, M.; Maes, I.; D’Haenens, E.; Rai, K.; Rijal, S.; et al. Genomes of Leishmania parasites directly sequenced from patients with visceral leishmaniasis in the Indian subcontinent. PLoS Negl. Trop. Dis. 2019, 13, e0007900. [Google Scholar] [CrossRef] [Green Version]
- Xue-Franzen, Y.; Kjaerulff, S.; Holmberg, C.; Wright, A.; Nielsen, O. Genomewide identification of pheromone-targeted transcription in fission yeast. BMC Genom. 2006, 7, 303. [Google Scholar] [CrossRef] [Green Version]
- Troell, K.; Hallstrom, B.; Divne, A.M.; Alsmark, C.; Arrighi, R.; Huss, M.; Beser, J.; Bertilsson, S. Cryptosporidium as a testbed for single cell genome characterization of unicellular eukaryotes. BMC Genom. 2016, 17, 471. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tucker, M.S.; Khan, A.; Jenkins, M.C.; Dubey, J.P.; Rosenthal, B.M. Hastening Progress in Cyclospora Requires Studying Eimeria Surrogates. Microorganisms 2022, 10, 1977. https://doi.org/10.3390/microorganisms10101977
Tucker MS, Khan A, Jenkins MC, Dubey JP, Rosenthal BM. Hastening Progress in Cyclospora Requires Studying Eimeria Surrogates. Microorganisms. 2022; 10(10):1977. https://doi.org/10.3390/microorganisms10101977
Chicago/Turabian StyleTucker, Matthew S., Asis Khan, Mark C. Jenkins, Jitender P. Dubey, and Benjamin M. Rosenthal. 2022. "Hastening Progress in Cyclospora Requires Studying Eimeria Surrogates" Microorganisms 10, no. 10: 1977. https://doi.org/10.3390/microorganisms10101977
APA StyleTucker, M. S., Khan, A., Jenkins, M. C., Dubey, J. P., & Rosenthal, B. M. (2022). Hastening Progress in Cyclospora Requires Studying Eimeria Surrogates. Microorganisms, 10(10), 1977. https://doi.org/10.3390/microorganisms10101977







