Identification of Bacterial Communities in Laboratory-Adapted Glyptotendipes tokunagai and Wild-Stream-Inhabiting Chironomus flaviplumus
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Assessment of Environmental Factors in the Studied Area
2.3. DNA Extraction and High-Throughput Sequencing
2.4. Bioinformatics
2.5. Statistical Analysis
3. Results
3.1. Environmental Conditions of the Studied Sites
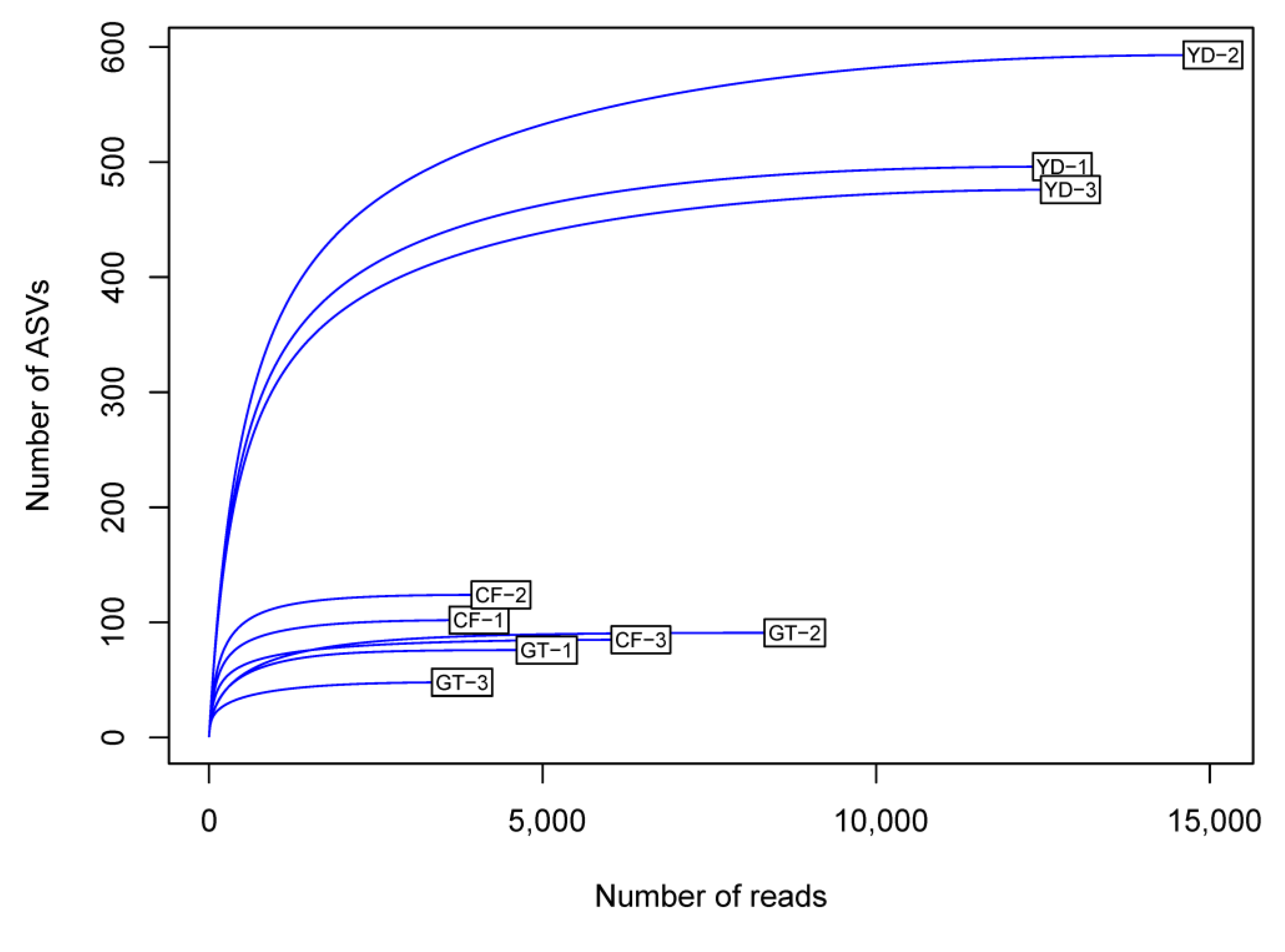
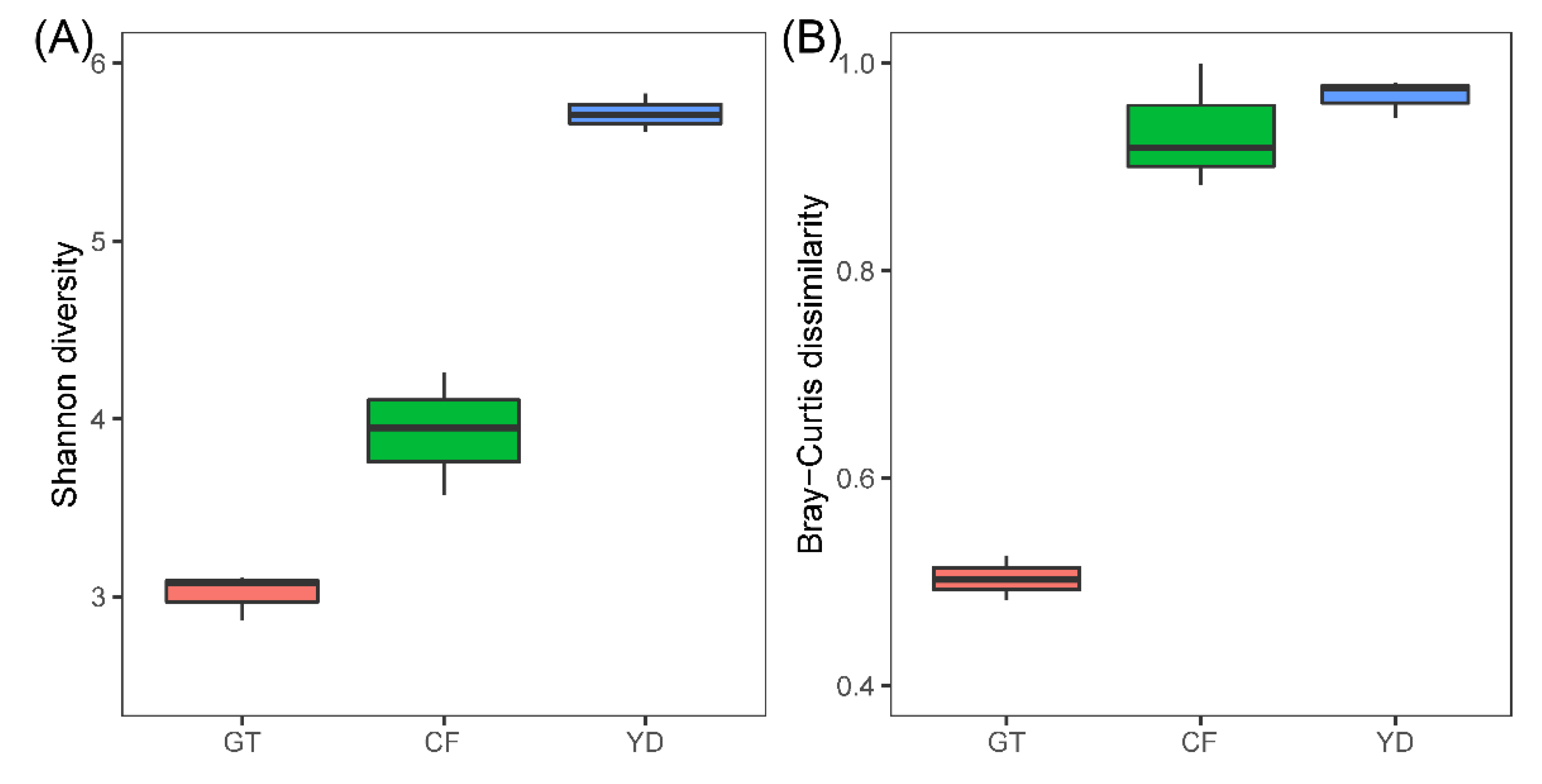
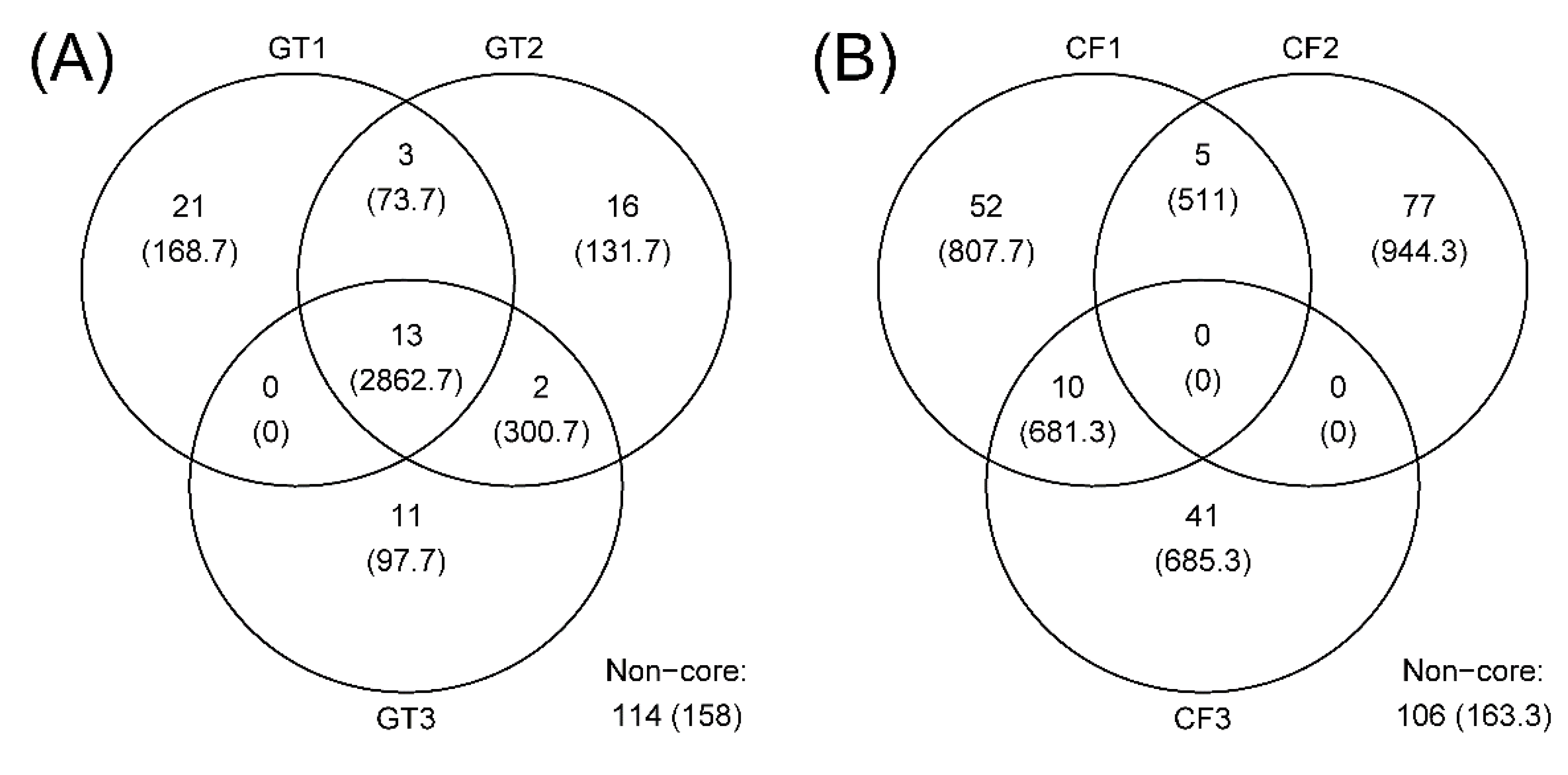
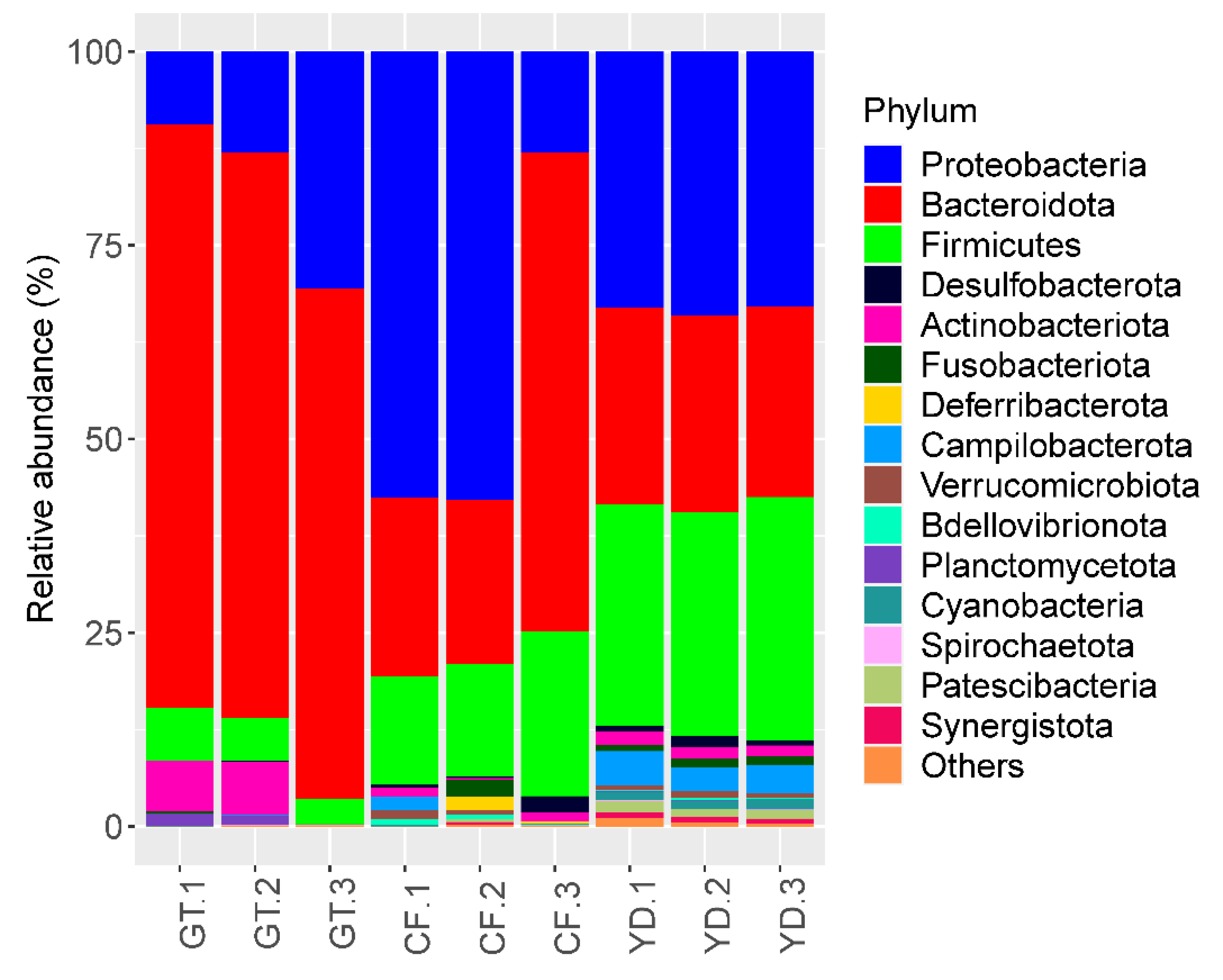
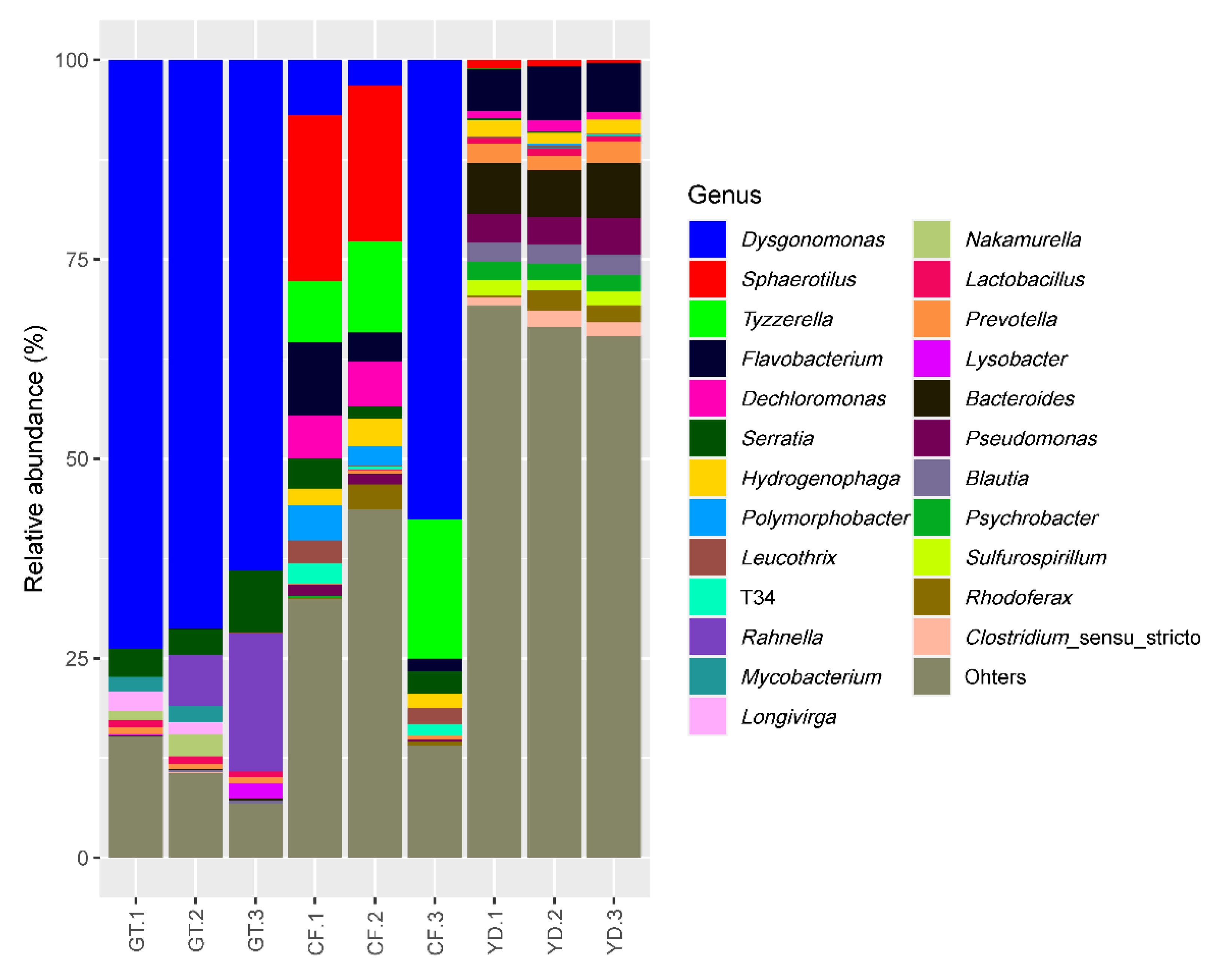
3.2. Microbial Community Composition
3.3. Functional Potential of Microbial Communities
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Armitage, P.D.; Pinder, L.; Cranston, P. The Chironomidae: Biology and Ecology of Non-Biting Midges; Springer Science & Business Media: Berlin/Heidelberg, Germany, 1995. [Google Scholar]
- Mezgebu, A.; Lakew, A.; Lemma, B.; Beneberu, G. The potential use of chironomids (Insecta: Diptera) as bioindicators in streams and rivers around Sebeta, Ethiopia. Afr. J. Aquat. Sci. 2019, 44, 369–376. [Google Scholar] [CrossRef]
- Molineri, C.; Tejerina, E.G.; Torrejón, S.E.; Pero, E.J.I.; Hankel, G.E. Indicative value of different taxonomic levels of Chironomidae for assessing the water quality. Ecol. Indic. 2020, 108, 105703. [Google Scholar] [CrossRef]
- Al-Shami, S.A.; Rawi, C.S.M.; HassanAhmad, A.; Nor, S.A.M. Distribution of Chironomidae (Insecta: Diptera) in polluted rivers of the Juru River Basin, Penang, Malaysia. J. Environ. Sci. 2010, 22, 1718–1727. [Google Scholar] [CrossRef]
- Sela, R.; Laviad-Shitrit, S.; Thorat, L.; Nath, B.B.; Halpern, M. Chironomus ramosus Larval Microbiome Composition Provides Evidence for the Presence of Detoxifying Enzymes. Microorganisms 2021, 9, 1571. [Google Scholar] [CrossRef]
- Sun, X.; Liu, W.; Li, R.; Zhao, C.; Pan, L.; Yan, C. A chromosome level genome assembly of Propsilocerus akamusi to understand its response to heavy metal exposure. Mol. Ecol. Resour. 2021, 21, 1996–2012. [Google Scholar] [CrossRef]
- Nath, B.B. Extracellular hemoglobin and environmental stress tolerance in Chironomus larvae. J. Limnol. 2018, 77, 104–112. [Google Scholar] [CrossRef]
- Senderovich, Y.; Halpern, M. The protective role of endogenous bacterial communities in chironomid egg masses and larvae. ISME J. 2013, 7, 2147–2158. [Google Scholar] [CrossRef]
- Cranston, P. Morphology. In The Chironomidae. The Biology and Ecology of Non-Biting Midges; Armitage, P., Cranston, P.S., Pinder, L.C.V., Eds.; Chapman and Hall: London, UK, 1995. [Google Scholar]
- Baek, M.J.; Yoon, T.J.; Bae, Y.J. Development of Glyptotendipes tokunagai (Diptera: Chironomidae) Under Different Temperature Conditions. Environ. Entomol. 2012, 41, 950–958. [Google Scholar] [CrossRef]
- Baek, M.J.; Yoon, T.J.; Kang, H.J.; Bae, Y.J. Biological and Genetic Characteristics of Glyptotendipes tokunagai (Diptera: Chironomidae) on the Basis of Successive Rearing of Forty-Two Generations Over Seven Years Under Laboratory Conditions. Environ. Entomol. 2014, 43, 1406–1418. [Google Scholar] [CrossRef]
- Park, K.; Kim, W.-S.; Park, J.-W.; Kwak, I.-S. Complete mitochondrial genome of Chironomus flaviplumus (Diptera: Chironomidae) collected in Korea. Mitochondrial DNA Part B 2021, 6, 2843–2844. [Google Scholar] [CrossRef]
- Kwak, I.-S.; Park, J.-W.; Kim, W.-S.; Park, K. Morphological and genetic species identification in the Chironomus larvae (Diptera: Chironomidae) found in domestic tap water purification plants. Korean J. Ecol. Environ. 2020, 53, 286–294. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- OECD. Test No. 233: Sediment-Water Chironomid Life-Cycle Toxicity Test Using Spiked Water or Spiked Sediment; Organisation for Economic Co-Operation and Development Publishing: Paris, France, 2010. [Google Scholar]
- Hamilton, E. A Manual of Chemical & Biological Methods for Seawater Analysis; Parsons, T.R., Maita, Y., Lalli, C.M., Eds.; Pergamon Press: Oxford, UK, 1984; p. 184. ISBN 0-08-030287-4. [Google Scholar]
- Carlson, R.E. A trophic state index for lakes1. Limnol. Oceanogr. 1977, 22, 361–369. [Google Scholar] [CrossRef]
- Djurhuus, A.; Closek, C.J.; Kelly, R.P.; Pitz, K.J.; Michisaki, R.P.; Starks, H.A.; Walz, K.R.; Andruszkiewicz, E.A.; Olesin, E.; Hubbard, K.; et al. Environmental DNA reveals seasonal shifts and potential interactions in a marine community. Nat. Commun. 2020, 11, 254. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’hara, R.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Package ‘Vegan’. Community Ecology Package, version 2; R Core Team: Vienna, Austria, 2013; Volume 2, pp. 1–295. [Google Scholar]
- Andersen, K.S.; Kirkegaard, R.H.; Karst, S.M.; Albertsen, M. ampvis2: An R package to analyse and visualise 16S rRNA amplicon data. bioRxiv 2018, 299537. [Google Scholar] [CrossRef]
- Chernitsyna, S.M.; Khalzov, I.A.; Sitnikova, T.Y.; Naumova, T.V.; Khabuev, A.V.; Zemskaya, T.I. Microbial Communities Associated with Bentic Invertebrates of Lake Baikal. Curr. Microbiol. 2021, 78, 3020–3031. [Google Scholar] [CrossRef]
- Laviad-Shitrit, S.; Sharaby, Y.; Sela, R.; Thorat, L.; Nath, B.B.; Halpern, M. Copper and chromium exposure affect chironomid larval microbiota composition. Sci. Total Environ. 2021, 771, 145330. [Google Scholar] [CrossRef]
- Bridges, C.M.; Gage, D.J. Development and application of aerobic, chemically defined media for Dysgonomonas. Anaerobe 2021, 67, 102302. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Chua, H.; Lo, W.-h.; Lawford, H.; Yu, P.H.-F. Sphaerotilus natans isolated from activated sludge and its production of poly (3-hydroxybutyrate-co-3-hydroxyvalerate). In Biotechnology for Fuels and Chemicals; Springer: Champs, Switzerland, 2002; pp. 1061–1073. [Google Scholar]
- Ferreira, R.; Amado, R.; Padrão, J.; Ferreira, V.; Dias, N.M.; Melo, L.D.R.; Santos, S.B.; Nicolau, A. The first sequenced Sphaerotilus natans bacteriophage– characterization and potential to control its filamentous bacterium host. FEMS Microbiol. Ecol. 2021, 97, fiab029. [Google Scholar] [CrossRef] [PubMed]
- Pellegrin, V.; Juretschko, S.; Wagner, M.; Cottenceau, G. Morphological and biochemical properties of a Sphaerotilus sp. Isolated from paper mill slimes. Appl. Env. Microbiol. 1999, 65, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Cerqueda-García, D.; Améndola-Pimenta, M.; Zamora-Briseño, J.A.; González-Penagos, C.E.; Árcega-Cabrera, F.; Ceja-Moreno, V.; Rodríguez-Canul, R. Effects of chronic exposure to water accommodated fraction (WAF) of light crude oil on gut microbiota composition of the lined sole (Achirus lineatus). Mar. Environ. Res. 2020, 161, 105116. [Google Scholar] [CrossRef]
- Shao, M.; Zhu, Y. Long-term metal exposure changes gut microbiota of residents surrounding a mining and smelting area. Sci. Rep. 2020, 10, 4453. [Google Scholar] [CrossRef]
- Salinero, K.K.; Keller, K.; Feil, W.S.; Feil, H.; Trong, S.; Di Bartolo, G.; Lapidus, A. Metabolic analysis of the soil microbe Dechloromonas aromatica str. RCB: Indications of a surprisingly complex life-style and cryptic anaerobic pathways for aromatic degradation. BMC Genom. 2009, 10, 351. [Google Scholar] [CrossRef]
- Zhang, S.; Amanze, C.; Sun, C.; Zou, K.; Fu, S.; Deng, Y.; Liu, X.; Liang, Y. Evolutionary, genomic, and biogeographic characterization of two novel xenobiotics-degrading strains affiliated with Dechloromonas. Heliyon 2021, 7, e07181. [Google Scholar] [CrossRef]
- Husain, R.; Vikram, N.; Pandey, S.; Yadav, G.; Bose, S.K.; Ahamad, A.; Shamim, M.; Kumar, K.G.; Khan, N.A.; Zuhaib, M.; et al. Microorganisms: An eco-friendly tools for the waste management and environmental safety. In Biotechnological Innovations for Environmental Bioremediation; Arora, S., Kumar, A., Ogita, S., Yau, Y.Y., Eds.; Springer: Singapore, 2022; pp. 949–981. [Google Scholar]
- Eiler, A.; Bertilsson, S. Flavobacteria blooms in four eutrophic lakes: Linking population dynamics of freshwater bacterioplankton to resource availability. Appl. Environ. Microbiol. 2007, 73, 3511–3518. [Google Scholar] [CrossRef]
- Shao, K.; Yao, X.; Wu, Z.; Jiang, X.; Hu, Y.; Tang, X.; Xu, Q.; Gao, G. The bacterial community composition and its environmental drivers in the rivers around eutrophic Chaohu Lake, China. BMC Microbiol. 2021, 21, 179. [Google Scholar] [CrossRef]
- Kim, W.-S.; Kim, R.; Park, K.; Chamilani, N.; Kwak, I.-S. The molecular biomarker genes expressions of rearing species Chironomus riparious and field species Chironomus plumosus exposure to heavy metals. Korean J. Ecol. Environ. 2015, 48, 86–94. [Google Scholar] [CrossRef]

| (n = 3) | ||||||
| NO2 (μg/L) | NO3 (μg/L) | NH4 (μg/L) | TN (μg/L) | |||
| 47.2 ± 0.1 | 4354.7 ± 15.7 | 557.2 ± 13.7 | 5607.8 ± 28.2 | |||
| PO4 (μg/L) | TP (μg/L) | SS (mg/L) | Chl-a (μg/L) | |||
| 112.6 ± 0.9 | 140.7 ± 2 | 2.6 ± 0.5 | 7.8 ± 0.6 | |||
| (n = 7) | ||||||
| Water Temperature (°C) | DO (mg/L) | SPC (μS/cm) | PSU | pH | ||
| 6.5 ± 0 | 16.4 ± 0.7 | 623.9 ± 3 | 0.3 ± 0 | 7.2 ± 0.2 | ||
| KEGG ID | Gene | Function | GT (%) | CF (%) |
|---|---|---|---|---|
| K07665 | cusR, copR, silR | Two-component system, OmpR family, copper resistance phosphate regulon response regulator CusR | 0.006 | 0.033 |
| K03327 | TC.MATE, SLC47A, norM, mdtK, dinF | Multidrug resistance protein, MATE family | 0.072 | 0.047 |
| K03297 | emrE, qac, mmr, smr | Small multidrug resistance pump | 0.011 | 0.019 |
| K11741 | sugE | Quaternary ammonium compound-resistance protein SugE | 0.066 | 0.036 |
| K11811 | arsH | Arsenical resistance protein ArsH | 0.001 | 0.014 |
| K07245 | pcoD | Copper resistance protein D | 0.011 | 0.002 |
| K07156 | copC, pcoC | Copper resistance protein C | 0.01 | 0.002 |
| K07233 | pcoB, copB | Copper resistance protein B | 0.001 | 0.005 |
| K02617 | paaY | Phenylacetic acid degradation protein | 0.001 | 0.003 |
| K07803 | zraP | Zinc resistance-associated protein | 0 | 0.001 |
| K15726 | czcA | Cobalt–zinc–cadmium resistance protein CzcA | 0.056 | 0.049 |
| K16264 | czcD, zitB | Cobalt–zinc–cadmium efflux system protein | 0.066 | 0.031 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, H.; Kim, W.-S.; Park, J.-W.; Kwak, I.-S. Identification of Bacterial Communities in Laboratory-Adapted Glyptotendipes tokunagai and Wild-Stream-Inhabiting Chironomus flaviplumus. Microorganisms 2022, 10, 2107. https://doi.org/10.3390/microorganisms10112107
Song H, Kim W-S, Park J-W, Kwak I-S. Identification of Bacterial Communities in Laboratory-Adapted Glyptotendipes tokunagai and Wild-Stream-Inhabiting Chironomus flaviplumus. Microorganisms. 2022; 10(11):2107. https://doi.org/10.3390/microorganisms10112107
Chicago/Turabian StyleSong, Hokyung, Won-Seok Kim, Jae-Won Park, and Ihn-Sil Kwak. 2022. "Identification of Bacterial Communities in Laboratory-Adapted Glyptotendipes tokunagai and Wild-Stream-Inhabiting Chironomus flaviplumus" Microorganisms 10, no. 11: 2107. https://doi.org/10.3390/microorganisms10112107
APA StyleSong, H., Kim, W.-S., Park, J.-W., & Kwak, I.-S. (2022). Identification of Bacterial Communities in Laboratory-Adapted Glyptotendipes tokunagai and Wild-Stream-Inhabiting Chironomus flaviplumus. Microorganisms, 10(11), 2107. https://doi.org/10.3390/microorganisms10112107







