Stieleria sedimenti sp. nov., a Novel Member of the Family Pirellulaceae with Antimicrobial Activity Isolated in Portugal from Brackish Sediments
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation
2.2. Partial 16S rRNA Gene Sequence-Based Phylogeny
2.3. Genome-Based Phylogeny and Genomic Analyses
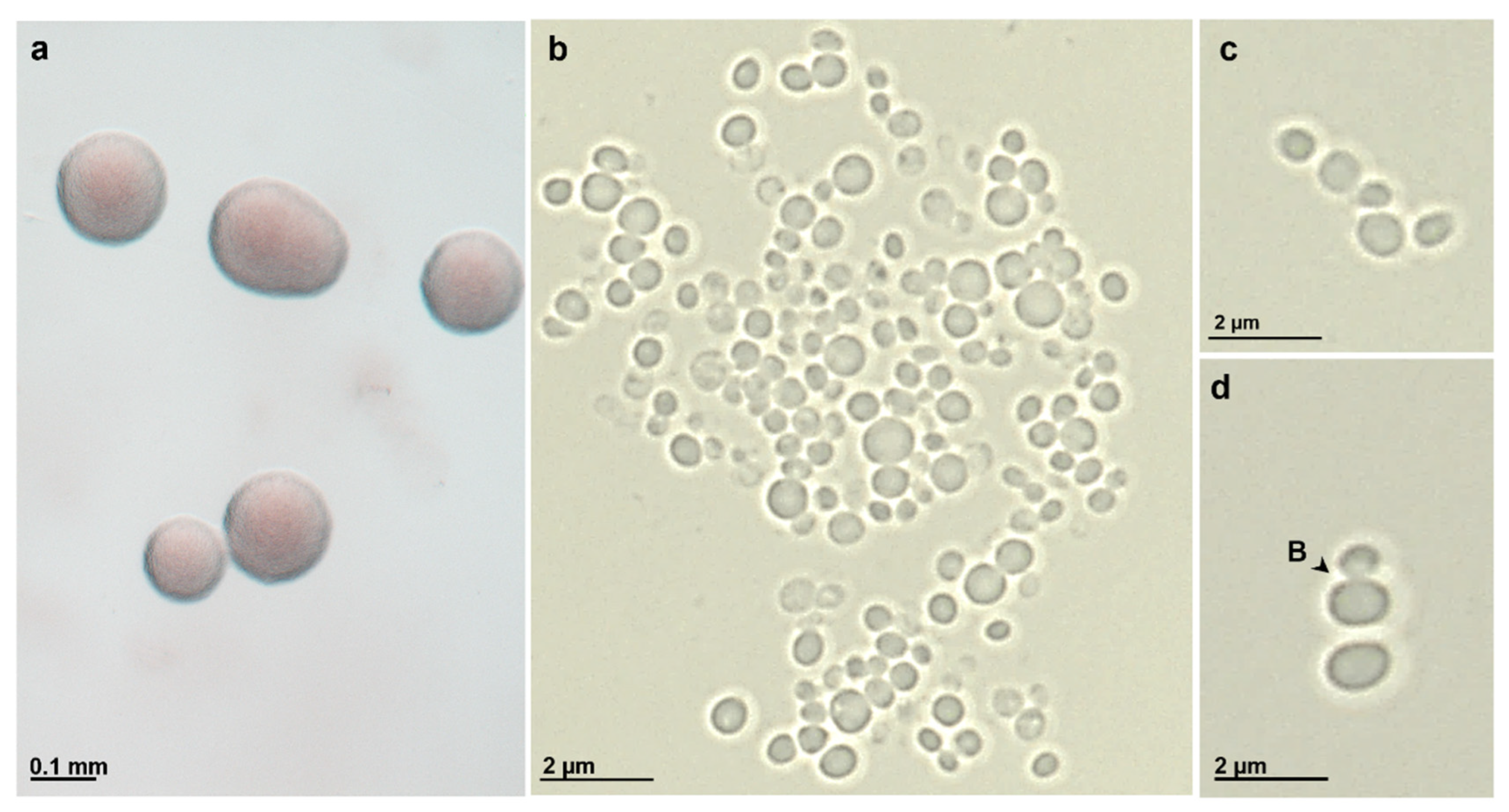
2.4. Morphological and Physiological Characterization
2.5. Extraction of Metabolites and Antimicrobial Screening
3. Results and Discussion
3.1. Phylogeny of Strain ICT_E10.1T and Genomic Features
3.2. Ecology
3.3. Physiological and Morphological Features
3.4. Genome Mining for BGCs and Antimicrobial Screening
4. Conclusions
5. Description of Stieleria sedimenti sp. nov.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Hofer, U. The cost of antimicrobial resistance. Nat. Rev. Microbiol. 2019, 17, 3. [Google Scholar] [CrossRef] [PubMed]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, J.D.; Vitorino, I.; Reyes, F.; Vicente, F.; Lage, O.M. From Ocean to Medicine: Pharmaceutical Applications of Metabolites from Marine Bacteria. Antibiotics 2020, 9, 455. [Google Scholar] [CrossRef] [PubMed]
- Rivas-Marín, E.; Devos, D.P. The Paradigms They Are a-Changin’: Past, present and future of PVC bacteria research. Antonie Leeuwenhoek 2018, 111, 785–799. [Google Scholar] [CrossRef] [Green Version]
- Wagner, M.; Horn, M. The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr. Opin. Biotechnol. 2006, 17, 241–249. [Google Scholar] [CrossRef]
- Cho, J.-C.; Vergin, K.L.; Morris, R.M.; Giovannoni, S.J. Lentisphaera araneosa gen. nov., sp. nov, a transparent exopolymer producing marine bacterium, and the description of a novel bacterial phylum, Lentisphaerae. Environ. Microbiol. 2004, 6, 611–621. [Google Scholar] [CrossRef]
- Oren, A.; Garrity, G.M. Valid publication of the names of forty-two phyla of prokaryotes. Int. J. Syst. Evol. Microbiol. 2021, 71, 005056. [Google Scholar] [CrossRef]
- Wiegand, S.; Jogler, M.; Jogler, C. On the maverick Planctomycetes. FEMS Microbiol. Rev. 2018, 42, 739–760. [Google Scholar] [CrossRef] [Green Version]
- Lage, O.M.; van Niftrik, L.; Jogler, C.; Devos, D.P. Planctomycetes. In Encyclopedia of Microbiology, 4th ed.; Schmidt, T.M., Ed.; Academic Press: Oxford, UK, 2019; pp. 614–626. [Google Scholar]
- Wiegand, S.; Jogler, M.; Boedeker, C.; Pinto, D.; Vollmers, J.; Rivas-Marin, E.; Kohn, T.; Peeters, S.H.; Heuer, A.; Rast, P.; et al. Cultivation and functional characterization of 79 planctomycetes uncovers their unique biology. Nat. Microbiol. 2020, 5, 126–140. [Google Scholar] [CrossRef]
- Acehan, D.; Santarella-Mellwig, R.; Devos, D.P. A bacterial tubulovesicular network. J. Cell Sci. 2014, 127, 277–280. [Google Scholar] [CrossRef]
- Boedeker, C.; Schuler, M.; Reintjes, G.; Jeske, O.; van Teeseling, M.C.; Jogler, M.; Rast, P.; Borchert, D.; Devos, D.P.; Kucklick, M.; et al. Determining the bacterial cell biology of Planctomycetes. Nat. Commun. 2017, 8, 14853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivas-Marin, E.; Canosa, I.; Devos, D.P. Evolutionary Cell Biology of Division Mode in the Bacterial Planctomycetes-Verrucomicrobia-Chlamydiae Superphylum. Front. Microbiol. 2016, 7, 1964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lage, O.M.; Bondoso, J.; Lobo-da-Cunha, A. Insights into the ultrastructural morphology of novel Planctomycetes. Antonie Leeuwenhoek 2013, 104, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Rivas-Marin, E.; Peeters, S.H.; Claret Fernandez, L.; Jogler, C.; van Niftrik, L.; Wiegand, S.; Devos, D.P. Non-essentiality of canonical cell division genes in the planctomycete Planctopirus limnophila. Sci. Rep. 2020, 10, 66. [Google Scholar] [CrossRef] [Green Version]
- Devos, D.P. PVC bacteria: Variation of, but not exception to, the Gram-negative cell plan. Trends Microbiol. 2014, 22, 14–20. [Google Scholar] [CrossRef] [Green Version]
- Vitorino, I.; Albuquerque, L.; Wiegand, S.; Kallscheuer, N.; da Costa, M.S.; Lobo-da-Cunha, A.; Jogler, C.; Lage, O.M. Alienimonas chondri sp. nov., a novel planctomycete isolated from the biofilm of the red alga Chondrus crispus. Syst. Appl. Microbiol. 2020, 43, 126083. [Google Scholar] [CrossRef]
- Kulichevskaya, I.S.; Ivanova, A.A.; Suzina, N.E.; Sinninghe Damsté, J.S.; Dedysh, S.N. Anatilimnocola floriformis sp. nov., a novel member of the family Pirellulaceae from a boreal lake, and emended description of the genus Anatilimnocola. Antonie Leeuwenhoek 2022, 115, 1253–1264. [Google Scholar] [CrossRef]
- Godinho, O.; Botelho, R.; Albuquerque, L.; Wiegand, S.; Kallscheuer, N.; da Costa, M.S.; Lobo-da-Cunha, A.; Jogler, C.; Lage, O.M. Bremerella alba sp. nov., a novel planctomycete isolated from the surface of the macroalga Fucus spiralis. Syst. Appl. Microbiol. 2021, 44, 126189. [Google Scholar] [CrossRef]
- Kumar, D.; Kumar, G.; Jagadeeshwari, U.; Sasikala, C.; Ramana, C.V. “Candidatus Laterigemmans baculatus” gen. nov. sp. nov., the first representative of rod shaped planctomycetes with lateral budding in the family Pirellulaceae. Syst. Appl. Microbiol. 2021, 44, 126188. [Google Scholar] [CrossRef]
- Kumar, G.; Kumar, D.; Jagadeeshwari, U.; Sreya, P.K.; Shabbir, A.; Sasikala, C.; Ramana, C.V. Crateriforma spongiae sp. nov., isolated from a marine sponge and emended description of the genus “Crateriforma”. Antonie Leeuwenhoek 2021, 114, 341–353. [Google Scholar] [CrossRef]
- Storesund, J.E.; Ovreas, L. Diversity of Planctomycetes in iron-hydroxide deposits from the Arctic Mid Ocean Ridge (AMOR) and description of Bythopirellula goksoyri gen. nov., sp. nov., a novel Planctomycete from deep sea iron-hydroxide deposits. Antonie Leeuwenhoek 2013, 104, 569–584. [Google Scholar] [CrossRef] [PubMed]
- Kulichevskaya, I.S.; Ivanova, A.A.; Baulina, O.I.; Rijpstra, W.I.C.; Sinninghe Damste, J.S.; Dedysh, S.N. Fimbriiglobus ruber gen. nov., sp. nov., a Gemmata-like planctomycete from Sphagnum peat bog and the proposal of Gemmataceae fam. nov. Int. J. Syst. Evol. Microbiol. 2017, 67, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Kulichevskaya, I.S.; Ivanova, A.A.; Naumoff, D.G.; Beletsky, A.V.; Rijpstra, W.I.C.; Sinninghe Damste, J.S.; Mardanov, A.V.; Ravin, N.V.; Dedysh, S.N. Frigoriglobus tundricola gen. nov., sp. nov., a psychrotolerant cellulolytic planctomycete of the family Gemmataceae from a littoral tundra wetland. Syst. Appl. Microbiol. 2020, 43, 126129. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, A.A.; Kulichevskaya, I.S.; Dedysh, S.N. Gemmata palustris sp. nov., a Novel Planctomycete from a Fen in Northwestern Russia. Microbiology 2021, 90, 598–606. [Google Scholar] [CrossRef]
- Vitorino, I.R.; Lobo-da Cunha, A.; Vasconcelos, V.; Vicente, F.; Lage, O.M. Isolation, diversity and antimicrobial activity of planctomycetes from the Tejo river estuary (Portugal). FEMS Microbiol. Ecol. 2022, 98, fiac066. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Kulichevskaya, I.S.; Beletsky, A.V.; Ivanova, A.A.; Rijpstra, W.I.C.; Damste, J.S.S.; Mardanov, A.V.; Ravin, N.V. Lacipirellula parvula gen. nov., sp. nov., representing a lineage of planctomycetes widespread in low-oxygen habitats, description of the family Lacipirellulaceae fam. nov. and proposal of the orders Pirellulales ord. nov., Gemmatales ord. nov. and Isosphaerales ord. nov. Syst. Appl. Microbiol. 2020, 43, 126050. [Google Scholar] [CrossRef]
- Lage, O.M.; Albuquerque, L.; Lobo-da Cunha, A.; da Costa, M.S. Mariniblastus fucicola gen. nov., sp. nov. a novel planctomycete associated with macroalgae. Int. J. Syst. Evol. Microbiol. 2017, 67, 1571–1576. [Google Scholar] [CrossRef]
- Vitorino, I.; Santos, J.D.N.; Godinho, O.; Vicente, F.; Vasconcelos, V.; Lage, O.M. Novel and Conventional Isolation Techniques to Obtain Planctomycetes from Marine Environments. Microorganisms 2021, 9, 2078. [Google Scholar] [CrossRef]
- Kulichevskaya, I.S.; Ivanova, A.A.; Suzina, N.E.; Rijpstra, W.I.C.; Sinninghe Damste, J.S.; Dedysh, S.N. Paludisphaera borealis gen. nov., sp. nov., a hydrolytic planctomycete from northern wetlands, and proposal of Isosphaeraceae fam. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 837–844. [Google Scholar] [CrossRef]
- Kaushik, R.; Sharma, M.; Gaurav, K.; Jagadeeshwari, U.; Shabbir, A.; Sasikala, C.; Ramana, C.V.; Pandit, M.K. Paludisphaera soli sp. nov., a new member of the family Isosphaeraceae isolated from high altitude soil in the Western Himalaya. Antonie Leeuwenhoek 2020, 113, 1663–1674. [Google Scholar] [CrossRef]
- Gaurav, K.; Kumar, D.; Jagadeeshwari, U.; Shabbir, A.; Sasikala, C.; Ramana, C.V. Phylo-taxogenomics of the genus Tautonia with descriptions of Tautonia marina sp. nov., Tautonia rosea sp. nov., and emended description of the genus. Syst. Appl. Microbiol. 2021, 44, 126229. [Google Scholar] [CrossRef] [PubMed]
- Bondoso, J.; Albuquerque, L.; Lobo-da-Cunha, A.; da Costa, M.S.; Harder, J.; Lage, O.M. Rhodopirellula lusitana sp. nov. and Rhodopirellula rubra sp. nov., isolated from the surface of macroalgae. Syst. Appl. Microbiol. 2014, 37, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Gaurav, K.; Jagadeeshwari, U.; Deepshikha, G.; Ch, S. Roseimaritima sediminicola sp. nov., a new member of Planctomycetaceae isolated from Chilika lagoon. Int. J. Syst. Evol. Microbiol. 2020, 70, 2616–2623. [Google Scholar] [CrossRef]
- Bondoso, J.; Albuquerque, L.; Nobre, M.F.; Lobo-da-Cunha, A.; da Costa, M.S.; Lage, O.M. Roseimaritima ulvae gen. nov., sp. nov. and Rubripirellula obstinata gen. nov., sp. nov. two novel planctomycetes isolated from the epiphytic community of macroalgae. Syst. Appl. Microbiol. 2015, 38, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Vitorino, I.R.; Lobo-da-Cunha, A.; Vasconcelos, V.; Lage, O.M. Rubinisphaera margarita sp. nov., a novel planctomycete isolated from marine sediments collected in the Portuguese north coast. Int. J. Syst. Evol. Microbiol. 2022, 72. [Google Scholar] [CrossRef] [PubMed]
- Kovaleva, O.L.; Elcheninov, A.G.; Toshchakov, S.V.; Novikov, A.A.; Bonch-Osmolovskaya, E.A.; Kublanov, I.V. Tautonia sociabilis gen. nov., sp. nov., a novel thermotolerant planctomycete, isolated from a 4000 m deep subterranean habitat. Int. J. Syst. Evol. Microbiol. 2019, 69, 2299–2304. [Google Scholar] [CrossRef]
- Kulichevskaya, I.S.; Serkebaeva, Y.M.; Kim, Y.; Rijpstra, W.I.; Damste, J.S.; Liesack, W.; Dedysh, S.N. Telmatocola sphagniphila gen. nov., sp. nov., a novel dendriform planctomycete from northern wetlands. Front. Microbiol. 2012, 3, 146. [Google Scholar] [CrossRef] [Green Version]
- Kulichevskaya, I.S.; Ivanova, A.A.; Detkova, E.N.; Rijpstra, W.I.C.; Sinninghe Damste, J.S.; Dedysh, S.N. Tundrisphaera lichenicola gen. nov., sp. nov., a psychrotolerant representative of the family Isosphaeraceae from lichen-dominated tundra soils. Int. J. Syst. Evol. Microbiol. 2017, 67, 3583–3589. [Google Scholar] [CrossRef]
- Seeger, C.; Butler, M.K.; Yee, B.; Mahajan, M.; Fuerst, J.A.; Andersson, S.G.E. Tuwongella immobilis gen. nov., sp. nov., a novel non-motile bacterium within the phylum Planctomycetes. Int. J. Syst. Evol. Microbiol. 2017, 67, 4923–4929. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Krieg, N.R.; Staley, J.T.; Brown, D.R.; Hedlund, B.P.; Paster, B.J.; Ward, N.L.; Ludwig, W.; Whitman, W.B. Phylum XXV. Planctomycetes Garrity and Holt 2001 137 emend. Ward. In Bergey’s Manual of Systematic Bacteriology: The Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes; Springer: New York, NY, USA, 2010; Volume 4. [Google Scholar]
- Fukunaga, Y.; Kurahashi, M.; Sakiyama, Y.; Ohuchi, M.; Yokota, A.; Harayama, S. Phycisphaera mikurensis gen. nov., sp. nov., isolated from a marine alga, and proposal of Phycisphaeraceae fam. nov., Phycisphaerales ord. nov. and Phycisphaerae classis nov. in the phylum Planctomycetes. J. Gen. Appl. Microbiol. 2009, 55, 267–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lodha, T.; Narvekar, S.; Karodi, P. Classification of uncultivated anammox bacteria and Candidatus Uabimicrobium into new classes and provisional nomenclature as Candidatus Brocadiia classis nov. and Candidatus Uabimicrobiia classis nov. of the phylum Planctomycetes and novel family Candidatus Scalinduaceae fam. nov to accommodate the genus Candidatus Scalindua. Syst. Appl. Microbiol. 2021, 44, 126272. [Google Scholar] [CrossRef] [PubMed]
- Jetten, M.; Op den Camp, H.; Kuenen, J.G.; Strous, M. Description of the order brocadiales. Mitochondrion 2010. [Google Scholar]
- van Niftrik, L.; Jetten, M.S. Anaerobic ammonium-oxidizing bacteria: Unique microorganisms with exceptional properties. Microbiol. Mol. Biol. Rev. 2012, 76, 585–596. [Google Scholar] [CrossRef] [Green Version]
- Vitorino, I.R.; Lage, O.M. The Planctomycetia: An overview of the currently largest class within the phylum Planctomycetes. Antonie Leeuwenhoek 2022, 115, 169–201. [Google Scholar] [CrossRef]
- Kallscheuer, N.; Jogler, C. The bacterial phylum Planctomycetes as novel source for bioactive small molecules. Biotechnol. Adv. 2021, 53, 107818. [Google Scholar] [CrossRef]
- Belova, S.E.; Saltykova, V.A.; Dedysh, S.N. Antimicrobial Activity of a Novel Freshwater Planctomycete Lacipirellula parvula PX69T. Microbiology 2020, 89, 503–509. [Google Scholar] [CrossRef]
- Jeske, O.; Surup, F.; Ketteniss, M.; Rast, P.; Forster, B.; Jogler, M.; Wink, J.; Jogler, C. Developing Techniques for the Utilization of Planctomycetes As Producers of Bioactive Molecules. Front. Microbiol. 2016, 7, 1242. [Google Scholar] [CrossRef] [Green Version]
- Graca, A.P.; Calisto, R.; Lage, O.M. Planctomycetes as Novel Source of Bioactive Molecules. Front. Microbiol. 2016, 7, 1241. [Google Scholar] [CrossRef] [Green Version]
- Sandargo, B.; Jeske, O.; Boedeker, C.; Wiegand, S.; Wennrich, J.P.; Kallscheuer, N.; Jogler, M.; Rohde, M.; Jogler, C.; Surup, F. Stieleriacines, N-Acyl Dehydrotyrosines From the Marine Planctomycete Stieleria neptunia sp. nov. Front. Microbiol. 2020, 11, 1408. [Google Scholar] [CrossRef]
- Fedorenko, V.; Genilloud, O.; Horbal, L.; Marcone, G.L.; Marinelli, F.; Paitan, Y.; Ron, E.Z. Antibacterial Discovery and Development: From Gene to Product and Back. Biomed. Res. Int. 2015, 2015, 591349. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.; Vaddavalli, R.; Siripuram, S.; Eedara, R.V.V.; Yadav, S.; Rabishankar, O.; Lodha, T.; Chintalapati, S.; Chintalapati, V. Planctopirus hydrillae sp. nov., an antibiotic producing Planctomycete isolated from the aquatic plant Hydrilla and its whole genome shotgun sequence analysis. J. Antibiot. 2018, 71, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Jeske, O.; Jogler, M.; Petersen, J.; Sikorski, J.; Jogler, C. From genome mining to phenotypic microarrays: Planctomycetes as source for novel bioactive molecules. Antonie Leeuwenhoek 2013, 104, 551–567. [Google Scholar] [CrossRef] [PubMed]
- Kallscheuer, N.; Jeske, O.; Sandargo, B.; Boedeker, C.; Wiegand, S.; Bartling, P.; Jogler, M.; Rohde, M.; Petersen, J.; Medema, M.H.; et al. The planctomycete Stieleria maiorica Mal15(T) employs stieleriacines to alter the species composition in marine biofilms. Commun. Biol. 2020, 3, 303. [Google Scholar] [CrossRef] [PubMed]
- Kallscheuer, N.; Moreira, C.; Airs, R.; Llewellyn, C.A.; Wiegand, S.; Jogler, C.; Lage, O.M. Pink- and orange-pigmented Planctomycetes produce saproxanthin-type carotenoids including a rare C45 carotenoid. Environ. Microbiol. Rep. 2019, 11, 741–748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panter, F.; Garcia, R.; Thewes, A.; Zaburannyi, N.; Bunk, B.; Overmann, J.; Gutierrez, M.V.; Krug, D.; Muller, R. Production of a Dibrominated Aromatic Secondary Metabolite by a Planctomycete Implies Complex Interaction with a Macroalgal Host. ACS Chem. Biol. 2019, 14, 2713–2719. [Google Scholar] [CrossRef]
- Surup, F.; Wiegand, S.; Boedeker, C.; Heuer, A.; Peeters, S.H.; Jogler, M.; Jetten, M.S.M.; Rohde, M.; Jogler, C.; Kallscheuer, N. Stieleria varia sp. nov., isolated from wood particles in the Baltic Sea, constitutes a novel species in the family Pirellulaceae within the phylum Planctomycetes. Antonie Leeuwenhoek 2020, 113, 1953–1963. [Google Scholar] [CrossRef]
- Lage, O.M.; Bondoso, J. Planctomycetes diversity associated with macroalgae. FEMS Microbiol. Ecol. 2011, 78, 366–375. [Google Scholar] [CrossRef] [Green Version]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; Wiley: Hoboken, NJ, USA, 1991; pp. 115–175. [Google Scholar]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [Green Version]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Lee, I.; Chalita, M.; Ha, S.M.; Na, S.I.; Yoon, S.H.; Chun, J. ContEst16S: An algorithm that identifies contaminated prokaryotic genomes using 16S RNA gene sequences. Int. J. Syst. Evol. Microbiol. 2017, 67, 2053–2057. [Google Scholar] [CrossRef]
- Kallscheuer, N.; Wiegand, S.; Peeters, S.H.; Jogler, M.; Boedeker, C.; Heuer, A.; Rast, P.; Jetten, M.S.M.; Rohde, M.; Jogler, C. Description of three bacterial strains belonging to the new genus Novipirellula gen. nov., reclassificiation of Rhodopirellula rosea and Rhodopirellula caenicola and readjustment of the genus threshold of the phylogenetic marker rpoB for Planctomycetaceae. Antonie Leeuwenhoek 2020, 113, 1779–1795. [Google Scholar] [CrossRef]
- Bondoso, J.; Harder, J.; Lage, O.M. rpoB gene as a novel molecular marker to infer phylogeny in Planctomycetales. Antonie Leeuwenhoek 2013, 104, 477–488. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [Green Version]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Goker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [Green Version]
- Lee, I.; Ouk Kim, Y.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Medlar, A.J.; Törönen, P.; Holm, L. AAI-profiler: Fast proteome-wide exploratory analysis reveals taxonomic identity, misclassification and contamination. Nucleic Acids Res. 2018, 46, W479–W485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alanjary, M.; Steinke, K.; Ziemert, N. AutoMLST: An automated web server for generating multi-locus species trees highlighting natural product potential. Nucleic Acids Res. 2019, 47, W276–W282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [Green Version]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef] [Green Version]
- Santos, J.D.; Vitorino, I.; De la Cruz, M.; Diaz, C.; Cautain, B.; Annang, F.; Perez-Moreno, G.; Gonzalez Martinez, I.; Tormo, J.R.; Martin, J.M.; et al. Bioactivities and Extract Dereplication of Actinomycetales Isolated From Marine Sponges. Front. Microbiol. 2019, 10, 727. [Google Scholar] [CrossRef] [Green Version]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glockner, F.O.; Ludwig, W.; Schleifer, K.H.; Whitman, W.B.; Euzeby, J.; Amann, R.; Rossello-Mora, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef]
- Kohn, T.; Wiegand, S.; Boedeker, C.; Rast, P.; Heuer, A.; Jetten, M.S.M.; Schuler, M.; Becker, S.; Rohde, C.; Muller, R.W.; et al. Planctopirus ephydatiae, a novel Planctomycete isolated from a freshwater sponge. Syst. Appl. Microbiol. 2020, 43, 126022. [Google Scholar] [CrossRef]
- Wiegand, S.; Jogler, M.; Boedeker, C.; Heuer, A.; Rast, P.; Peeters, S.H.; Jetten, M.S.M.; Kaster, A.K.; Rohde, M.; Kallscheuer, N.; et al. Additions to the genus Gimesia: Description of Gimesia alba sp. nov., Gimesia algae sp. nov., Gimesia aquarii sp. nov., Gimesia aquatilis sp. nov., Gimesia fumaroli sp. nov. and Gimesia panareensis sp. nov., isolated from aquatic habitats of the Northern Hemisphere. Antonie Leeuwenhoek 2020, 113, 1999–2018. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Rodriguez, R.L.; Konstantinidis, K.T. MyTaxa: An advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Res. 2014, 42, e73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunath, B.J.; Bremges, A.; Weimann, A.; McHardy, A.C.; Pope, P.B. Metagenomics and CAZyme Discovery. Methods Mol. Biol. 2017, 1588, 255–277. [Google Scholar] [CrossRef] [PubMed]
- Izumi, H.; Sagulenko, E.; Webb, R.I.; Fuerst, J.A. Isolation and diversity of planctomycetes from the sponge Niphates sp., seawater, and sediment of Moreton Bay, Australia. Antonie Leeuwenhoek 2013, 104, 533–546. [Google Scholar] [CrossRef] [PubMed]
- Winkelmann, N.; Harder, J. An improved isolation method for attached-living Planctomycetes of the genus Rhodopirellula. J. Microbiol. Methods 2009, 77, 276–284. [Google Scholar] [CrossRef]
- Kallscheuer, N.; Rast, P.; Jogler, M.; Wiegand, S.; Kohn, T.; Boedeker, C.; Jeske, O.; Heuer, A.; Quast, C.; Glockner, F.O.; et al. Analysis of bacterial communities in a municipal duck pond during a phytoplankton bloom and isolation of Anatilimnocola aggregata gen. nov., sp. nov., Lacipirellula limnantheis sp. nov. and Urbifossiella limnaea gen. nov., sp. nov. belonging to the phylum Planctomycetes. Environ. Microbiol. 2020, 23, 1379–1396. [Google Scholar] [CrossRef]
- Schlesner, H.; Hirsch, P. Rejection of the genus name Pirella for pear-shaped budding bacteria and proposal to create the genus Pirellula gen. nov. Int. J. Syst. Bacteriol. 1987, 37, 441. [Google Scholar] [CrossRef] [Green Version]
- Santana-Molina, C.; Henriques, V.; Hornero-Méndez, D.; Devos, D.P.; Rivas-Marin, E. The ‘squalene route’ to carotenoid biosynthesis is widespread in Bacteria. bioRxiv 2021. [Google Scholar] [CrossRef]
- Romano, S.; Jackson, S.A.; Patry, S.; Dobson, A.D.W. Extending the “One Strain Many Compounds” (OSMAC) Principle to Marine Microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef] [Green Version]
- Santos, J.D.N.; João, S.A.; Martín, J.; Vicente, F.; Reyes, F.; Lage, O.M. iChip-Inspired Isolation, Bioactivities and Dereplication of Actinomycetota from Portuguese Beach Sediments. Microorganisms 2022, 10, 1471. [Google Scholar] [CrossRef]






| Characteristics | ICT_E10.1T | a Enr13T | b Mal15T | c Pla52nT |
|---|---|---|---|---|
| Similarity (%) of the complete 16S rRNA gene sequence | na | 98.8 | 98.4 | 95.8 |
| Genome size (Mb) | 9.8 | 11.0 * | 9.9 * | 9.6 * |
| G+C content (mol%) | 58.8 | 58.9 * | 59.3 * | 56.0 * |
| Number of protein-encoding genes | 6964 | 7799 | 6936 | 7000 |
| Number of hypothetical proteins | 4578 | 5220 | 4522 | 4848 |
| dDDH estimated with strain ICT_E10.1T (%) | na | 37.0 | 24.0 | 23.2 |
| ANI value with strain ICT_E10.1T (%) | na | 88.5 | 79.9 | 71.4 |
| Similarity (%) of the rpoB gene with strain ICT_E10.1T | na | 94.1 | 88.9 | 82.0 |
| AAI value with strain ICT_E10.1T (%) | na | 90.6 | 83.1 | 67.8 |
| Family of CAZymes | Number of Enzymes | |||
|---|---|---|---|---|
| ICT_E10.1T | a Enr13T | b Mal15T | c Pla52nT | |
| Glycoside hydrolases | 119 | 155 | 146 | 96 |
| Carbohydrate esterases | 34 | 45 | 43 | 46 |
| Glycosyltransferases | 118 | 124 | 130 | 113 |
| Auxiliary activities | 7 | 8 | 6 | 7 |
| Carbohydrate-binding modules | 100 | 94 | 84 | 79 |
| Polysaccharide lyases | 14 | 16 | 10 | 6 |
| Unknown | 15 | 13 | 15 | 14 |
| Cohesins | 1 | 1 | 1 | 0 |
| Total | 438 | 456 | 435 | 361 |
| Characteristics | ICT_E10.1T | Enr13T | Mal15T | Pla52nT |
| Cell shape | Spherical to ovoid | Spherical to ovoid | Spherical to pear-shaped | Ovoid to grain rice-shaped |
| Cell size (µm) | 1.7 ± 0.3 × 1.4 ± 0.3 | 1.6 ± 0.1 × 1.1 ± 0.1 | 1.9 ± 0.2 × 1.4 ± 0.2 | 1.8 ± 0.3 × 0.9 ± 0.2 |
| Main form of aggregation between cells | Aggregates and short chains | Aggregates | Rosettes | Rosettes and short chains |
| Reproduction | Budding | Budding | Budding | Budding |
| Motility | Yes | Yes | Yes | Yes |
| Crateriform structures | No | Yes | Yes | Yes |
| Colony color | Pink | Pink | Pink | Light orange |
| Isolation source (location) | Brackish sediments (Portugal) | Posidonia sp. (Italy) | Sediments (Spain) | Wood particles in sea water (Germany) |
| Temperature range (°C) | 20–30 | 9–35 | 11–37 | 20–45 |
| pH range | 6.5–11 | 6.5–9.0 | 5.5–9.0 | 6.0–8.0 |
| % (w/v) NaCl tolerance | 0.5–3 | NDA | NDA | NDA |
| Vitamin B12 requirement | No | NDA | NDA | NDA |
| Carbon sources | NAG, cellobiose, galactose, fructose, lactose, arabinose, xylose and glucose | NDA | NAG, arabinose, cellobiose, fucose, fructose, galactose, gentiobiose, glucose, gluconic acid, glucuronamide, glucuronic acid, lactose, lactulose, mannose, melibiose, glucoside, draffinose, rhamnose, sucrose, trehalose, turanose, psicose | NDA |
| Nitrogen sources | NAG, peptone, yeast extract, ammonium sulfate, casamino acids, urea, sodium nitrate, asparagine, glutamine, histidine, phenylalanine, tryptophan, and alanine | NDA | NDA | NDA |
| Respiration | Aerobic | Aerobic | Aerobic | Aerobic |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vitorino, I.R.; Klimek, D.; Calusinska, M.; Lobo-da-Cunha, A.; Vasconcelos, V.; Lage, O.M. Stieleria sedimenti sp. nov., a Novel Member of the Family Pirellulaceae with Antimicrobial Activity Isolated in Portugal from Brackish Sediments. Microorganisms 2022, 10, 2151. https://doi.org/10.3390/microorganisms10112151
Vitorino IR, Klimek D, Calusinska M, Lobo-da-Cunha A, Vasconcelos V, Lage OM. Stieleria sedimenti sp. nov., a Novel Member of the Family Pirellulaceae with Antimicrobial Activity Isolated in Portugal from Brackish Sediments. Microorganisms. 2022; 10(11):2151. https://doi.org/10.3390/microorganisms10112151
Chicago/Turabian StyleVitorino, Inês Rosado, Dominika Klimek, Magdalena Calusinska, Alexandre Lobo-da-Cunha, Vítor Vasconcelos, and Olga Maria Lage. 2022. "Stieleria sedimenti sp. nov., a Novel Member of the Family Pirellulaceae with Antimicrobial Activity Isolated in Portugal from Brackish Sediments" Microorganisms 10, no. 11: 2151. https://doi.org/10.3390/microorganisms10112151
APA StyleVitorino, I. R., Klimek, D., Calusinska, M., Lobo-da-Cunha, A., Vasconcelos, V., & Lage, O. M. (2022). Stieleria sedimenti sp. nov., a Novel Member of the Family Pirellulaceae with Antimicrobial Activity Isolated in Portugal from Brackish Sediments. Microorganisms, 10(11), 2151. https://doi.org/10.3390/microorganisms10112151









