Virulence Potential and Antibiotic Susceptibility of S. aureus Strains Isolated from Food Handlers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Processing
2.2. Storage and Regeneration of Bacterial Cultures
2.3. DNA Extraction
2.4. S. aureus Genotyping
2.5. Detection of Toxin Genes, Virulence Factors and Methicillin Resistance by RT-PCR
2.6. Electrophoretic Analysis of PCR Products
2.7. Preparation of Culture Medium and Inoculum Suspension for the Antibiotic Susceptibility Tests
2.8. Antibiotic Susceptibility Testing by Disc Diffusion Method
2.9. Determination of Vancomycin Resistance by Dilution (MIC)
2.10. Biochemical Tests
2.10.1. Detection of DNase-Producing Strains
2.10.2. Detection of Strains with Proteolytic Activity
2.10.3. Haemolysis on Blood Agar
2.10.4. Starch Hydrolysis
3. Results and Discussion
3.1. S. aureus—Prevalence on Food Handlers
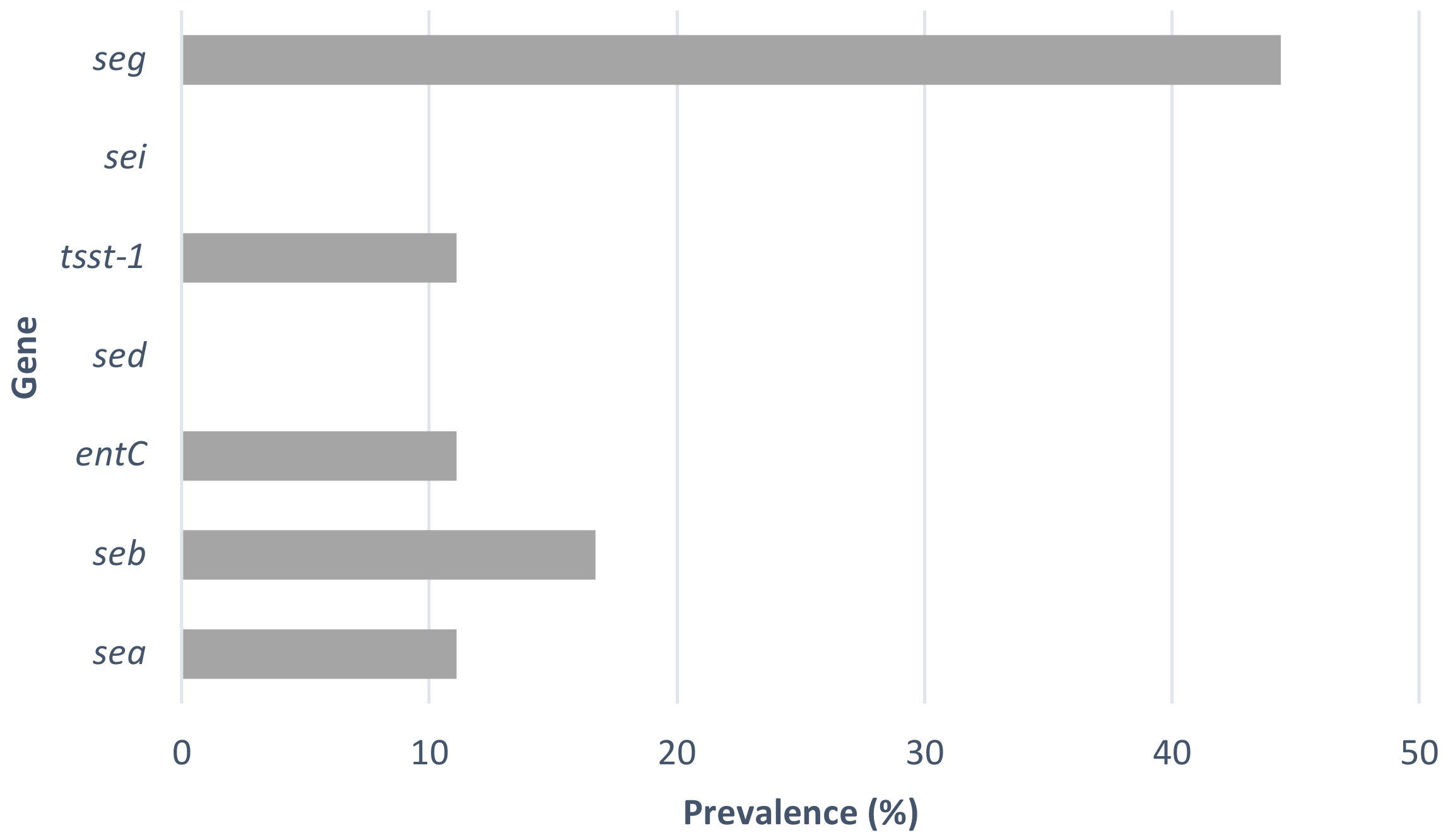
3.2. Detection of Enterotoxin Genes
3.3. Antimicrobial Susceptibility
3.4. Virulence Factors
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lambrechts, A.; Human, I.S.; Doughari, J.H.; Lues, J.F.R. Bacterial contamination of the hands of food handlers as indicator of hand washing efficacy in some convenient food industries. Pak. J. Med. Sci. 2014, 30, 755–758. [Google Scholar]
- ECDC. The European Union One Health 2020 Zoonoses Report. EFSA J. 2021, 12, 324. [Google Scholar]
- EFSA, E.F.S.A. and ECDC, European Centre for DiseasePrevention and Control. The European Union summary report on trends and sources ofzoonoses, zoonotic agents and food-borne outbreaks in 2015. EFSA J. 2016, 14, 231. [Google Scholar]
- Greig, J.D.; Todd, E.C.D.; Bartleson, C.A.; Michaels, B.S. Outbreaks where food workers have been implicated in the spread of foodborne disease. Part 1. Description of the problem, methods, and agents involved. J. Food Prot. 2007, 70, 1752–1761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kluytmans, J.; van Belkum, A.; Verbrugh, H. Nasal carriage of Staphylococcus aureus: Epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 1997, 10, 505–520. [Google Scholar] [CrossRef]
- Argudín, M.Á.; Mendoza, M.C.; Rodicio, M.R. Food Poisoning and Staphylococcus aureus Enterotoxins. Toxins 2010, 2, 1751–1773. [Google Scholar] [CrossRef]
- Van Tonder, I.; Lues, J.F.; Theron, M.M. The personal and general hygiene practices of food handlers in the delicatessen sections of retail outlets in South Africa. J. Environ. Health 2007, 70, 33–38. [Google Scholar]
- Host Institution: Department of Food Hygiene and Technology, Institute of Food Science and Technology, Universidad de León, León, Spain; Likotrafiti, E.; Oniciuc, E.A.; Prieto, M.; Santos, J.A.; López, S.; Alvarez-Ordóñez, A. Risk assessment of antimicrobial resistance along the food chain through culture-independent methodologies. EFSA J. 2018, 16, e160811. [Google Scholar]
- Deurenberg, R.H.; Stobberingh, E.E. The evolution of Staphylococcus aureus. Infect. Genet. Evol. 2008, 8, 747–763. [Google Scholar] [CrossRef]
- Dinges, M.M.; Orwin, P.M.; Schlievert, P.M. Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 2000, 13, 16–34. [Google Scholar] [CrossRef]
- Štěpán, J.; Pantůček, R.; Doškař, J. Molecular diagnostics of clinically important staphylococci. Folia Microbiol. 2004, 49, 353–386. [Google Scholar] [CrossRef] [PubMed]
- Gordon, R.J.; Lowy, F.D. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 2008, 46 (Suppl. S5), S350–S359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plata, K.; Rosato, A.E.; Wegrzyn, G. Staphylococcus aureus as an infectious agent: Overview of biochemistry and molecular genetics of its pathogenicity. Acta Biochim. Pol. 2009, 56, 597–612. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.; Hensel, M. Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 2004, 17, 14–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsen, A.R.; Stegger, M.; Sørum, M. Spa typing directly from a mecA, spa and pvl multiplex PCR assay—a cost-effective improvement for methicillin-resistant Staphylococcus aureus surveillance. Clin. Microbiol. Infect. 2008, 14, 611–614. [Google Scholar] [CrossRef]
- AL-Tam, F.; Brunel, A.-S.; Bouzinbi, N.; Corne, P.; Bañuls, A.-L.; Shahbazkia, H.R. DNAGear- a free software for spa type identification in Staphylococcus aureus. BMC Res. Notes 2012, 5, 642. [Google Scholar] [CrossRef] [Green Version]
- Sharma, N.K.; Rees, C.E.; Dodd, C.E. Development of a single-reaction multiplex PCR toxin typing assay for Staphylococcus aureus strains. Appl. Environ. Microbiol. 2000, 66, 1347–1353. [Google Scholar] [CrossRef] [Green Version]
- Wongboot, W.; Chomvarin, C.; Engchanil, C.; Chaimanee, P. Multiplex PCR for detection of superantigenic toxin genes in methicillin-sensitive and methicillin-resistant Staphylococcus aureus isolated from patients and carriers of a hospital in northeast Thailand. Southeast Asian J. Trop. Med. Public Health 2013, 44, 660–671. [Google Scholar]
- Rall, V.L.; Vieira, F.; Rall, R.; Vieitis, R.; Fernandes, A.; Candeias, J.; Cardoso, K.; Araújo, J. PCR detection of staphylococcal enterotoxin genes in Staphylococcus aureus strains isolated from raw and pasteurized milk. Vet. Microbiol. 2008, 132, 408–413. [Google Scholar] [CrossRef]
- Thomas, L.C.; Gidding, H.; Ginn, A.; Olma, T.; Iredell, J. Development of a real-time Staphylococcus aureus and MRSA (SAM-) PCR for routine blood culture. J. Microbiol. Methods 2007, 68, 296–302. [Google Scholar] [CrossRef]
- DTU Food National Food Institute; European Union Reference Laboratory-Antibiotic Resistance. Protocol for PCR Amplification of mecA, mecC (mecALGA251), Spa and Pvl. 2nd Version, September 2012. Available online: https://www.eurl-ar.eu/CustomerData/Files/Folders/21-protocols/279_pcr-spa-pvl-meca-mecc-sept12.pdf (accessed on 17 December 2019).
- Testing T.E.C.o.A.S. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 12.0. 2022. Available online: http://www.eucast.org (accessed on 19 April 2022).
- Steward, C.D.; Raney, P.M.; Morrell, A.K.; Williams, P.P.; McDougal, L.K.; Jevitt, L.; McGowan, J.E.; Tenover, F.C. Testing for Induction of Clindamycin Resistance in Erythromycin-Resistant Isolates of Staphylococcus aureus. J. Clin. Microbiol. 2005, 43, 1716–1721. [Google Scholar] [CrossRef] [Green Version]
- Kateete, D.P.; Kimani, C.N.; Katabazi, F.A.; Okeng, A.; Okee, M.S.; Nanteza, A.; Joloba, M.L.; Najjuka, F.C. Identification of Staphylococcus aureus: DNase and Mannitol salt agar improve the efficiency of the tube coagulase test. Ann. Clin. Microbiol. Antimicrob. 2010, 9, 23. [Google Scholar] [CrossRef] [Green Version]
- Saran, S.; Isar, J.; Saxena, R.K. A modified method for the detection of microbial proteases on agar plates using tannic acid. J Biochem. Biophys. Methods 2007, 70, 697–699. [Google Scholar] [CrossRef]
- Adams, M.R.; Moss, M.O. Food Microbiology; The Royal Society of Chemistry: Cambridge, UK, 2008; pp. 18, 19, 160, 161, 252–257, 418–426. [Google Scholar]
- Wertheim, H.; Melles, D.C.; Vos, M.C.; van Leeuwen, W.; van Belkum, A.; Verbrugh, H.A.; Nouwen, J.L. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 2005, 5, 751–762. [Google Scholar] [CrossRef]
- Fawzi, M.; Gomaa, N.F.; Bakr, W.M. Assessment of hand washing facilities, personal hygiene and the bacteriological quality of hand washes in some grocery and dairy shops in Alexandria, Egypt. J. Egypt Public Health Assoc. 2009, 84, 71–93. [Google Scholar]
- Lues, J.F.R.; van Tonder, I. The occurrence of indicator bacteria on hands and aprons of food handlers in the delicatessen sections of a retail group. Food Control 2007, 18, 326–332. [Google Scholar] [CrossRef]
- Kluytmans, J.; Wertheim, H. Nasal Carriage of Staphylococcus aureus and Prevention of Nosocomial Infections. Infection 2005, 33, 3–8. [Google Scholar] [CrossRef]
- Bencardino, D.; Amagliani, G.; Brandi, G. Carriage of Staphylococcus aureus among food handlers: An ongoing challenge in public health. Food Control 2021, 130, 108362. [Google Scholar]
- Balaban, N.; Rasooly, A. Staphylococcal enterotoxins. Int. J. Food Microbiol. 2000, 61, 1–10. [Google Scholar] [CrossRef]
- Sadat, A.; Shata, R.R.; Farag, A.M.M.; Ramadan, H.; Alkhedaide, A.; Soliman, M.M.; Elbadawy, M.; Abugomaa, A.; Awad, A. Prevalence and Characterization of PVL-Positive Staphylococcus aureus Isolated from Raw Cow’s Milk. Toxins 2022, 14, 97. [Google Scholar] [CrossRef]
- Xu, S.X.; McCormick, J.K. Staphylococcal superantigens in colonization and disease. Front Cell Infect. Microbiol. 2012, 2, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ertas, N.; Gönülalan, Z.; Yildirim, Y.; Kum, E. Detection of Staphylococcus aureus enterotoxins in sheep cheese and dairy desserts by multiplex PCR technique. Int. J. Food Microbiol. 2010, 142, 74–77. [Google Scholar] [CrossRef] [PubMed]
- Pinchuk, I.; Beswick, E.; Reyes, V. Staphylococcal Enterotoxins. Toxins 2010, 2, 2177–2197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, R.; Pinho, E.; Almeida, G.; Azevedo, N.F.; Almeida, C. Prevalence and Diversity of Staphylococcus aureus and Staphylococcal Enterotoxins in Raw Milk From Northern Portugal. Front. Microbiol. 2022, 13, 703. [Google Scholar] [CrossRef] [PubMed]
- Fooladvand, S.; Sarmadian, H.; Habibi, D.; van Belkum, A.; Ghaznavi-Rad, E. High prevalence of methicillin resistant and enterotoxin gene-positive Staphylococcus aureus among nasally colonized food handlers in central Iran. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 87–92. [Google Scholar] [CrossRef]
- Pereira, V.; Lopes, C.; Castro, A.; Silva, J.; Gibbs, P.; Teixeira, P. Characterization for enterotoxin production, virulence factors, and antibiotic susceptibility of Staphylococcus aureus isolates from various foods in Portugal. Food Microbiol. 2009, 26, 278–282. [Google Scholar] [CrossRef]
- Heritage, J.; Evans, E.G.V.; Killington, R.A. Microbiology in Action; Cambridge University Press: Cambridge, UK, 1999; pp. 111–112, 271–288. [Google Scholar]
- Crago, B.; Ferrato, C.; Drews, S.J.; Svenson, L.W.; Tyrrell, G.; Louie, M. Prevalence of Staphylococcus aureus and Methicillin-Resistant S. aureus (MRSA) in Food Samples Associated with Foodborne Illness in Alberta, Canada from 2007 to 2010 Food Microbiology; Elsevier: Amsterdam, The Netherlands, 2012; Volume 32, pp. 202–205. [Google Scholar]
- Loir, Y.L.; Baron, F.; Gautier, M. Staphylococcus aureus and food poisoning. Genet. Mol. Res. 2003, 2, 63–76. [Google Scholar]
- Jacob, M. Safe Food Handling: A Training Guide for Managers in Food Service Establishments; World Health Organization: Geneva, Switzerland, 1989. [Google Scholar]
- Fey, P.; Saiïd-Salim, B.; Rupp, M.E.; Hinrichs, S.H.; Boxrud, D.J.; Davis, C.C.; Kreiswirth, B.N.; Schlievert, P.M. Comparative Molecular Analysis of Community- or Hospital-Acquired Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2003, 47, 196–203. [Google Scholar] [CrossRef] [Green Version]
- Bamberger, D. Bacteremia and endocarditis due to methicillinresistant Staphylococcus aureus: The potential role of daptomycin. Ther. Clin. Risk Manag. 2007, 3, 675–684. [Google Scholar]
- Herold, B.; Immergluck, L.C.; Maranan, M.C.; Lauderdale, D.S.; Gaskin, R.E.; Boyle-Vavra, S.; Leitch, C.D.; Daum, R.S. Community-Acquired Methicillin-Resistant Staphylococcus aureus in children with no identified predisposing risk. J. Am. Med. Assoc. 1998, 279, 593–598. [Google Scholar] [CrossRef]
- Sharma, K.D.; Saini, R.P.; Karthik, L. Current trends of antibiotic resistance in clinical isolates of Staphylococcus aureus. Front. Biol. 2014, 9, 287–290. [Google Scholar] [CrossRef]
- Fowler, V.; Olsen, M.K.; Corey, G.R.; Woods, C.W.; Cabell, C.H.; Reller, L.B.; Cheng, A.C.; Dudley, T.; Oddone, E.Z. Clinical Identifiers of Complicated Staphylococcus aureus Bacteremia. Antimicrob. Agents Chemother. 2003, 163, 2066–2072. [Google Scholar] [CrossRef] [Green Version]
- EFSA Antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in the EU in 2013. EFSA J. 2015, 13, 4036.
- Piątkowska, E.; Piątkowski, J.; Przondo-Mordarska, A. The strongest resistance of Staphylococcus aureus to erythromycin is caused by decreasing uptake of the antibiotic into the cells. Cell Mol Biol Lett. 2012, 17, 633–645. [Google Scholar] [CrossRef]
- Bruce, S.A.; Smith, J.T.; Mydosh, J.L.; Ball, J.; Needle, D.B.; Gibson, R.; Andam, C.P. Shared antibiotic resistance and virulence genes in Staphylococcus aureus from diverse animal hosts. Sci. Rep. 2022, 12, 4413. [Google Scholar] [CrossRef]
- Uehara, Y. Current Status of Staphylococcal Cassette Chromosome mec (SCCmec). Antibiotics 2022, 11, 86. [Google Scholar] [CrossRef]
- Cunha, M.; Rugolo, L.; Lopes, C. Study of virulence factors in coagulase-negative staphylococci isolated from newborns. Mem. Inst. Oswaldo Cruz 2006, 101, 661–668. [Google Scholar] [CrossRef] [Green Version]
- Berends, E.T.; Horswill, A.R.; Haste, N.M.; Monestier, M.; Nizet, V.; Von Köckritz-Blickwede, M. Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J. Innate Immun. 2010, 2, 576–586. [Google Scholar] [CrossRef] [Green Version]
- Deplanche, M.; Mouhali, N.; Nguyen, M.-T.; Cauty, C.; Ezan, F.; Diot, A.; Raulin, L.; Dutertre, S.; Langouet, S.; Legembre, P.; et al. . Staphylococcus aureus induces DNA damage in host cell. Sci. Rep. 2019, 9, 7694. [Google Scholar] [CrossRef] [Green Version]
- Berube, B.J.; Wardenburg, J.B. Staphylococcus aureus α-toxin: Nearly a century of intrigue. Toxins 2013, 5, 1140–1166. [Google Scholar] [CrossRef] [Green Version]
- Shaw, L.; Golonka, E.; Potempa, J.; Foster, S.J. The role and regulation of the extracellular proteases of Staphylococcus aureus. Microbiology 2004, 150, 217–228. [Google Scholar] [CrossRef]
- Arvidson, S. Extracellular enzymes. In Gram-Positive Pathogens; Fischetti, V.A., Novick, R.P., Ferretti, J.J., Portnoy, D.A., Rood, J.I., Eds.; ASM Press: Washington, DC, USA, 2006; pp. 478–485. [Google Scholar]
- Pietrocola, G.; Nobile, G.; Rindi, S.; Speziale, P. Staphylococcus aureus Manipulates Innate Immunity through Own and Host-Expressed Proteases. Front. Cell. Infect. Microbiol. 2017, 7, 166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lakshmi, H.P.; Prasad, U.V.; Yeswanth, S.; Swarupa, V.; Prasad, O.H.; Narasu, M.L.; Sarma, P.V.G.K. Molecular characterization of α-amylase from Staphylococcus aureus. Bioinformation 2013, 9, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Lina, G.; Piémont, Y.; Godail-Gamot, F.; Bes, M.; Peter, M.-O.; Gauduchon, V.; Vandenesch, F.; Etienne, J. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 1999, 29, 1128–1132. [Google Scholar] [CrossRef] [PubMed]
- Gillet, Y.; Issartel, B.; Vanhems, P.; Fournet, J.-C.; Lina, G.; Bes, M.; Vandenesch, F.; Piémont, Y.; Brousse, N.; Floret, D.; et al. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet 2002, 359, 753–759. [Google Scholar] [CrossRef]
| Primer Int. Ref. | Obs. | ||
|---|---|---|---|
| SA-U | 5′-TGTATGTATGGAGGTGTAAC-3′ | Forward primer used to amplify toxin genes together with primers SA-A, SA-B, SA-C, ENTC, SA-D and SA-E | [17] |
| SA-A | 5′-ATTAACCGAAGGTTCTGT-3′ | Reverse primer for sea gene | |
| SA-B | 5′-ATAGTGACGAGTTAGGTA-3′ | Reverse primer for seb gene | |
| SA-C | 5′-AAGTACATTTTGTAAGTTCC-3′ | Reverse primer for sec gene | |
| ENT-C | 5′-AATTGTGTTTCTTTTATTTTCATAA-3′ | Reverse primer for sec gene | |
| SA-D | 5′-TTCGGGAAAATCACCCTTAA-3′ | Reverse primer for sed gene | |
| SA-E | 5′-GCCAAAGCTGTCTGAG-3′ | Reverse primer for see gene | |
| SA-tst1-R | 5′-GGCAGCATCAGCCTTATAATTT-3′ | Reverse primer for tst1 gene | [18] |
| SA-tst1-F | 5′-GTGGATCCGTCATTCATTGTT-3′ | Forward primer for tst1 gene | |
| SEG-1 | 5′-AAGTAGACATTTTTGGCGTTCC-3′ | Forward primer for see gene | [19] |
| SEG-2 | 5′-AGAACCATCAAACTCGTATAGC-3′ | Reverse primer for see gene | |
| SEI-1 | 5′-GGTGATATTGGTGTAGGTAAC-3′ | Forward primer for see gene | |
| SEI-2 | 5′-ATCCATATTCTTTGCCTTTACCAG-3′ | Reverse primer for see gene | |
| mecA-1 | 5′-AAAATCGATGGTAAAGGTTGGC-3′ | Forward primer for see gene | [20] |
| mecA-2 | 5′-AGTTCTGCAGTACCGGATTTGC-3′ | Reverse primer for see gene | |
| mecC-F | 5′-GAAAAAAAGGCTTAGAACGCCTC-3′ | Forward primer for mecC gene | [21] |
| mecC-R | 5′-GAAGATCTTTTCCGTTTTCAGC-3′ | Reverse primer for mecC gene | |
| pvl-F | 5’-GCTGGACAAAACTTCTTGGAATAT-3’ | Forward primer for pvl gene | |
| pvl-R | 5’-GATAGGACACCAATAAATTCTGGATTG-3’ | Reverse primer for pvl gene |
| Strain Code | spa Type | Haemolysis | DNase Activity | Starch Hydrolysis | Proteolytic Activity | Toxin Genes Detected | Antibiotic Resistance | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sea | seb | sec | sed | tsst-1 | sei | seg | |||||||
| B863 | t267 | β | + | - | - | - | |||||||
| B864 | t267 | β | + | - | - | x | - | ||||||
| B865 | t571 | β | + | - | + | Erythromycin | |||||||
| B870 | t162 | β | + | - | - | x | - | ||||||
| B904 | t530 | β | + | - | - | x | - | ||||||
| B928 | t148 | β | + | - | - | x | x | - | |||||
| B929 | t2540 | α | + | - | - | x | x | x | x | - | |||
| B937 | t571 | β | + | - | + | Erythromycin | |||||||
| B938 | t130 | β | + | - | - | x | Erythromycin | ||||||
| B939 | t359 | α | + | - | - | - | |||||||
| B1014 | t084 | α | + | - | - | - | |||||||
| B1198 | t002 | β | + | - | - | x | Erythromycin | ||||||
| B1207 | t2164 | β | + | - | - | Erythromycin | |||||||
| B1209 | t148 | β | + | - | - | x | x | - | |||||
| B1252 | t127 | α | + | - | - | x | x | - | |||||
| B1258 | t571 | β | + | - | + | Erythromycin | |||||||
| B1265 | t148 | α | + | - | - | x | Erythromycin | ||||||
| B1270 | t148 | α | + | - | - | x | Erythromycin | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandes, A.; Ramos, C.; Monteiro, V.; Santos, J.; Fernandes, P. Virulence Potential and Antibiotic Susceptibility of S. aureus Strains Isolated from Food Handlers. Microorganisms 2022, 10, 2155. https://doi.org/10.3390/microorganisms10112155
Fernandes A, Ramos C, Monteiro V, Santos J, Fernandes P. Virulence Potential and Antibiotic Susceptibility of S. aureus Strains Isolated from Food Handlers. Microorganisms. 2022; 10(11):2155. https://doi.org/10.3390/microorganisms10112155
Chicago/Turabian StyleFernandes, Adriana, Carla Ramos, Victor Monteiro, Joana Santos, and Paulo Fernandes. 2022. "Virulence Potential and Antibiotic Susceptibility of S. aureus Strains Isolated from Food Handlers" Microorganisms 10, no. 11: 2155. https://doi.org/10.3390/microorganisms10112155
APA StyleFernandes, A., Ramos, C., Monteiro, V., Santos, J., & Fernandes, P. (2022). Virulence Potential and Antibiotic Susceptibility of S. aureus Strains Isolated from Food Handlers. Microorganisms, 10(11), 2155. https://doi.org/10.3390/microorganisms10112155






