Insight into the Taxonomic and Functional Diversity of Bacterial Communities Inhabiting Blueberries in Portugal
Abstract
1. Introduction
2. Materials and Methods
2.1. Metabarcoding of V. myrtillus Populations in Portugal
2.2. Isolation of Bacteria from V. myrtillus
2.3. Identification and PGP Characterization of Isolated Bacterial Strains
2.3.1. MALDI-TOF
2.3.2. Identification Based on Sequencing of 16S rRNA
2.3.3. Analysis of the Plant Growth-Promoting (PGP) Potential of Isolated Strains
2.4. In-Plant Evaluation of PGP Ability
2.5. Colonization Ability of Isolated Bacteria
3. Results
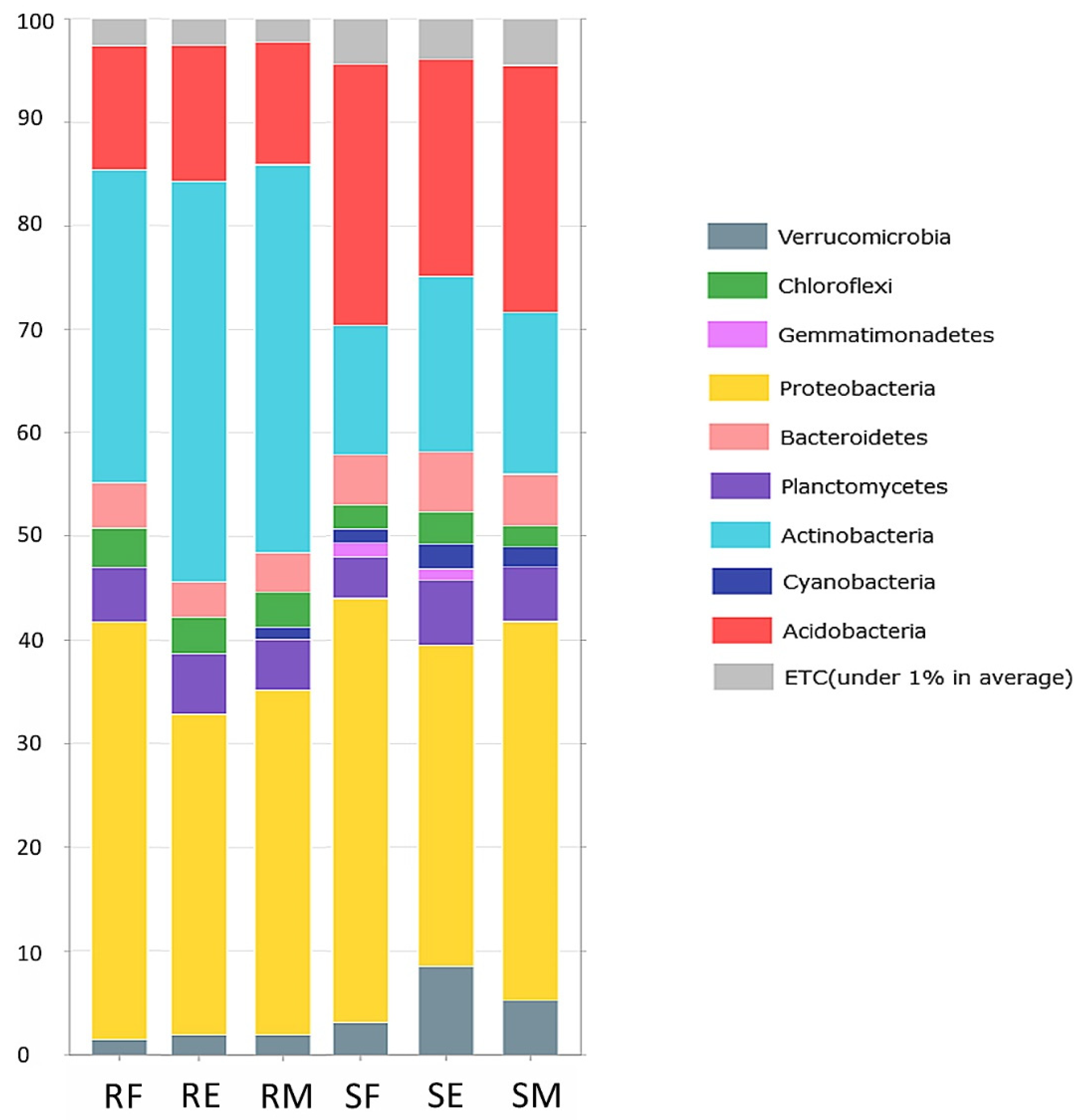
3.1. The Composition of Bacterial Communities Associated with V. myrtillus in Portugal
3.2. Culturomic Analysis of the Bacterial Communities of Portuguese Blueberries
3.3. Evaluation of the PGP Potential of the Isolated Strains
3.4. Analysis of PGP on Plant Assays
3.5. Analysis of Root Colonization in Blueberry Seedlings
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tundis, R.; Tenuta, M.C.; Loizzo, M.R.; Bonesi, M.; Finetti, F.; Trabalzini, L.; Deguin, B. Vaccinium Species (Ericaceae): From Chemical Composition to Bio-Functional Activities. Appl. Sci. 2021, 11, 5655. [Google Scholar] [CrossRef]
- Caspersen, S.; Svensson, B.; Håkansson, T.; Winter, C.; Khalil, S.; Asp, H. Blueberry-Soil interactions from an organic perspective. Sci. Hortic. 2015, 208, 78–91. [Google Scholar] [CrossRef]
- Die, J.V.; Rowland, L.J. Elucidating cold acclimation pathway in blueberry by transcriptome profiling. Environ. Exp. Bot. 2014, 106, 87–98. [Google Scholar] [CrossRef]
- Schoebitz, M.; López, M.D.; Serri, H.; Aravena, V.; Zagal, E.; Roldán, A. Characterization of Bioactive Compounds in Blueberry and Their Impact on Soil Properties in Response to Plant Biostimulants. Commun. Soil Sci. Plant Anal. 2019, 50, 2482–2494. [Google Scholar] [CrossRef]
- Pato, J.; Obeso, J.R.; Ploquin, E.F.; Jiménez-Alfaro, B. Experimental evidence from Cantabrian mountain heathlands suggests new recommendations for management of Vaccinium myrtillus L. Plant Ecol. Divers. 2016, 9, 199–206. [Google Scholar] [CrossRef]
- Gil, J.A.; Gómez-Serrano, M.Á.; López-López, P. Population Decline of the Capercaillie Tetrao urogallus aquitanicus in the Central Pyrenees. Ardeola 2020, 67, 285–306. [Google Scholar] [CrossRef]
- Eckerter, T.; Buse, J.; Förschler, M.; Pufal, G. Additive positive effects of canopy openness on European bilberry (Vaccinium myrtillus) fruit quantity and quality. For. Ecol. Manag. 2019, 433, 122–130. [Google Scholar] [CrossRef]
- Carvalho, M.; Matos, M.; Carnide, V. Short communication: Identification of cultivated and wild Vaccinium species grown in Portugal. Spanish J. Agric. Res. 2018, 16, e07SC01. [Google Scholar] [CrossRef]
- Jansen, J.; Den Nijs, H.C.M.; Paiva, J.A.R. Some Notes on Vaccinium uliginosum L. subsp. gaultherioides (bigelow) s.b. young, a new taxon to the flora of portugal. Port. Acta Biol. 2000, 19, 177–186. [Google Scholar]
- Jiménez-Gómez, A.; Saati-Santamaría, Z.; Kostovcik, M.; Rivas, R.; Velázquez, E.; Mateos, P.F.; Menéndez, E.; García-Fraile, P. Selection of the Root Endophyte Pseudomonas brassicacearum CDVBN10 as Plant Growth Promoter for Brassica napus L. Crops. Agronomy 2020, 10, 1788. [Google Scholar] [CrossRef]
- Pang, Z.; Chen, J.; Wang, T.; Gao, C.; Li, Z.; Guo, L.; Xu, J.; Cheng, Y. Linking Plant Secondary Metabolites and Plant Microbiomes: A Review. Front. Plant Sci. 2021, 12, 300. [Google Scholar] [CrossRef] [PubMed]
- Ku, Y.S.; Rehman, H.M.; Lam, H.M. Possible Roles of Rhizospheric and Endophytic Microbes to Provide a Safe and Affordable Means of Crop Biofortification. Agronomy 2019, 9, 764. [Google Scholar] [CrossRef]
- Flores-Félix, J.D.; Velázquez, E.; Martínez-Molina, E.; González-Andrés, F.; Squartini, A.; Rivas, R. Connecting the lab and the field: Genome analysis of phyllobacterium and rhizobium strains and field performance on two vegetable crops. Agronomy 2021, 11, 1124. [Google Scholar] [CrossRef]
- Morvan, S.; Meglouli, H.; Lounès-Hadj Sahraoui, A.; Hijri, M. Into the wild blueberry (Vaccinium angustifolium) rhizosphere microbiota. Environ. Microbiol. 2020, 22, 3803–3822. [Google Scholar] [CrossRef]
- Timonen, S.; Sinkko, H.; Sun, H.; Sietiö, O.M.; Rinta-Kanto, J.M.; Kiheri, H.; Heinonsalo, J. Ericoid Roots and Mycospheres Govern Plant-Specific Bacterial Communities in Boreal Forest Humus. Microb. Ecol. 2017, 73, 939–953. [Google Scholar] [CrossRef]
- Vohník, M.; Mrnka, L.; Lukešová, T.; Bruzone, M.C.; Kohout, P.; Fehrer, J. The cultivable endophytic community of Norway spruce ectomycorrhizas from microhabitats lacking ericaceous hosts is dominated by ericoid mycorrhizal Meliniomyces variabilis. Fungal Ecol. 2013, 6, 281–292. [Google Scholar] [CrossRef]
- Herlemann, D.P.R.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5, 1571–1579. [Google Scholar] [CrossRef]
- Vierna, J.; Doña, J.; Vizcaíno, A.; Serrano, D.; Jovani, R.; Chain, F. PCR cycles above routine numbers do not compromise high-throughput DNA barcoding results. Genome 2017, 60, 868–873. [Google Scholar] [CrossRef] [PubMed]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; Sankaran, K.; Fukuyama, J.A.; McMurdie, P.J.; Holmes, S.P. Bioconductor Workflow for Microbiome Data Analysis: From raw reads to community analyses. F1000Research 2016, 5, 1492. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Gómez, A.; García-Estévez, I.; García-Fraile, P.; Escribano-Bailón, M.T.; Rivas, R. Increase in phenolic compounds of Coriandrum sativum L. after the application of a Bacillus halotolerans biofertilizer. J. Sci. Food Agric. 2020, 100, 2742–2749. [Google Scholar] [CrossRef]
- Flores-Félix, J.D.; Marcos-García, M.; Silva, L.R.; Menéndez, E.; Martínez-Molina, E.; Mateos, P.F.; Velázquez, E.; García-Fraile, P.; Andrade, P.; Rivas, R. Rhizobium as plant probiotic for strawberry production under microcosm conditions. Symbiosis 2015, 67, 25–32. [Google Scholar] [CrossRef]
- Nunes, A.R.; Sánchez-Juanes, F.; Gonçalves, A.C.; Alves, G.; Silva, L.R.; Flores-Félix, J.D. Evaluation of Raw Cheese as a Novel Source of Biofertilizer with a High Level of Biosecurity for Blueberry. Agronomy 2022, 12, 1150. [Google Scholar] [CrossRef]
- Bal, A.; Anand, R.; Berge, O.; Chanway, C.P. Isolation and identification of diazotrophic bacteria from internal tissues of Pinus contorta and Thuja plicata. Can. J. For. Res. 2012, 42, 807–813. [Google Scholar] [CrossRef]
- Sánchez-juanes, F.; Teixeira-martín, V.; González-buitrago, J.M.; Velázquez, E.; Flores-félix, J.D. Identification of species and subspecies of lactic acid bacteria present in spanish cheeses type “torta” by maldi-tof ms and phes gene analyses. Microorganisms 2020, 8, 301. [Google Scholar] [CrossRef] [PubMed]
- Doan, N.T.L.; Van Hoorde, K.; Cnockaert, M.; De Brandt, E.; Aerts, M.; Le Thanh, B.; Vandamme, P. Validation of MALDI-TOF MS for rapid classification and identification of lactic acid bacteria, with a focus on isolates from traditional fermented foods in Northern Vietnam. Lett. Appl. Microbiol. 2012, 55, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Flores-Félix, J.D.; Silva, L.R.; Rivera, L.P.; Marcos-García, M.; García-Fraile, P.; Martínez-Molina, E.; Mateos, P.F.; Velázquez, E.; Andrade, P.; Rivas, R. Plants Probiotics as a Tool to Produce Highly Functional Fruits: The Case of Phyllobacterium and Vitamin C in Strawberries. PLoS ONE 2015, 10, e0122281. [Google Scholar] [CrossRef]
- Anadon-Rosell, A.; Palacio, S.; Nogués, S.; Ninot, J.M. Vaccinium myrtillus stands show similar structure and functioning under different scenarios of coexistence at the Pyrenean treeline. Plant Ecol. 2016, 217, 1115–1128. [Google Scholar] [CrossRef]
- Mažeikienė, I.; Frercks, B.; Burokienė, D.; Mačionienė, I.; Šalaševičienė, A. Endophytic Community Composition and Genetic-Enzymatic Features of Cultivable Bacteria in Vaccinium myrtillus L. in Forests of the Baltic-Nordic Region. Forests 2021, 12, 1647. [Google Scholar] [CrossRef]
- Ebadzadsahrai, G.; Soby, S. 16S rRNA Amplicon Profiling of Cranberry (Vaccinium macrocarpon Ait.) Flower and Berry Surfaces. Microbiol. Resour. Announc. 2019, 8, e01479-18. [Google Scholar] [CrossRef] [PubMed]
- Perotto, S.; Daghino, S.; Martino, E.; Voyron, S. Metabarcoding Reveals Diverse Endophytic Fungal Communities in Vaccinium Myrtillus Plant Organs and Suggests Systemic Distribution of Some Ericoid Mycorrhizal and DSE Fungi. Sci. Rep. 2022, 12, 11013. [Google Scholar]
- Casarrubia, S.; Daghino, S.; Kohler, A.; Morin, E.; Khouja, H.-R.; Daguerre, Y.; Veneault-Fourrey, C.; Martin, F.M.; Perotto, S.; Martino, E. The Hydrophobin-Like OmSSP1 May Be an Effector in the Ericoid Mycorrhizal Symbiosis. Front. Plant Sci. 2018, 9, 546. [Google Scholar] [CrossRef] [PubMed]
- Merino-Martín, L.; Griffiths, R.I.; Gweon, H.S.; Furget-Bretagnon, C.; Oliver, A.; Mao, Z.; Le Bissonnais, Y.; Stokes, A. Rhizosphere bacteria are more strongly related to plant root traits than fungi in temperate montane forests: Insights from closed and open forest patches along an elevational gradient. Plant Soil 2020, 450, 183–200. [Google Scholar] [CrossRef]
- Toju, H.; Peay, K.G.; Yamamichi, M.; Narisawa, K.; Hiruma, K.; Naito, K.; Fukuda, S.; Ushio, M.; Nakaoka, S.; Onoda, Y.; et al. Core microbiomes for sustainable agroecosystems. Nat. Plants 2018, 4, 247–257. [Google Scholar] [CrossRef]
- Durán, P.; Thiergart, T.; Garrido-Oter, R.; Agler, M.; Kemen, E.; Schulze-Lefert, P.; Hacquard, S. Microbial Interkingdom Interactions in Roots Promote Arabidopsis Survival. Cell 2018, 175, 973–983.e14. [Google Scholar] [CrossRef]
- Coleman-Derr, D.; Desgarennes, D.; Fonseca-Garcia, C.; Gross, S.; Clingenpeel, S.; Woyke, T.; North, G.; Visel, A.; Partida-Martinez, L.P.; Tringe, S.G. Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol. 2016, 209, 798–811. [Google Scholar] [CrossRef]
- Reis, F.; Soares-Castro, P.; Costa, D.; Tavares, R.M.; Baptista, P.; Santos, P.M.; Lino-Neto, T. Climatic impacts on the bacterial community profiles of cork oak soils. Appl. Soil Ecol. 2019, 143, 89–97. [Google Scholar] [CrossRef]
- Ofek-Lalzar, M.; Sela, N.; Goldman-Voronov, M.; Green, S.J.; Hadar, Y.; Minz, D. Niche and host-associated functional signatures of the root surface microbiome. Nat. Commun. 2014, 5, 4950. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Krug, L.; Yang, H.; Li, H.; Yang, M.; Berg, G.; Cernava, T. Nicotiana tabacum seed endophytic communities share a common core structure and genotype-specific signatures in diverging cultivars. Comput. Struct. Biotechnol. J. 2020, 18, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Chaparro, J.M.; Sheflin, A.M.; Manter, D.K.; Vivanco, J.M. Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fertil. Soils 2012, 48, 489–499. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Vivanco, J.M. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014, 8, 790–803. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; Mattupalli, C.; Eversole, K.; Leach, J.E. Enabling sustainable agriculture through understanding and enhancement of microbiomes. New Phytol. 2021, 230, 2129–2147. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.X.; Guo, X.; Qin, Y.; Garrido-Oter, R.; Schulze-Lefert, P.; Bai, Y. High-throughput cultivation and identification of bacteria from the plant root microbiota. Nat. Protoc. 2021, 16, 988–1012. [Google Scholar] [CrossRef]
- Diakite, A.; Dubourg, G.; Dione, N.; Afouda, P.; Bellali, S.; Ngom, I.I.; Valles, C.; Tall, M.L.; Lagier, J.C.; Raoult, D. Optimization and standardization of the culturomics technique for human microbiome exploration. Sci. Rep. 2020, 10, 1–7. [Google Scholar] [CrossRef]
- Xiong, J.; Lu, J.; Li, X.; Qiu, Q.; Chen, J.; Yan, C. Effect of rice (Oryza sativa L.) genotype on yield: Evidence from recruiting spatially consistent rhizosphere microbiome. Soil Biol. Biochem. 2021, 161, 108395. [Google Scholar] [CrossRef]
- Mahapatra, S.; Rayanoothala, P.; Solanki, M.K.; Das, S. Wheat Microbiome: Present Status and Future Perspective. In Phytobiomes: Current Insights and Future Vistas; Springer: Singapore, 2020; pp. 191–223. [Google Scholar] [CrossRef]
- Ashfaq, M.Y.; Da’na, D.A.; Al-Ghouti, M.A. Application of MALDI-TOF MS for identification of environmental bacteria: A review. J. Environ. Manage. 2022, 305, 114359. [Google Scholar] [CrossRef]
- Sánchez-Juanes, F.; García Señán, A.I.; Egido, S.H.; Siller Ruiz, M.; González Buitrago, J.M.; Muñoz Bellido, J.L. Use of MALDI-TOF Techniques in the Diagnosis of Urinary Tract Pathogens. In The Use of Mass Spectrometry Technology (MALDI-TOF) in Clinical Microbiology; Academic Press: London, UK, 2018; pp. 145–158. [Google Scholar] [CrossRef]
- Keswani, C.; Prakash, O.; Bharti, N.; Vílchez, J.I.; Sansinenea, E.; Lally, R.D.; Borriss, R.; Singh, S.P.; Gupta, V.K.; Fraceto, L.F.; et al. Re-addressing the biosafety issues of plant growth promoting rhizobacteria. Sci. Total Environ. 2019, 690, 841–852. [Google Scholar] [CrossRef]
- Glick, B.R. Beneficial Plant-Bacterial Interactions; Bernard, R.G., Ed.; Department of Biology University of Waterloo: Waterloo, ON, Canada, 2015; ISBN 9783319139203. [Google Scholar]
- Nobahar, A.; Sarikhani, M.R.; Chalabianlou, N. Buffering capacity affects phosphorous solubilization assays in rhizobacteria. Rhizosphere 2017, 4, 119–125. [Google Scholar] [CrossRef]
- Blanco-Vargas, A.; Rodríguez-Gacha, L.M.; Sánchez-Castro, N.; Garzón-Jaramillo, R.; Pedroza-Camacho, L.D.; Poutou-Piñales, R.A.; Rivera-Hoyos, C.M.; Díaz-Ariza, L.A.; Pedroza-Rodríguez, A.M. Phosphate-solubilizing Pseudomonas sp., and Serratia sp., co-culture for Allium cepa L. growth promotion. Heliyon 2020, 6, e05218. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Bahena, M.H.; Cuesta, M.J.; Flores-Félix, J.D.; Mulas, R.; Rivas, R.; Castro-Pinto, J.; Brañas, J.; Mulas, D.; González-Andrés, F.; Velázquez, E.; et al. Pseudomonas helmanticensis sp. nov., isolated from a forest soil. Int. J. Syst. Evol. Microbiol. 2014, 64, 2338–2345. [Google Scholar] [CrossRef] [PubMed]
- da Silva, A.V.; de Oliveira, A.J.; Tanabe, I.S.B.; Silva, J.V.; da Silva Barros, T.W.; da Silva, M.K.; França, P.H.B.; Leite, J.; Putzke, J.; Montone, R.; et al. Antarctic lichens as a source of phosphate-solubilizing bacteria. Extremophiles 2021, 25, 181–191. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, S.J.; Nam, I.H. Effect of Treating Acid Sulfate Soils with Phosphate Solubilizing Bacteria on Germination and Growth of Tomato (Lycopersicon esculentum L.). Int. J. Environ. Res. Public Heal. 2021, 18, 8919. [Google Scholar] [CrossRef]
- Kim, S.J.; Jung, G.Y.; Nam, I.H. Draft genome sequence of Caballeronia jiangsuensis EK, a phosphate-solubilizing bacterium isolated from the rhizosphere of reed. Korean J. Microbiol. 2021, 57, 106–108. [Google Scholar] [CrossRef]
- Marcotte, A.R.; Anbar, A.D.; Majestic, B.J.; Herckes, P. Mineral Dust and Iron Solubility: Effects of Composition, Particle Size, and Surface Area. Atmosphere 2020, 11, 533. [Google Scholar] [CrossRef]
- Maymon, M.; Martínez-Hidalgo, P.; Tran, S.S.; Ice, T.; Craemer, K.; Anbarchian, T.; Sung, T.; Hwang, L.H.; Chou, M.; Fujishige, N.A.; et al. Mining the phytomicrobiome to understand how bacterial coinoculations enhance plant growth. Front. Plant Sci. 2015, 6, 1–14. [Google Scholar] [CrossRef]
- Złoch, M.; Thiem, D.; Gadzała-Kopciuch, R.; Hrynkiewicz, K. Synthesis of siderophores by plant-associated metallotolerant bacteria under exposure to Cd2+. Chemosphere 2016, 156, 312–325. [Google Scholar] [CrossRef]
- Pajuelo, E.; Arjona, S.; Rodríguez-Llorente, I.D.; Mateos-Naranjo, E.; Redondo-Gómez, S.; Merchán, F.; Navarro-Torre, S. Coastal Ecosystems as Sources of Biofertilizers in Agriculture: From Genomics to Application in an Urban Orchard. Front. Mar. Sci. 2021, 8, 1162. [Google Scholar] [CrossRef]
- De Hita, D.; Fuentes, M.; Zamarreño, A.M.; Ruiz, Y.; Garcia-Mina, J.M. Culturable Bacterial Endophytes From Sedimentary Humic Acid-Treated Plants. Front. Plant Sci. 2020, 11, 837. [Google Scholar] [CrossRef] [PubMed]
- Arruda, L.; Beneduzi, A.; Martins, A.; Lisboa, B.; Lopes, C.; Bertolo, F.; Passaglia, L.M.P.; Vargas, L.K. Screening of rhizobacteria isolated from maize (Zea mays L.) in Rio Grande do Sul State (South Brazil) and analysis of their potential to improve plant growth. Appl. Soil Ecol. 2013, 63, 15–22. [Google Scholar] [CrossRef]
- Salomon, M.V.; Bottini, R.; de Souza Filho, G.A.; Cohen, A.C.; Moreno, D.; Gil, M.; Piccoli, P. Bacteria isolated from roots and rhizosphere of Vitis vinifera retard water losses, induce abscisic acid accumulation and synthesis of defense-related terpenes in in vitro cultured grapevine. Physiol. Plant. 2014, 151, 359–374. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.L.; Halo, B.A.; Elyassi, A.; Ali, S.; Al-Hosni, K.; Hussain, J.; Al-Harrasi, A.; Lee, I.J. Indole acetic acid and ACC deaminase from endophytic bacteria improves the growth of Solanum lycopersicum. Electron. J. Biotechnol. 2016, 21, 58–64. [Google Scholar] [CrossRef]
- Gumiere, T.; Ribeiro, C.M.; Vasconcellos, R.L.F.; Cardoso, E.J.B.N. Indole-3-acetic acid producing root-associated bacteria on growth of Brazil Pine (Araucaria angustifolia) and Slash Pine (Pinus elliottii). Antonie Van Leeuwenhoek 2014, 105, 663–669. [Google Scholar] [CrossRef]
- Pereira, S.I.A.; Monteiro, C.; Vega, A.L.; Castro, P.M.L. Endophytic culturable bacteria colonizing Lavandula dentata L. plants: Isolation, characterization and evaluation of their plant growth-promoting activities. Ecol. Eng. 2016, 87, 91–97. [Google Scholar] [CrossRef]
- Ahemad, M.; Kibret, M. Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective. J. King Saud Univ.-Sci. 2014, 26, 1–20. [Google Scholar] [CrossRef]
- Bartel, B.; LeClere, S.; Magidin, M.; Zolman, B.K. Inputs to the Active Indole-3-Acetic Acid Pool: De Novo Synthesis, Conjugate Hydrolysis, and Indole-3-Butyric Acid b-Oxidation. J. Plant Growth Regul. 2001, 20, 198–216. [Google Scholar] [CrossRef]
- Guo, J.-M.; Xin, Y.-Y.; Yin, X.-B. Selective differentiation of indoleacetic acid and indolebutyric acid using colorimetric recognition after Ehrlich reaction. J. Agric. Food Chem. 2010, 58, 6556–6561. [Google Scholar] [CrossRef]
- Jiménez-Gómez, A.; Flores-Félix, J.D.; García-Fraile, P.; Mateos, P.F.; Menéndez, E.; Velázquez, E.; Rivas, R. Probiotic activities of Rhizobium laguerreae on growth and quality of spinach. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Ważny, R.; Jędrzejczyk, R.J.; Rozpądek, P.; Domka, A.; Turnau, K. Biotization of highbush blueberry with ericoid mycorrhizal and endophytic fungi improves plant growth and vitality. Appl. Microbiol. Biotechnol. 2022, 106, 4775–4786. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wan, Y.; Shakeel, M.; Wang, D.; Xiao, L. Effect of mycorrhizal fungi inoculation on bacterial diversity, community structure and fruit yield of blueberry. Rhizosphere 2021, 19, 100360. [Google Scholar] [CrossRef]
- Contreras-Pérez, M.; Hernández-Salmerón, J.; Rojas-Solís, D.; Rocha-Granados, C.; Orozco-Mosqueda, M.D.C.; Parra-Cota, F.I.; de los Santos-Villalobos, S.; Santoyo, G. Draft genome analysis of the endophyte, Bacillus toyonensis COPE52, a blueberry (Vaccinium spp. var. Biloxi) growth-promoting bacterium. 3 Biotech 2019, 9, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Schoebitz, M.; López, M.D.; Serrí, H.; Martínez, O.; Zagal, E. Combined application of microbial consortium and humic substances to improve the growth performance of blueberry seedlings. J. soil Sci. plant Nutr. 2016, 16, 1010–1023. [Google Scholar] [CrossRef]
- Sawana, A.; Adeolu, M.; Gupta, R.S. Molecular signatures and phylogenomic analysis of the genus burkholderia: Proposal for division of this genus into the emended genus burkholderia containing pathogenic organisms and a new genus paraburkholderia gen. nov. harboring environmental species. Front. Genet. 2014, 5, 429. [Google Scholar] [CrossRef]
- Dobritsa, A.P.; Samadpour, M. Transfer of eleven species of the genus Burkholderia to the genus Paraburkholderia and proposal of Caballeronia gen. nov. to accommodate twelve species of the genera Burkholderia and Paraburkholderia. Int. J. Syst. Evol. Microbiol. 2016, 66, 2836–2846. [Google Scholar] [CrossRef]
- Miotto-Vilanova, L.; Courteaux, B.; Padilla, R.; Rabenoelina, F.; Jacquard, C.; Clément, C.; Comte, G.; Lavire, C.; Ait Barka, E.; Kerzaon, I.; et al. Impact of Paraburkholderia phytofirmans PsJN on Grapevine Phenolic Metabolism. Int. J. Mol. Sci. 2019, 20, 5775. [Google Scholar] [CrossRef]
- Rahman, M.; Rahman, M.; Sabir, A.A.; Mukta, J.A.; Khan, M.M.A.; Mohi-Ud-Din, M.; Miah, M.G.; Islam, M.T. Plant probiotic bacteria Bacillus and Paraburkholderia improve growth, yield and content of antioxidants in strawberry fruit. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Beukes, C.W.; Venter, S.N.; Steenkamp, E.T. The history and distribution of nodulating Paraburkholderia, a potential inoculum for Fynbos forage species. Grass Forage Sci. 2021, 76, 10–32. [Google Scholar] [CrossRef]
- Daghino, S.; Martino, E.; Voyron, S.; Perotto, S. Metabarcoding of fungal assemblages in Vaccinium myrtillus endosphere suggests colonization of above-ground organs by some ericoid mycorrhizal and DSE fungi. Sci. Rep. 2022, 12, 1–11. [Google Scholar] [CrossRef]
- Compant, S.; Clément, C.; Sessitsch, A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 2010, 42, 669–678. [Google Scholar] [CrossRef]
- Hassen, A.I.; Labuschagne, N. Root colonization and growth enhancement in wheat and tomato by rhizobacteria isolated from the rhizoplane of grasses. World J. Microbiol. Biotechnol. 2010, 26, 1837–1846. [Google Scholar] [CrossRef]
- Weid, I.; Artursson, V.; Seldin, L.; Jansson, J.K. Antifungal and Root Surface Colonization Properties of GFP-Tagged Paenibacillus brasilensis PB177. World J. Microbiol. Biotechnol. 2005, 21, 1591–1597. [Google Scholar] [CrossRef]
- Vio, S.A.; García, S.S.; Casajus, V.; Arango, J.S.; Galar, M.L.; Bernabeu, P.R.; Luna, M.F. Paraburkholderia. Benef. Microbes Agro-Ecology Bact. Fungi 2020, 271–311. [Google Scholar] [CrossRef]
- Hsu, P.C.L.; O’Callaghan, M.; Condron, L.; Hurst, M.R.H. Use of a gnotobiotic plant assay for assessing root colonization and mineral phosphate solubilization by Paraburkholderia bryophila Ha185 in association with perennial ryegrass (Lolium perenne L.). Plant Soil 2018, 425, 43–55. [Google Scholar] [CrossRef]
- Neal, A.L.; Ahmad, S.; Gordon-Weeks, R.; Ton, J. Benzoxazinoids in root exudates of maize attract pseudomonas putida to the rhizosphere. PLoS ONE 2012, 7, e35498. [Google Scholar] [CrossRef]
- Fournier, P.E.; Lagier, J.C.; Dubourg, G.; Raoult, D. From culturomics to taxonomogenomics: A need to change the taxonomy of prokaryotes in clinical microbiology. Anaerobe 2015, 36, 73–78. [Google Scholar] [CrossRef]
- Johnston-Monje, D.; Raizada, M.N. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE 2011, 6, e20396. [Google Scholar] [CrossRef]






| Strain | Bicalcium Phosphate (PSI) | Tricalcium Phosphate (PSI) | Hydroxyapatite (PSI) | Siderophore | IAA (µg/mL) |
|---|---|---|---|---|---|
| VMSES02 | - | 2.14 ± 0.02 | |||
| VMSES14 | 0.90 | 0.75 | - | 1.62 ± 0.01 | |
| VMSES 32 | 0.69 | + | 4.67 ± 0.02 | ||
| VMSES31 | 2.53 | 1.91 | + | 1.30 ± 0.01 | |
| VMSES36 | 0.21 | - | 0.52 ± 0.02 | ||
| VMSES63 | + | 0.76 ± 0.16 | |||
| VMSES74 | + | 8.81 ± 0.02 | |||
| VMFR04B | 0.55 | + | 12.35 ± 0.09 | ||
| VMFR45 | 0.46 | 0.39 | + | 2.80 ± 0.02 | |
| VMFR46 | 0.60 | 0.23 | + | 0.77 ± 0.01 | |
| VMFR53 | + | 1.17 ± 0.02 | |||
| VMMAR66 | + | 1.89 ± 0.02 | |||
| Mirt15 | 1.02 | 1.05 | 0.35 | + | 38.37 ± 0.08 |
| Mirt26 | + | 1.18 ± 0.11 | |||
| Mirt39A | 1.45 | 1.20 | 0.31 | + | 26.57 ± 0.02 |
| Tomato 7 dpi | Tomato 14 dpi | Blueberry 7 dpi | Blueberry 14 dpi | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Treatment | Root Length (cm) | Secondary roots | Aerial Length (cm) | Root Length (cm) | Secondary roots | Aerial Length (cm) | Root Length (cm) | Secondary roots | Aerial Length (cm) | Root Length (cm) | Secondary roots | Aerial Length (cm) |
| Control | 1.80 ± 0.14a | 2.60 ± 0.41a | 2.77 ± 0.32b | 3.50 ± 0.44a | 4.01 ± 0.60b | 3.70 ± 0.48b | 0.91 ± 0.24a | 2.00 ± 0.50a | 0.39 ± 0.10a | 1.32 ± 0.11a | 3.21 ± 0.75a | 0.62 ± 0.32a |
| MIRT15 | 2.47 ± 0.51a | 1.83 ± 0.64a | 1.80 ± 0.46a | 3.73 ± 0.78a | 3.03 ± 0.96 a | 2.68 ± 0.68a | 0.96 ± 0.17a | 2.00 ± 0.50a | 0.41 ± 0.07a | 1.38 ± 0.10a | 2.83 ± 1.00a | 1.02 ± 0.41ab |
| MIRT26 | 2.47 ± 0.46a | 2.20 ± 0.72a | 1.81 ± 0.43a | 3.71 ± 0.68a | 3.10 ± 1.08 a | 2.71 ± 0.64a | 0.87 ± 0.44a | 2.00 ± 0.50a | 0.44 ± 0.07a | 1.42 ± 0.09a | 3.01 ± 1.15a | 0.74 ± 0.22a |
| MIRT39A | 2.10 ± 0.50a | 2.01 ± 0.56a | 3.22 ± 0.45b | 3.10 ± 0.76a | 3.02 ± 0.84 a | 5.20 ± 0.67c | 1.02 ± 0.16a | 2.00 ± 0.50a | 0.43 ± 0.05a | 1.26 ± 0.20a | 2.87 ± 0.95a | 0.77 ± 0.36a |
| VMFR04B | 3.40 ± 0.38b | 6.01 ± 1.46bc | 3.11 ± 0.37b | 5.10 ± 0.57b | 9.09 ± 2.21d | 4.78 ± 0.56c | 1.05 ± 0.41a | 2.00 ± 0.10a | 0.41 ± 0.02a | 2.06 ± 0.13b | 2.71 ± 0.74a | 1.18 ± 0.34ab |
| VMFR45 | 3.92 ± 0.43b | 6.10 ± 1.20bc | 3.77 ± 0.29c | 6.11 ± 0.68c | 9.11 ± 1.79d | 5.71 ± 0.56c | 1.00 ± 0.43a | 2.00 ± 0.30a | 0.43 ± 0.04a | 2.01 ± 0.19b | 2.89 ± 0.72a | 1.29 ± 0.21b |
| VMFR46 | 3.80 ± 0.40b | 6.02 ± 1.41bc | 3.67 ± 0.46c | 6.12 ± 0.60c | 9.01 ± 2.12d | 5.88 ± 0.68c | 0.96 ± 0.22a | 2.00 ± 0.20a | 0.42 ± 0.09a | 2.16 ± 0.18b | 2.95 ± 0.52a | 1.42 ± 0.21b |
| VMFR53 | 3.60 ± 0.43b | 4.60 ± 0.56b | 3.27 ± 0.41b | 5.41 ± 0.64b | 7.04 ± 0.84c | 4.92 ± 0.60c | 1.10 ± 0.16a | 3.00 ± 0.20b | 0.45 ± 0.07a | 1.97 ± 0.17b | 3.21 ± 0.74a | 1.32 ± 0.37b |
| VMMAR66 | 3.27 ± 0.29b | 4.00 ± 1.27ab | 2.62 ± 0.28b | 4.89 ± 0.44b | 6.21 ± 1.90c | 3.91 ± 0.42b | 0.93 ± 0.44a | 2.00 ± 0.50a | 0.44 ± 0.051a | 2.01 ± 0.21b | 2.66 ± 0.12a | 1.11 ± 0.28ab |
| VMSES02 | 3.20 ± 0.49ab | 7.31 ± 1.18c | 2.33 ± 0.44ab | 4.78 ± 0.73b | 11.13 ± 1.92d | 3.58 ± 0.67b | 0.97 ± 0.42a | 2.00 ± 0.20a | 0.42 ± 0.03a | 1.99 ± 0.07b | 2.34 ± 0.88a | 1.09 ± 0.51ab |
| VMSES14 | 3.50 ± 0.35b | 4.30 ± 0.97b | 3.44 ± 0.48bc | 5.22 ± 0.73b | 7.32 ± 1.45c | 5.51 ± 0.67c | 0.88 ± 0.21a | 3.00 ± 0.50b | 0.42 ± 0.07a | 2.03 ± 0.14b | 3.47 ± 0.62a | 1.32 ± 0.42b |
| VMSES31 | 3.80 ± 0.49b | 5.30 ± 0.65b | 2.76 ± 0.45b | 5.71 ± 0.74bc | 8.04 ± 0.98cd | 4.21 ± 0.66b | 1.02 ± 0.41a | 2.00 ± 0.20a | 0.41 ± 0.07a | 1.85 ± 0.14b | 2.73 ± 0.65a | 1.27 ± 0.46ab |
| VMSES32 | 3.00 ± 0.41ab | 2.60 ± 0.74a | 3.22 ± 0.43b | 5.69 ± 0.69bc | 4.17 ± 1.12b | 4.80 ± 0.64c | 0.89 ± 0.2a | 2.00 ± 0.20a | 0.52 ± 0.06a | 1.89 ± 0.21b | 2.37 ± 0.62a | 1.12 ± 0.61ab |
| VMSES63 | 3.40 ± 0.34b | 5.10 ± 1.01b | 3.13 ± 0.44b | 5.09 ± 0.76b | 9.27 ± 1.63c | 4.71 ± 0.66c | 1.14 ± 0.16a | 2.00 ± 0.20a | 0.43 ± 0.06a | 1.95 ± 0.08b | 2.62 ± 0.32a | 1.28 ± 0.14b |
| VMSES74 | 3.62 ± 0.45b | 4.61 ± 0.56b | 3.07 ± 0.43b | 5.60 ± 0.68bc | 7.31 ± 0.84c | 4.59 ± 0.64bc | 0.87 ± 0.42a | 3.00 ± 0.30b | 0.42 ± 0.062a | 1.99 ± 0.16b | 3.20 ± 0.84a | 0.98 ± 0.32a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonçalves, A.C.; Sánchez-Juanes, F.; Meirinho, S.; Silva, L.R.; Alves, G.; Flores-Félix, J.D. Insight into the Taxonomic and Functional Diversity of Bacterial Communities Inhabiting Blueberries in Portugal. Microorganisms 2022, 10, 2193. https://doi.org/10.3390/microorganisms10112193
Gonçalves AC, Sánchez-Juanes F, Meirinho S, Silva LR, Alves G, Flores-Félix JD. Insight into the Taxonomic and Functional Diversity of Bacterial Communities Inhabiting Blueberries in Portugal. Microorganisms. 2022; 10(11):2193. https://doi.org/10.3390/microorganisms10112193
Chicago/Turabian StyleGonçalves, Ana C., Fernando Sánchez-Juanes, Sara Meirinho, Luís R. Silva, Gilberto Alves, and José David Flores-Félix. 2022. "Insight into the Taxonomic and Functional Diversity of Bacterial Communities Inhabiting Blueberries in Portugal" Microorganisms 10, no. 11: 2193. https://doi.org/10.3390/microorganisms10112193
APA StyleGonçalves, A. C., Sánchez-Juanes, F., Meirinho, S., Silva, L. R., Alves, G., & Flores-Félix, J. D. (2022). Insight into the Taxonomic and Functional Diversity of Bacterial Communities Inhabiting Blueberries in Portugal. Microorganisms, 10(11), 2193. https://doi.org/10.3390/microorganisms10112193











