Current Data on Oral Peri-Implant and Periodontal Microbiota and Its Pathological Changes: A Systematic Review
Abstract
:1. Introduction
2. Materials and Methods
- Population: dentate patients with at least 1 implant with a healthy site or with peri-implantitis or mucositis;
- Exposition: absence or presence of peri-implant disease (peri-implantitis and mucositis);
- Comparison: pathological and nonpathological implants or pathological implants and pathological or nonpathological teeth;
- Outcomes: The objectives were to compare the periodontal microbiota and the peri-implant microbiota in healthy and pathological conditions and to explore the microbiological characteristics of peri-implantitis. This was in order to determine the most frequently present and discriminating species of microorganisms by type of site and the key species by type of site and to see if there is a difference in micro-biological diversity;
- Study design: Clinical case–control studies;
- Question: Is the peri-implant microbiota different from the microbiota of periodontitis, and what are the characteristics of the peri-implant microbiota?
2.1. Information Sources and Search Strategy
2.2. Article Selection
2.3. Inclusion and Exclusion Criteria
2.3.1. Inclusion Criteria
2.3.2. Exclusion Criteria
2.4. Data Collection
2.5. Risk of Bias of Each Study and Data Synthesis
3. Results
3.1. Study Characteristics and Summary of Results
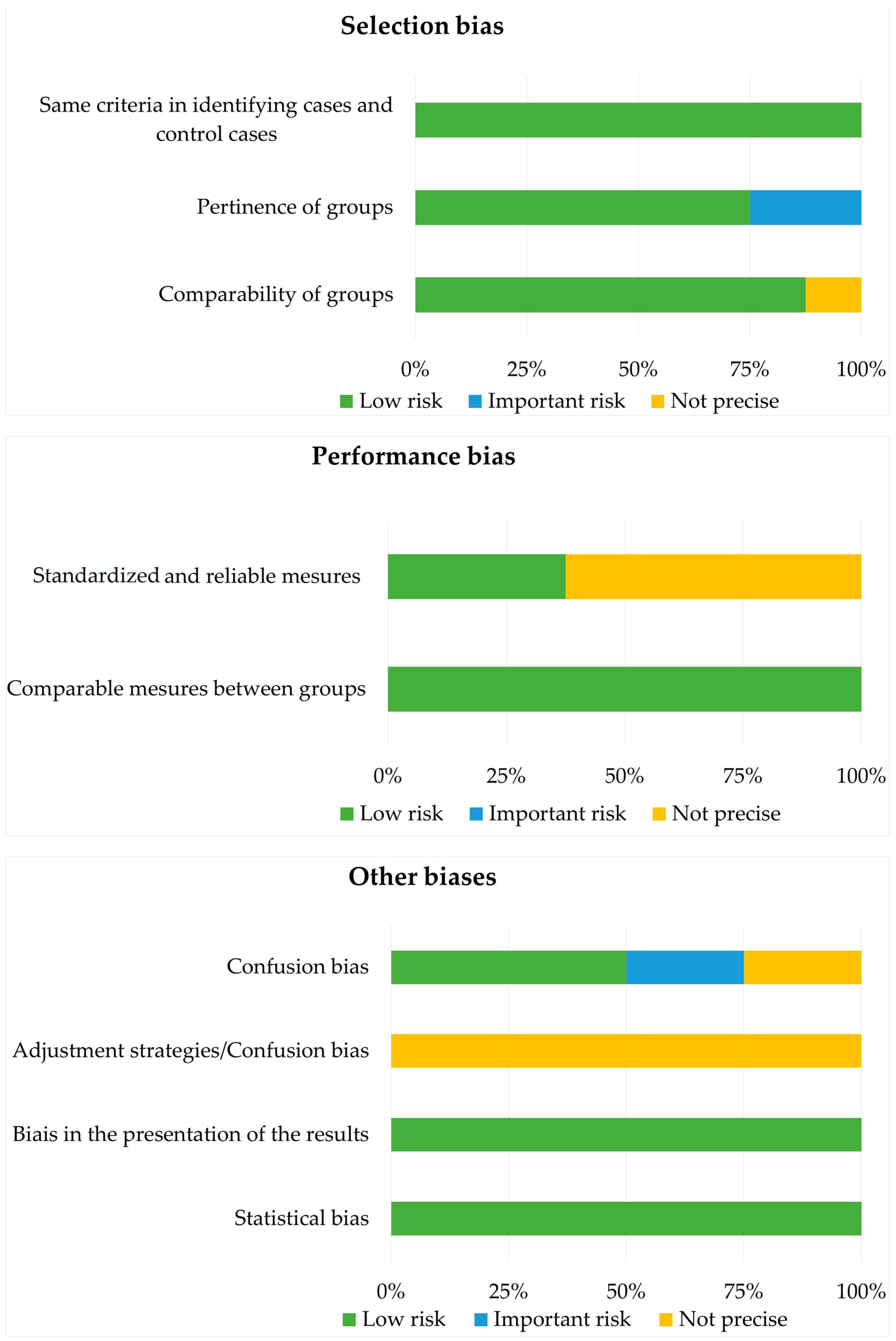
3.2. Risk of Bias Analysis with the Joanna Briggs Institute Case–Control Study Checklist (2017) [24]
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wade, W.G. The oral microbiome in health and disease. Pharmacol. Res. 2013, 69, 137–143. [Google Scholar] [CrossRef]
- Sahrmann, P.; Gilli, F.; Wiedemeier, D.B.; Attin, T.; Schmidlin, P.R.; Karygianni, L. The Microbiome of peri-implantitis: A systematic review and meta-Analysis. Microorganisms 2020, 8, 661. [Google Scholar] [CrossRef]
- Han, A.; Tsoi, J.K.H.; Rodrigues, F.P.; Leprince, J.G.; Palin, W.M. Bacterial adhesion mechanisms on dental implant surfaces and the influencing factors. Int. J. Adhes. Adhes. 2016, 69, 58–71. [Google Scholar] [CrossRef] [Green Version]
- Pokrowiecki, R.; Mielczarek, A.; Zaręba, T.; Tyski, S. Oral microbiome and peri-implant diseases: Where are we now? Ther. Clin. Risk Manag. 2017, 3, 1529–1542. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Tong, Z.; Zhang, Y.; Si, M.; He, F. Microbial profiles of peri-implant mucositis and peri-implantitis: Submucosal microbial dysbiosis correlates with disease severity. Clin. Oral Implant. Res. 2022, 33, 172–183. [Google Scholar] [CrossRef]
- Berglundh, T.; Armitage, G.; Araujo, M.G.; Avila-Ortiz, G.; Blanco, J.; Camargo, P.M.; Chen, S.; Cochran, D.; Derks, J.; Figuero, E.; et al. Peri-implant diseases and conditions: Consensus report of workgroup 4 of the 2017 world workshop on the classification of periodontal and peri-implant diseases and conditions. J. Periodontol. 2018, 89, S313–S318. [Google Scholar] [CrossRef]
- Charalampakis, G.; Belibasakis, G.N. Microbiome of peri-implant infections: Lessons from conventional, molecular and metagenomic analyses. Virulence 2015, 6, 183–187. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.S.; Mason, M.R.; Brooker, M.R.; O’Brien, K. Pyrosequencing reveals unique microbial signatures associated with healthy and failing dental implants. J. Clin. Periodontol. 2012, 39, 425–433. [Google Scholar] [CrossRef] [Green Version]
- Zheng, H.; Xu, L.; Wang, Z.; Li, L.; Zhang, J.; Zhang, Q.; Chen, T.; Lin, J.; Chen, F. Subgingival microbiome in patients with healthy and ailing dental implants. Sci. Rep. 2015, 5, 10948. [Google Scholar] [CrossRef]
- Jakobi, M.; Stumpp, S.; Stiesch, M.; Eberhard, J.; Heuer, W. The peri-implant and periodontal microbiota in patients with and without clinical signs of inflammation. Dent. J. 2015, 3, 24–42. [Google Scholar] [CrossRef]
- Sousa, V.; Nibali, L.; Spratt, D.; Dopico, J.; Mardas, N.; Petrie, A.; Donos, N. Peri-implant and periodontal microbiome diversity in aggressive periodontitis patients: A pilot study. Clin. Oral Implant. Res. 2017, 28, 558–570. [Google Scholar] [CrossRef]
- de Melo, F.; Milanesi, F.C.; Angst, P.D.M.; Oppermann, R.V. A systematic review of the microbiota composition in various peri-implant conditions: Data from 16s rrna gene sequencing. Arch. Oral Biol. 2020, 117, 104776. [Google Scholar] [CrossRef]
- Leonhardt, Å.; Gröndahl, K.; Bergström, C.; Lekholm, U. Long-term follow-up of osseointegrated titanium implants using clinical, radiographic and microbiological parameters: Microbiota and implants. Clin. Oral Implant. Res. 2002, 13, 127–132. [Google Scholar] [CrossRef]
- Botero, J.E.; González, A.M.; Mercado, R.A.; Olave, G.; Contreras, A. Subgingival microbiota in peri-implant mucosa lesions and adjacent teeth in partially edentulous patients. J. Periodontol. 2005, 76, 1490–1495. [Google Scholar] [CrossRef]
- Cortelli, S.C.; Cortelli, J.R.; Romeiro, R.L.; Costa, F.O.; Aquino, D.R.; Orzechowski, P.R.; Araújo, V.C.; Duarte, P.M. Frequency of periodontal pathogens in equivalent peri-implant and periodontal clinical statuses. Arch. Oral Biol. 2013, 58, 67–74. [Google Scholar] [CrossRef] [Green Version]
- Canullo, L.; Peñarrocha-Oltra, D.; Covani, U.; Rossetti, P. Microbiologic and clinical findings of implants in healthy condition and with peri-implantitis. Int. J. Oral Maxillofac. Implant. 2015, 30, 834–842. [Google Scholar] [CrossRef]
- Canullo, L.; Peñarrocha-Oltra, D.; Covani, U.; Botticelli, D.; Serino, G.; Penarrocha, M. Clinical and Microbiological findings in patients with peri-implantitis: A cross-sectional study. Clin. Oral Implant. Res. 2016, 27, 376–382. [Google Scholar] [CrossRef]
- Padial-Molina, M.; López-Martínez, J.; O’Valle, F.; Galindo-Moreno, P. Microbial profiles and detection techniques in peri-implant diseases: A systematic review. J. Oral Maxillofac. Res. 2016, 7, e10. [Google Scholar] [CrossRef] [Green Version]
- Teles, F.R.F. The microbiome of peri-implantitis: Is it unique? Compend. Contin. Educ. Dent. 2017, 38 (Suppl. 8), 22–25. [Google Scholar]
- Pérez-Chaparro, P.J.; Duarte, P.M.; Shibli, J.A.; Montenegro, S.; Lacerda Heluy, S.; Figueiredo, L.C.; Faveri, M.; Feres, M. The current weight of evidence of the microbiologic profile associated with peri-implantitis: A systematic review. J. Periodontol. 2016, 87, 1295–1304. [Google Scholar] [CrossRef]
- Wilson, D.J.; Middleton, J.R.; Adkins, P.R.F.; Goodell, G.M. Test agreement among biochemical methods, matrix-assisted laser desorption ionization–time of flight mass spectrometry, and 16s rrna sequencing for identification of microorganisms isolated from bovine milk. J. Clin. Microbiol. 2019, 57, e01381-18. [Google Scholar] [CrossRef]
- Rakic, M.; Grusovin, M.; Canullo, L. The microbiologic profile associated with peri-implantitis in humans: A systematic review. Int. J. Oral Maxillofac. Implant. 2016, 31, 359–368. [Google Scholar] [CrossRef]
- Lafaurie, G.I.; Sabogal, M.A.; Castillo, D.M.; Rincón, M.V.; Gómez, L.A.; Lesmes, Y.A.; Chambrone, L. Microbiome and microbial biofilm profiles of peri-implantitis: A systematic review. J. Periodontol. 2017, 88, 1066–1089. [Google Scholar] [CrossRef]
- Munn, Z.; Moola, S.; Riitano, D.; Lisy, K. The development of a critical appraisal tool for use in systematic reviews addressing questions of prevalence. Int. J. Health Policy Manag. 2014, 3, 23–28. [Google Scholar] [CrossRef] [Green Version]
- Sanz-Martin, I.; Doolittle-Hall, J.; Teles, R.P.; Patel, M.; Belibasakis, G.N.; Hämmerle, C.H.F.; Jung, R.E.; Teles, F.R.F. Exploring the microbiome of healthy and diseased peri-implant sites using illumina sequencing. J. Clin. Periodontol. 2017, 44, 1274–1284. [Google Scholar] [CrossRef] [Green Version]
- Apatzidou, D.; Lappin, D.F.; Hamilton, G.; Papadopoulos, C.A.; Konstantinidis, A.; Riggio, M.P. Microbiome of peri -implantitis affected and healthy dental sites in patients with a history of chronic periodontitis. Arch. Oral Biol. 2017, 83, 145–152. [Google Scholar] [CrossRef]
- Belkacemi, S.; Mazel, A.; Tardivo, D.; Tavitian, P.; Stephan, G.; Bianca, G.; Terrer, E.; Drancourt, M.; Aboudharam, G. Peri-implantitis-associated methanogens: A preliminary report. Sci. Rep. 2018, 8, 9447. [Google Scholar] [CrossRef] [Green Version]
- Al-Ahmad, A.; Muzafferiy, F.; Anderson, A.C.; Wölber, J.P.; Ratka-Krüger, P.; Fretwurst, T.; Nelson, K.; Vach, K.; Hellwig, E. Shift of microbial composition of peri-implantitis-associated oral biofilm as revealed by 16S RRNA gene cloning. J. Med. Microbiol. 2018, 67, 332–340. [Google Scholar] [CrossRef]
- Yu, X.; Chan, Y.; Zhuang, L.; Lai, H.; Lang, N.P.; Keung Leung, W.; Watt, R.M. Intra-oral single-site comparisons of periodontal and peri-implant microbiota in health and disease. Clin. Oral Implant. Res. 2019, 30, 760–776. [Google Scholar] [CrossRef]
- Rajasekar, A.; Varghese, S.S. Microbiological profile in periodontitis and peri-implantitis: A systematic review. J. Long Term Eff. Med. Implant. 2022, 32, 83–94. [Google Scholar] [CrossRef]
- Zhang, X.-M.; Li, Y.; Gu, Y.-X.; Zhang, C.-N.; Lai, H.-C.; Shi, J.-Y. Ta-coated titanium surface with superior bacteriostasis and osseointegration. Int. J. Nanamed. 2019, 14, 8693–8706. [Google Scholar] [CrossRef]

| Author, Year | Study Design | Participants | Study Group | Exploration Method | Results |
|---|---|---|---|---|---|
| Zheng, H. et al., 2015 [9] | case–control | 24 |
| 16S rRNA sequencing | Peri-implantitis is associated with more diversity of the microbiota. Healthy and pathological sites have distinct bacterial communities. Peri-implantitis is associated with Firmicutes, Fusobacteria, Proteobacteria, and Actinobacteria. |
| Jakobi, M. et al., 2015 [10] | case–control | 18 |
| 16S rRNA sequencing | Tendency to more diversity in affected sites without significant difference; sites with peri-implantitis associated with Neisseria and Kingella; sites with periodontitis associated with Rothia, Tannerella, and Parabacteroides; pathological sites (tooth and implant) associated with Enterococcus, Streptococci, Porphyromonas gingivalis, F. nucleatum, Fretibacterium, P. intermedia, and Bacillus |
| Sousa, V. et al., 2017 [11] | case–control | 24 |
| 16S rRNA sequencing | More microbial diversity at dental sites than at implant sites; some bacteria are implant-specific: Propionibacteria, Paludibacter, Staphylococci, Filifactor, and Mogibacterium; dominant genus of peri-implantitis: Firmicutes, dominant genus of nonpathological implant: Streptococci |
| Sanz-Martin, I. et al., 2017 [25] | case–control | 67 |
| 16S rRNA sequencing | Different pathological and nonpathological peri-implant microbiota; peri-implantitis is associated with more microbiota diversity; peri-implantitis is associated with the red complex and new pathogens: Filifactor alocis, Fretibacterium fastidiosum, and Treponema maltophilum; healthy sites are associated with Streptococci, Rothia, and Haemophilus |
| Apatzidou, D. et al., 2017 [26] | case–control | 10 |
| 16S rRNA sequencing | Methanobrevibacter oralis is present in more than 50% of the samples; no significant difference between the 2 groups; no association between Methanobrevibacter and peri-implantitis. |
| Belkacemi, S. et al., 2018 [27] | case–control | 28 |
| 16S rRNA sequencing | Different pathological and nonpathological peri-implant microbiota; bacterial drift associated with peri-implantitis with increased presence of the red complex; peri-implantitis is associated with Bacteroides (F. nucleatum), Porphyromonas gingivalis, and T. forsythia |
| Al-Ahmad, A. et al., 2018 [28] | case–control Intraoral comparison | 10 |
| 16S rRNA sequencing | Different pathological and nonpathological peri-implant microbiota; bacterial drift associated with peri-implantitis with increased presence of red complex; peri-implantitis is associated with Bacteroides (F. nucleatum), P. gingivalis and T. forsythia. |
| Yu, X. et al., 2019 [29] | case–control Intraoral comparison | 18 |
| 16S rRNA sequencing | Interindividual variability is more important than the variability related to pathological presence; peri-implantitis is characterized by P. gingivalis, T. forsythia, A. actinomycetemcomitans, Treponema spp.; and with rare species present in the mouth: Staphylocoques, Peptostreptococci, Enterobacteries, Helicobacter spp. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gazil, V.; Bandiaky, O.N.; Renard, E.; Idiri, K.; Struillou, X.; Soueidan, A. Current Data on Oral Peri-Implant and Periodontal Microbiota and Its Pathological Changes: A Systematic Review. Microorganisms 2022, 10, 2466. https://doi.org/10.3390/microorganisms10122466
Gazil V, Bandiaky ON, Renard E, Idiri K, Struillou X, Soueidan A. Current Data on Oral Peri-Implant and Periodontal Microbiota and Its Pathological Changes: A Systematic Review. Microorganisms. 2022; 10(12):2466. https://doi.org/10.3390/microorganisms10122466
Chicago/Turabian StyleGazil, Virginie, Octave Nadile Bandiaky, Emmanuelle Renard, Katia Idiri, Xavier Struillou, and Assem Soueidan. 2022. "Current Data on Oral Peri-Implant and Periodontal Microbiota and Its Pathological Changes: A Systematic Review" Microorganisms 10, no. 12: 2466. https://doi.org/10.3390/microorganisms10122466






