Bioprospecting for Novel Bacterial Sources of Hydrolytic Enzymes and Antimicrobials in the Romanian Littoral Zone of the Black Sea
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description, Sample Collection and Processing
2.2. Physicochemical Parameters Measurement
2.3. Isolation and Identification of Marine Bacterial Strains
2.4. Bacterial Growth Characterization
2.4.1. Halophily and Halotolerance Assessment
2.4.2. Growth Temperature Assessment
2.4.3. Growth Rate Determination
2.5. Functional Characterization of Bacterial Strains
2.5.1. Extracellular Hydrolytic Activities
2.5.2. Antimicrobial Activities
2.6. Bacterial Community Composition Assessment
2.6.1. Total DNA Extraction and Illumina Sequencing of 16S rRNA Amplicons
2.6.2. Sequence Analyses
2.7. Nucleotide Sequence Accession Numbers
3. Results
3.1. Physicochemical Characteristics of Seawater
3.2. Abundance of Cultured Marine Bacteria
3.3. Diversity and Taxonomic Profile of the Total Bacterial Community from Black Sea Water
3.4. Taxonomic Diversity and Growth Characteristics of Cultured Bacterial Strains
3.5. Functional Characteristics of Cultured Bacterial Strains
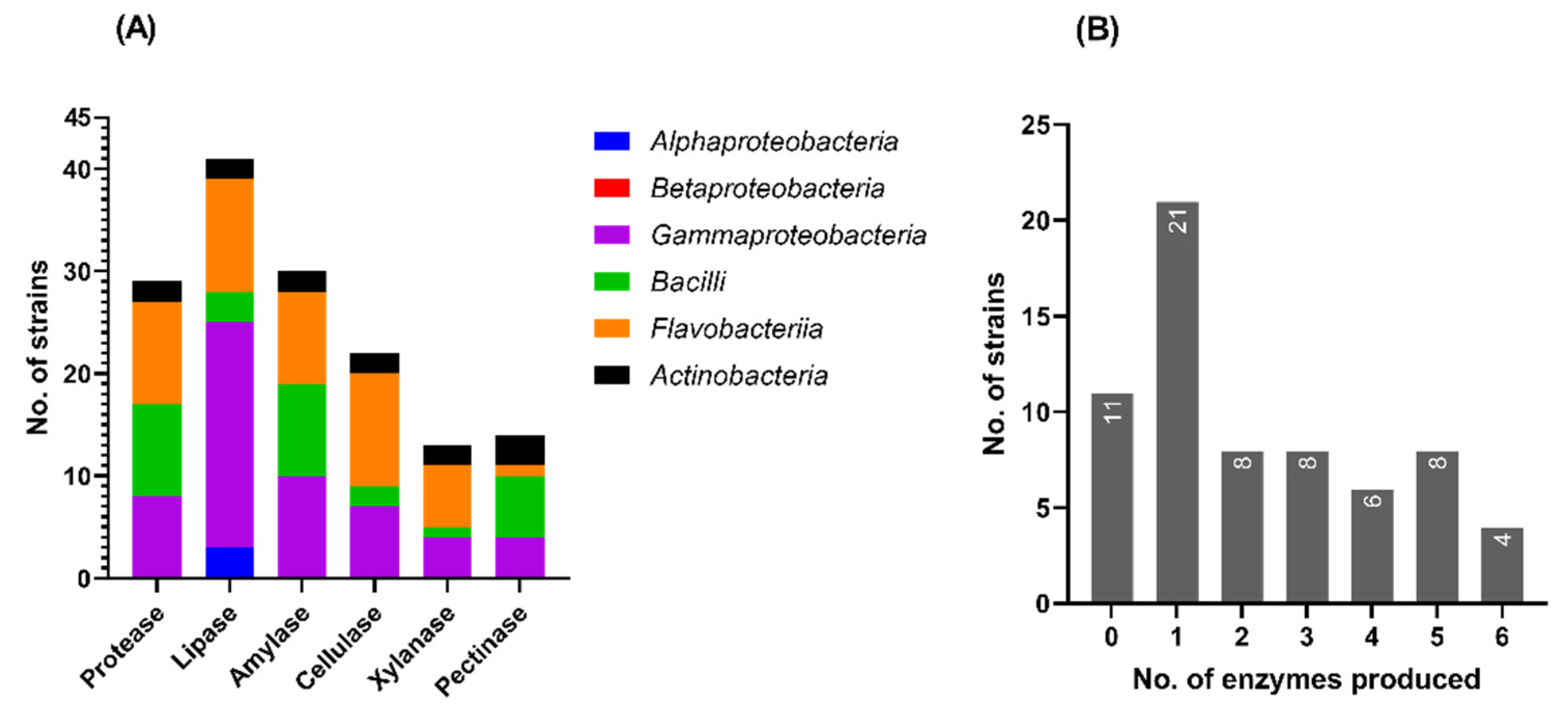
3.5.1. Production of Extracellular Hydrolases
3.5.2. Production of Antimicrobial Compounds
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Munn, C.B. Marine Microbiology: Ecology and Applications, 3rd ed.; CRC Press: Boca Raton, FL, USA, 2020; pp. 1–26, 65–109, 387–406. [Google Scholar]
- Arrigo, K.R. Marine microorganisms and global nutrient cycles. Nature 2005, 437, 349–355. [Google Scholar] [CrossRef]
- Zhang, J.; Jiang, L.; Chen, X.; Lv, K.; Basiony, M.; Zhu, G.; Karthik, L.; Ouyang, L.; Zhang, L.; Liu, X. Recent advances in biotechnology for marine enzymes and molecules. Curr. Opin. Biotechnol. 2021, 69, 308–315. [Google Scholar] [CrossRef]
- Birolli, W.G.; Lima, R.N.; Porto, A.L.M. Applications of Marine-Derived Microorganisms and Their Enzymes in Biocatalysis and Biotransformation, the Underexplored Potentials. Front. Microbiol. 2019, 10, 1453. [Google Scholar] [CrossRef]
- García-Davis, S.; Reyes, C.P.; Lagunes, I.; Padrón, J.M.; Fraile-Nuez, E.; Fernández, J.J.; Díaz-Marrero, A.R. Bioprospecting Antiproliferative Marine Microbiota From Submarine Volcano Tagoro. Front. Mar. Sci. 2021, 8, 687701. [Google Scholar] [CrossRef]
- BBC Research. Available online: https://www.bccresearch.com/market-research/biotechnology/global-markets-for-enzymes-in-industrial-applications.html (accessed on 12 September 2022).
- Trincone, A. Marine Biocatalysts: Enzymatic Features and Applications. Mar. Drugs 2011, 9, 478–499. [Google Scholar] [CrossRef] [Green Version]
- Di Donato, P.; Buono, A.; Poli, A.; Finore, I.; Abbamondi, G.R.; Nicolaus, B.; Lama, L. Exploring Marine Environments for the Identification of Extremophiles and Their Enzymes for Sustainable and Green Bioprocesses. Sustainability 2019, 11, 149. [Google Scholar] [CrossRef] [Green Version]
- De Santi, C.; Altermark, B.; de Pascale, D.; Willassen, N.-P. Bioprospecting around Arctic islands: Marine bacteria as rich source of biocatalysts. J. Basic Microbiol. 2016, 56, 238–253. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, J.; Marchesi, J.R.; Dobson, A.D. Marine metagenomics: Strategies for the discovery of novel enzymes with biotechnological applications from marine environments. Microb. Cell Factories 2008, 7, 27. [Google Scholar] [CrossRef] [Green Version]
- Trincone, A. Some Enzymes in Marine Environment: Prospective Applications Found in Patent Literature. Recent Pat. Biotechnol. 2012, 6, 134–148. [Google Scholar] [CrossRef]
- Dalmaso, G.Z.L.; Ferreira, D.; Vermelho, A.B. Marine Extremophiles: A Source of Hydrolases for Biotechnological Applications. Mar. Drugs 2015, 13, 1925–1965. [Google Scholar] [CrossRef]
- Srinivasan, R.; Kannappan, A.; Shi, C.; Lin, X. Marine Bacterial Secondary Metabolites: A Treasure House for Structurally Unique and Effective Antimicrobial Compounds. Mar. Drugs 2021, 19, 530. [Google Scholar] [CrossRef]
- Nweze, J.A.; Mbaoji, F.N.; Huang, G.; Li, Y.; Yang, L.; Zhang, Y.; Huang, S.; Pan, L.; Yang, D. Antibiotics Development and the Potentials of Marine-Derived Compounds to Stem the Tide of Multidrug-Resistant Pathogenic Bacteria, Fungi, and Protozoa. Mar. Drugs 2020, 18, 145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agrawal, S.; Acharya, D.; Adholeya, A.; Barrow, C.; Deshmukh, S.K. Nonribosomal Peptides from Marine Microbes and Their Antimicrobial and Anticancer Potential. Front. Pharmacol. 2017, 8, 828. [Google Scholar] [CrossRef]
- Schinke, C.; Martins, T.; Queiroz, S.C.N.; Melo, I.S.; Reyes, F.G.R. Antibacterial Compounds from Marine Bacteria, 2010–2015. J. Nat. Prod. 2017, 80, 1215–1228. [Google Scholar] [CrossRef]
- Murray, J.W.; Jannasch, H.W.; Honjo, S.; Anderson, R.F.; Reeburgh, W.S.; Top, Z.; Friederich, G.E.; Codispoti, L.A.; Izdar, E. Unexpected changes in the oxic/anoxic interface in the Black Sea. Nature 1989, 338, 411–413. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Pavlovska, M.; Stoica, E.; Prekrasna, I.; Yang, J.; Slobodnik, J.; Zhang, X.; Dykyi, E. Holistic pelagic biodiversity monitoring of the Black Sea via eDNA metabarcoding approach: From bacteria to marine mammals. Environ. Int. 2020, 135, 105307. [Google Scholar] [CrossRef]
- Jaiani, E.; Kusradze, I.; Kokashvili, T.; Geliashvili, N.; Janelidze, N.; Kotorashvili, A.; Kotaria, N.; Guchmanidze, A.; Tediashvili, M.; Prangishvili, D. Microbial Diversity and Phage–Host Interactions in the Georgian Coastal Area of the Black Sea Revealed by Whole Genome Metagenomic Sequencing. Mar. Drugs 2020, 18, 558. [Google Scholar] [CrossRef]
- Cabello-Yeves, P.J.; Callieri, C.; Picazo, A.; Mehrshad, M.; Haro-Moreno, J.M.; Roda-Garcia, J.J.; Dzhembekova, N.; Slabakova, V.; Slabakova, N.; Moncheva, S.; et al. The microbiome of the Black Sea water column analyzed by shotgun and genome centric metagenomics. Environ. Microbiome 2021, 16, 5. [Google Scholar] [CrossRef]
- Suominen, S.; Dombrowski, N.; Damsté, J.S.S.; Villanueva, L. A diverse uncultivated microbial community is responsible for organic matter degradation in the Black Sea sulphidic zone. Environ. Microbiol. 2021, 23, 2709–2728. [Google Scholar] [CrossRef] [Green Version]
- Vetriani, C.; Tran, H.V.; Kerkhof, L.J. Fingerprinting Microbial Assemblages from the Oxic/Anoxic Chemocline of the Black Sea. Appl. Environ. Microbiol. 2003, 69, 6481–6488. [Google Scholar] [CrossRef]
- Fuchsman, C.A.; Kirkpatrick, J.B.; Brazelton, W.J.; Murray, J.W.; Staley, J.T. Metabolic strategies of free-living and aggregate-associated bacterial communities inferred from biologic and chemical profiles in the Black Sea suboxic zone. FEMS Microbiol. Ecol. 2011, 78, 586–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Vliet, D.M.; von Meijenfeldt, F.B.; Dutilh, B.E.; Villanueva, L.; Damsté, J.S.S.; Stams, A.J.; Sánchez-Andrea, I. The bacterial sulfur cycle in expanding dysoxic and euxinic marine waters. Environ. Microbiol. 2021, 23, 2834–2857. [Google Scholar] [CrossRef] [PubMed]
- Çorbacı, C.; Özcan, K. Production of Alkaline Enzymes by Marine Actinobacteria Isolated from Black Sea Sediments. J. Inst. Sci. Technol. 2019, 9, 647–654. [Google Scholar] [CrossRef]
- Van Vliet, D.M.; Na Ayudthaya, S.P.; Diop, S.; Villanueva, L.; Stams, A.J.M.; Sánchez-Andrea, I. Anaerobic Degradation of Sulfated Polysaccharides by Two Novel Kiritimatiellales Strains Isolated From Black Sea Sediment. Front. Microbiol. 2019, 10, 253. [Google Scholar] [CrossRef]
- Ibryamova, S.; Arhangelova, N.; Koynova, T.; Dimitrov, D.; Dimitrova, Z.; Ivanov, R.; Kalchev, K.; Chipev, N.; Natchev, N.; Ignatova-Ivanova, T. Antifungal Activity of Lactic Acid Bacteria, Isolated from (Mytilus Galloprovincialis lam.) in The Bulgarian Black Sea Aquatory. J. IMAB 2020, 26, 2875–2882. [Google Scholar] [CrossRef]
- Ignatova-Ivanova, T.; Ibryamova, S.; Bachvarova, D.; Salim, S.; Valkova, S.; Simeonova, Y.; Dimitrov, D.; Ivanov, R.; Chipev, N.; Natchev, N. Determination of the antimicrobial activity of lactic acid bacteria isolated from the Black sea mussel Mytilus galloprovincialis Lamarck, 1819. Pharmacia 2022, 69, 637–644. [Google Scholar] [CrossRef]
- Bănaru, D.; Harmelin-Vivien, M.; Gomoiu, M.-T.; Onciu, T.-M. Influence of the Danube River inputs on C and N stable isotope ratios of the Romanian coastal waters and sediment (Black Sea). Mar. Pollut. Bull. 2007, 54, 1385–1394. [Google Scholar] [CrossRef]
- Ruginescu, R.; Gomoiu, I.; Popescu, O.; Cojoc, R.; Neagu, S.; Lucaci, I.; Batrinescu-Moteau, C.; Enache, M. Bioprospecting for Novel Halophilic and Halotolerant Sources of Hydrolytic Enzymes in Brackish, Saline and Hypersaline Lakes of Romania. Microorganisms 2020, 8, 1903. [Google Scholar] [CrossRef]
- Miller, C.S.; Handley, K.M.; Wrighton, K.; Frischkorn, K.R.; Thomas, B.C.; Banfield, J. Short-Read Assembly of Full-Length 16S Amplicons Reveals Bacterial Diversity in Subsurface Sediments. PLoS ONE 2013, 8, e56018. [Google Scholar] [CrossRef] [Green Version]
- Basic Local Alignment Search Tool. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 25 April 2020).
- Kushner, D.J. Growth and Nutrition of Halophilic Bacteria. In The Biology of Halophilic Bacteria; Vreeland, R.H., Hochstein, L.I., Eds.; CRC Press: Boca Raton, FL, USA, 1992; pp. 87–103. [Google Scholar]
- Cell Calculator++. Available online: https://www.doubling-time.com/compute_more.php (accessed on 18 July 2022).
- Hockett, K.L.; Baltrus, D.A. Use of the Soft-agar Overlay Technique to Screen for Bacterially Produced Inhibitory Compounds. J. Vis. Exp. 2017, 119, e55064. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Djurhuus, A.; Port, J.; Closek, C.; Yamahara, K.M.; Romero-Maraccini, O.; Walz, K.R.; Goldsmith, D.B.; Michisaki, R.; Breitbart, M.; Boehm, A.B.; et al. Evaluation of Filtration and DNA Extraction Methods for Environmental DNA Biodiversity Assessments across Multiple Trophic Levels. Front. Mar. Sci. 2017, 4, 314. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a Prokaryotic Universal Primer for Simultaneous Analysis of Bacteria and Archaea Using Next-Generation Sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Kondratev, S.I.; Medvedev, E.V.; Konovalov, S.K. Total Alkalinity and pH in the Black Sea Waters in 2010–2011. Phys. Oceanogr. 2017, 4, 35–45. [Google Scholar] [CrossRef] [Green Version]
- Shaltout, M.; Omstedt, A. Modelling the water and heat balances of the Mediterranean Sea using a two-basin model and available meteorological, hydrological, and ocean data. Oceanologia 2015, 57, 116–131. [Google Scholar] [CrossRef] [Green Version]
- Kalkan, S.; Altuğ, G. The composition of cultivable bacteria, bacterial pollution, and environmental variables of the coastal areas: An example from the Southeastern Black Sea, Turkey. Environ. Monit. Assess. 2020, 192, 356. [Google Scholar] [CrossRef]
- Arias, C.R.; Macian, M.C.; Aznar, R.; Garay, E.; Pujalte, M.J. Low incidence of Vibrio vulnificus among Vibrio isolates from sea water and shellfish of the western Mediterranean coast. J. Appl. Microbiol. 1999, 86, 125–134. [Google Scholar] [CrossRef]
- Zhao, Z.; Baltar, F.; Herndl, G.J. Linking extracellular enzymes to phylogeny indicates a predominantly particle-associated lifestyle of deep-sea prokaryotes. Sci. Adv. 2020, 6, eaaz4354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giebel, H.-A.; Kalhoefer, D.; Gahl-Janssen, R.; Choo, Y.-J.; Lee, K.; Cho, J.-C.; Tindall, B.J.; Rhiel, E.; Beardsley, C.; Aydogmus, O.; et al. Planktomarina temperata gen. nov., sp. nov., belonging to the globally distributed RCA cluster of the marine Roseobacter clade, isolated from the German Wadden Sea. Int. J. Syst. Evol. Microbiol. 2013, 63, 4207–4217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joint, I.; Mühling, M.; Querellou, J. Culturing marine bacteria—An essential prerequisite for biodiscovery. Microb. Biotechnol. 2010, 3, 564–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilbert, J.A.; Field, D.; Swift, P.; Newbold, L.; Oliver, A.; Smyth, T.; Somerfield, P.J.; Huse, S.; Joint, I. The seasonal structure of microbial communities in the Western English Channel. Environ. Microbiol. 2009, 11, 3132–3139. [Google Scholar] [CrossRef] [PubMed]
- World Register of Marine Species. Available online: https://www.marinespecies.org/index.php (accessed on 9 September 2022).
- Marquez, M.C.; Ventosa, A.; Ruiz-Berraquero, F. Marinococcus hispanicus, a New Species of Moderately Halophilic Gram-Positive Cocci. Int. J. Syst. Evol. Microbiol. 1990, 40, 165–169. [Google Scholar] [CrossRef] [Green Version]
- Yoon, J.-H.; Kim, I.-G.; Schumann, P.; Oh, T.-K.; Park, Y.-H. Marinibacillus campisalis sp. nov., a moderate halophile isolated from a marine solar saltern in Korea, with emended description of the genus Marinibacillus. Int. J. Syst. Evol. Microbiol. 2004, 54, 1317–1321. [Google Scholar] [CrossRef] [Green Version]
- Spring, S.; Ludwig, W.; Marquez, M.C.; Ventosa, A.; Schleifer, K.-H. Halobacillus gen. nov., with Descriptions of Halobacillus litoralis sp. nov. and Halobacillus trueperi sp. nov., and Transfer of Sporosarcina halophila to Halobacillus halophilus comb. nov. Int. J. Syst. Evol. Microbiol. 1996, 46, 492–496. [Google Scholar] [CrossRef] [Green Version]
- Al Khudary, R.; Stösser, N.I.; Qoura, F.; Antranikian, G. Pseudoalteromonas arctica sp. nov., an aerobic, psychrotolerant, marine bacterium isolated from Spitzbergen. Int. J. Syst. Evol. Microbiol. 2008, 58, 2018–2024. [Google Scholar] [CrossRef]
- Joung, Y.; Kim, H.; Joh, K. Flavobacterium jumunjinense sp. nov., isolated from a lagoon, and emended descriptions of Flavobacterium cheniae, Flavobacterium dongtanense and Flavobacterium gelidilacus. Int. J. Syst. Evol. Microbiol. 2013, 63, 3937–3943. [Google Scholar] [CrossRef] [Green Version]
- Romanenko, L.A.; Zhukova, N.; Rohde, M.; Lysenko, A.M.; Mikhailov, V.V.; Stackebrandt, E. Glaciecola mesophila sp. nov., a novel marine agar-digesting bacterium. Int. J. Syst. Evol. Microbiol. 2003, 53, 647–651. [Google Scholar] [CrossRef]
- Nedashkovskaya, O.I.; Kim, S.-G.; Balabanova, L.A.; Zhukova, N.V.; Bakunina, I.Y.; Mikhailov, V.V. Polaribacter staleyi sp. nov., a polysaccharide-degrading marine bacterium isolated from the red alga Ahnfeltia tobuchiensis. Int. J. Syst. Evol. Microbiol. 2018, 68, 623–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nedashkovskaya, O.I.; Kim, S.B.; Lysenko, A.M.; Frolova, G.M.; Mikhailov, V.V.; Lee, K.H.; Bae, K.S. Description of Aquimarina muelleri gen. nov., sp. nov., and proposal of the reclassification of [Cytophaga] latercula Lewin 1969 as Stanierella latercula gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 2005, 55, 225–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansen, J.E.; Nielsen, P.; Sjøholm, C. Description of Cellulophaga baltica gen. nov., sp. nov. and Cellulophaga fucicola gen. nov., sp. nov. and reclassification of [Cytophaga] lytica to Cellulophaga lytica gen. nov., comb. nov. Int. J. Syst. Bacteriol. 1999, 49, 1231–1240. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.-Q.; Schumann, P.; Li, W.-J.; Chen, G.-Z.; Tian, X.-P.; Stackebrandt, E.; Xu, L.-H.; Jiang, C.-L. Isoptericola halotolerans sp. nov., a novel actinobacterium isolated from saline soil from Qinghai Province, north-west China. Int. J. Syst. Evol. Microbiol. 2005, 55, 1867–1870. [Google Scholar] [CrossRef] [Green Version]
- Kemung, H.M.; Tan, L.T.-H.; Khan, T.M.; Chan, K.-G.; Pusparajah, P.; Goh, B.-H.; Lee, L.-H. Streptomyces as a Prominent Resource of Future Anti-MRSA Drugs. Front. Microbiol. 2018, 9, 2221. [Google Scholar] [CrossRef]
- Esteves, A.I.; Hardoim, C.C.; Xavier, J.R.; Gonçalves, J.M.; Costa, R. Molecular richness and biotechnological potential of bacteria cultured from Irciniidae sponges in the north-east Atlantic. FEMS Microbiol. Ecol. 2013, 85, 519–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, S.G.; Paula, P.; da Silva, J.P.; Mil-Homens, D.; Teixeira, M.C.; Fialho, A.M.; Costa, R.; Keller-Costa, T. Insights into the Antimicrobial Activities and Metabolomes of Aquimarina (Flavobacteriaceae, Bacteroidetes) Species from the Rare Marine Biosphere. Mar. Drugs 2022, 20, 423. [Google Scholar] [CrossRef]
- Helfrich, E.J.N.; Ueoka, R.; Dolev, A.; Rust, M.; Meoded, R.A.; Bhushan, A.; Califano, G.; Costa, R.; Gugger, M.; Steinbeck, C.; et al. Automated structure prediction of trans-acyltransferase polyketide synthase products. Nat. Chem. Biol. 2019, 15, 813–821. [Google Scholar] [CrossRef]
- Dieterich, C.L.; Probst, S.I.; Ueoka, R.; Sandu, I.; Schäfle, D.; Molin, M.D.; Minas, H.A.; Costa, R.; Oxenius, A.; Sander, P.; et al. Aquimarins, Peptide Antibiotics with Amino-Modified C-Termini from a Sponge-Derived Aquimarina sp. Bacterium. Angew. Chem. Int. Ed. 2022, 61, e202115802. [Google Scholar] [CrossRef]
- Zhou, X.; Willems, R.J.L.; Friedrich, A.W.; Rossen, J.W.A.; Bathoorn, E. Enterococcus faecium: From microbiological insights to practical recommendations for infection control and diagnostics. Antimicrob. Resist. Infect. Control 2020, 9, 130. [Google Scholar] [CrossRef]





| Locations | Temperature (°C) | pH | DO (mg·L−1) | Salinity (g·kg−1) |
|---|---|---|---|---|
| EN | 9.3 ± 0.2 | 8.1 ± 0.1 | 4.8 ± 1 | 18.01 ± 0.08 |
| CA | 11.3 ± 0.4 | 8.1 ± 0.1 | 9.9 ± 0.9 | 17.9 ± 0.14 |
| Aquimarina muelleri SWA EN P3.6 | Streptomyces sp. SWA CA P3.9 | ||
|---|---|---|---|
| Reference strains | S. aureus ATCC 25923 | + | + |
| E. coli ATCC 25922 | − | − | |
| P. aeruginosa ATCC 15442 | − | − | |
| S. enterica ATCC 14028 | − | − | |
| L. monocytogenes ATCC 1911 | + | + | |
| Clinical isolates | MRSA 388 | + | + |
| MRSA S1 | + | + | |
| MRSA F1 | + | + | |
| Acinetobacter sp. 19047 CNE3 | − | − | |
| Enterobacter asburiae 19069 ONE1 | − | w+ | |
| Enterobacter cloacae 19069 ONE2 | − | − | |
| Enterococcus faecium 19040 E1 | + | + | |
| Klebsiella sp. 19094 CK1 | − | − | |
| P. aeruginosa 19053 CNE5 | − | − |
| Genus | Species | Strain | Hydrolysis of: | Ref. | |||||
|---|---|---|---|---|---|---|---|---|---|
| Casein | Tween 80 | Starch | CMC | Xylan | Pectin | ||||
| Paraglaciecola | mesophila | SWA EN P3.5 | - | + | + | + | - | + | This study |
| MA CA P3.7 | + | + | + | + | − | − | This study | ||
| KMM 241T | − | + | + | nd | nd | nd | [56] | ||
| Pseudoalteromonas | sp. | SWA CA P1.16 | − | + | + | + | + | + | This study |
| sp. | MA CA P1.5 | + | + | + | + | + | − | This study | |
| arctica | A 37-1-2T | + | + | − a | − b | − | + | [54] | |
| Polaribacter | SWA EN P2.1 | + | + | + | + | + | − | This study | |
| staleyi | SWA CA P1.18 | + | + | + | + | + | + | This study | |
| 10Alg 139T | − | + | + | − | nd | nd | [57] | ||
| Aquimarina | muelleri | SWA EN P3.6 | + | + | + | + | − | − | This study |
| KMM 6020T | + | + | + | − | nd | nd | [58] | ||
| Cellulophaga | baltica | SWA EN P1.16 | − | + | − | + | + | − | This study |
| SWA CA P1.23 | + | + | − | + | + | − | This study | ||
| NN015840T | + | − | + | + | nd | nd | [59] | ||
| Flavobacterium | sp. | SWA EN P2.6 | + | + | + | + | + | − | This study |
| sp. | SWA CA P2.5 | + | + | + | + | + | − | This study | |
| jumunjinense | HME7102T | + | − | − | − | nd | nd | [55] | |
| Isoptericola | sp. | MA EN P3.8 | + | + | + | + | + | + | This study |
| halotolerans | YIM 70177T | − | − | − | nd | nd | nd | [60] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruginescu, R.; Lavin, P.; Iancu, L.; Menabit, S.; Purcarea, C. Bioprospecting for Novel Bacterial Sources of Hydrolytic Enzymes and Antimicrobials in the Romanian Littoral Zone of the Black Sea. Microorganisms 2022, 10, 2468. https://doi.org/10.3390/microorganisms10122468
Ruginescu R, Lavin P, Iancu L, Menabit S, Purcarea C. Bioprospecting for Novel Bacterial Sources of Hydrolytic Enzymes and Antimicrobials in the Romanian Littoral Zone of the Black Sea. Microorganisms. 2022; 10(12):2468. https://doi.org/10.3390/microorganisms10122468
Chicago/Turabian StyleRuginescu, Robert, Paris Lavin, Lavinia Iancu, Selma Menabit, and Cristina Purcarea. 2022. "Bioprospecting for Novel Bacterial Sources of Hydrolytic Enzymes and Antimicrobials in the Romanian Littoral Zone of the Black Sea" Microorganisms 10, no. 12: 2468. https://doi.org/10.3390/microorganisms10122468
APA StyleRuginescu, R., Lavin, P., Iancu, L., Menabit, S., & Purcarea, C. (2022). Bioprospecting for Novel Bacterial Sources of Hydrolytic Enzymes and Antimicrobials in the Romanian Littoral Zone of the Black Sea. Microorganisms, 10(12), 2468. https://doi.org/10.3390/microorganisms10122468







