Pseudomonas protegens FJKB0103 Isolated from Rhizosphere Exhibits Anti-Methicillin-Resistant Staphylococcus aureus Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Isolation and Assays for Antagonistic Capacity of Strain FJKB0103
2.3. Genome Sequencing and Annotation
2.4. Identification of Strain FJKB0103 Based on Genome
2.5. Genome Analysis of Putative Secondary Metabolite Clusters
2.6. Construction of P. protegens FJKB0103 Deleted Mutants and Complementary Strain
2.7. Extraction and Detection of DAPG
2.8. Determination of the Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) of DAPG, MAPG, PG, or Vancomycin on S. aureus
2.9. Bacteriolytic Assay
2.10. Biofilm Formation and Swarming Motility
3. Results
3.1. Antibiotic Activity of Isolates from Rhizosphere
3.2. General Genome Characteristics of Strain FJKB0103
3.3. Identification of Strain FJKB0103 Based on Polyphasic Taxonomy
3.4. Genome Mining to Reveal the Putative Secondary Metabolite Clusters
3.5. DAPG as the Anti-MRSA Compound in P. protegens FJKB0103
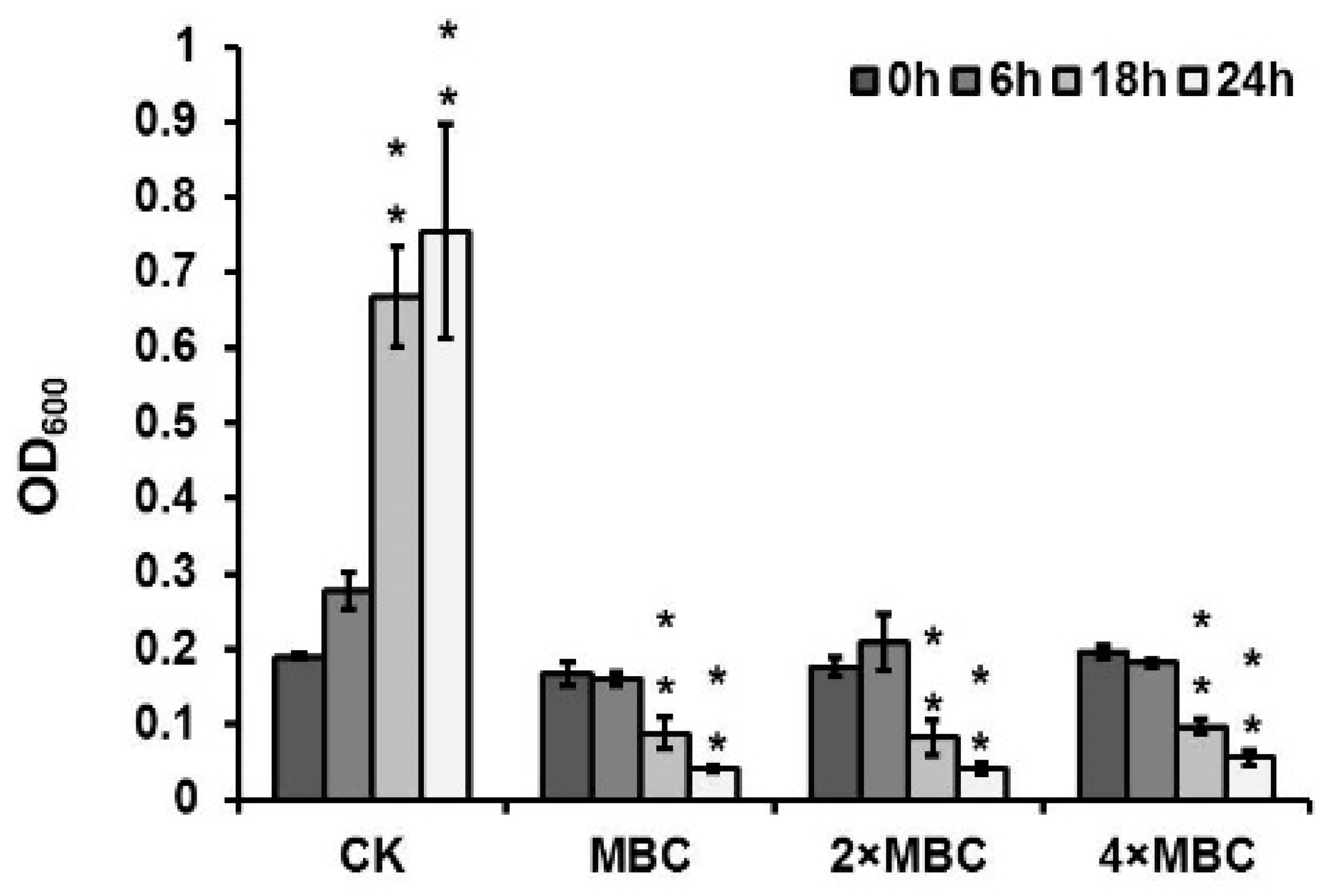
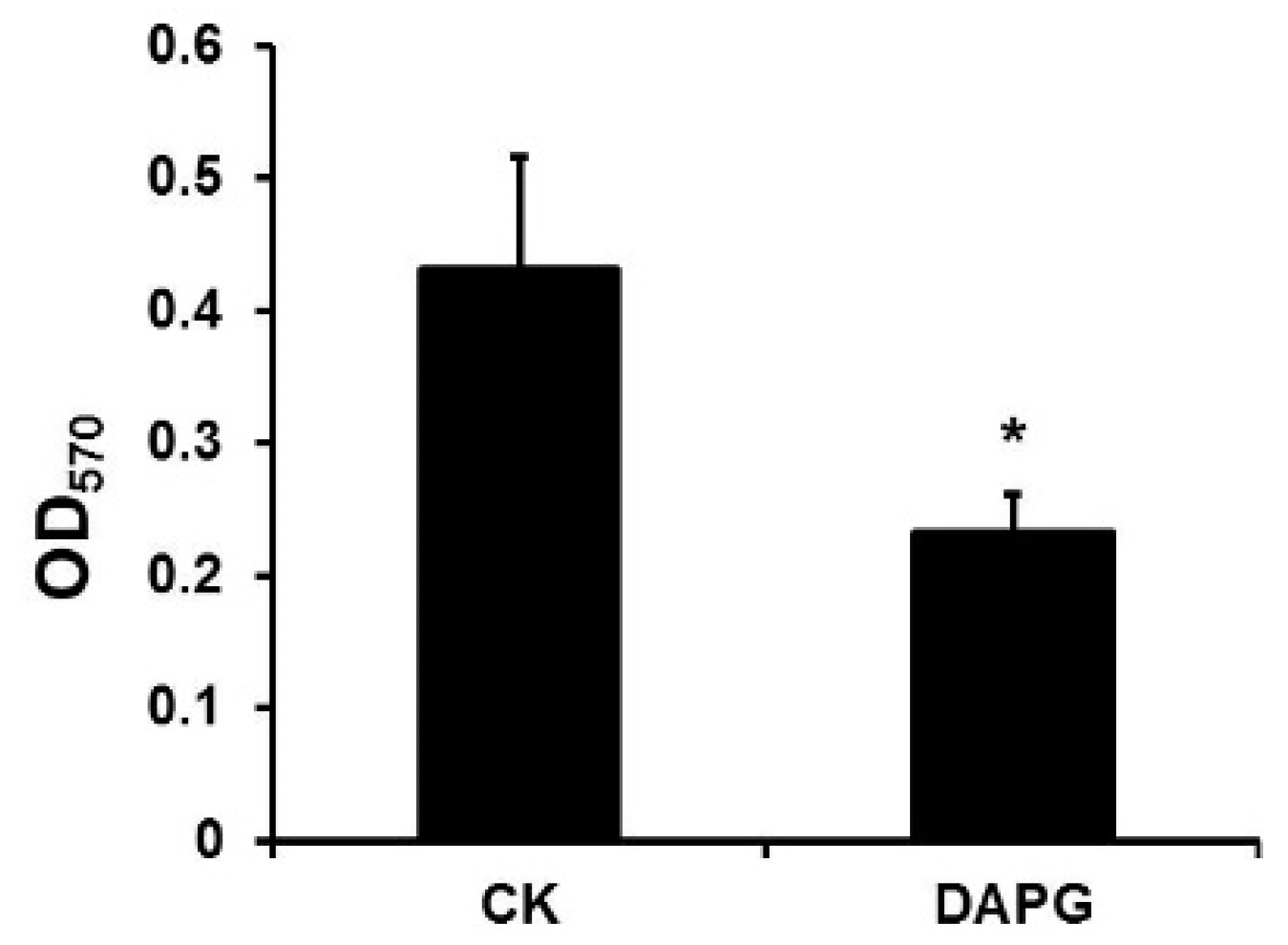
3.6. DAPG Affected the Biofilm Formation of S. aureus
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.; Fowler, V.G. Staphylococcus aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chambers, H.F.; DeLeo, F.R. Waves of Resistance: Staphylococcus aureus in the Antibiotic Era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Jevons, M.P. “Celbenin”—Resistant Staphylococci. Br. Med. J. 1961, 1, 124–125. [Google Scholar] [CrossRef]
- Witte, W. Antibiotic Resistance in Gram-Positive Bacteria: Epidemiological Aspects. J. Antimicro. B Chemoth. 1999, 44, 1–9. [Google Scholar] [CrossRef]
- Craft, K.M.; Nguyen, J.M.; Berg, L.J.; Townsend, S.D. Methicillin-Resistant Staphylococcus aureus (MRSA): Antibiotic-Resistance and the Biofilm Phenotype. Med. Chem. Commun. 2019, 1, 1231–1241. [Google Scholar] [CrossRef]
- Gross, H.; Loper, J.E. Genomics of Secondary Metabolite Production by Pseudomonas spp. Nat. Prod. Rep. 2009, 26, 1408–1446. [Google Scholar] [CrossRef]
- Iessy, A.; Filion, M. Phloroglucinol Derivatives in Plant-Beneficial Pseudomonas spp.: Biosynthesis, Regulation, and Functions. Metabolites 2021, 11, 182. [Google Scholar]
- Gleeson, O.; Gara, F.O.; Morrissey, J.P. The Pseudomonas fluorescens Secondary Metabolite 2,4 Diacetylphloroglucinol Impairs Mitochondrial Function in Saccharomyces cerevisiae. Anton. Leeuw. Int. J. G 2010, 97, 261–273. [Google Scholar] [CrossRef]
- Julian, W.T.; Vasilchenko, A.V.; Shpindyuk, D.D.; Poshvina, D.V.; Vasilchenko, A.S. Bacterial-Derived Plant Protection Metabolite 2,4-Diacetylphloroglucinol: Effects on Bacterial Cells at Inhibitory and Subinhibitory Concentrations. Biomolecules 2021, 11, 13. [Google Scholar] [CrossRef]
- De Souza, J.T.; Arnould, C.; Deulvot, C.; Lemanceau, P.; Gianinazzi-Pearson, V.; Raaijmakers, J.R. Effect of 2,4-Diacetylphloroglucinol on Pythium: Cellular Responses and Variation in Sensitivity Among Propagules and Species. Biol. Control. 2003, 93, 966–975. [Google Scholar] [CrossRef] [Green Version]
- Almario, J.; Bruto, M.; Vacheron, J.; Prigent-Combaret, C.; Moënne-Loccoz, Y.; Muller, D. Distribution of 2,4-Diacetylphloroglucinol Biosynthetic Genes among the Pseudomonas spp. Reveals Unexpected Polyphyletism. Front. Microbiol. 2017, 8, 1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Achkar, J.; Xian, M.; Zhao, H.; Frost, J.W. Biosynthesis of Phloroglucinol. J. Am. Chem. Soc. 2005, 127, 5332–5333. [Google Scholar] [CrossRef] [PubMed]
- Pavkov-Keller, T.; Schmidt, N.G.; Zadlo-Dobrowolska, A.; Kroutil, W.; Gruber, K. Structure and Catalytic Mechanism of a Bacterial Friedel-Crafts Acylase. Chembiochem 2019, 20, 88–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbas, A.; McGuire, J.E.; Crowley, D.; Baysse, C.; Dow, M.; O’Gara, F. The Putative Permease PhlE of F113 Has a Role in 2,4-Diacetylphloroglucinol Resistance and in General Stress Tolerance. Microbiology 2004, 150, 2443–2450. [Google Scholar] [CrossRef] [Green Version]
- Abbas, A.; Morrissey, J.P.; Marquez, P.C.; Sheehan, M.M.; Delany, I.R.; O’Gara, F. Characterization of Interactions between the Transcriptional Repressor PhlF and Its Binding Site at the phlA Promoter in Pseudomonas fluorescens F113. J. Bacteriol. 2002, 184, 3008–3016. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Yang, R.; Zhao, R.X.; Han, J.T.; Jia, W.J.; Li, D.Y.; Wang, Y.; Zhang, N.; Wu, Y.; Zhang, L.Q.; et al. Transcriptional Regulator PhlH Modulates 2,4-Diacetylphloroglucinol Biosynthesis in Response to the Biosynthetic Intermediate and End Product. Appl. Environ. Microbiol. 2017, 83, e01419-17. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Huang, J.H.; Zhang, F.; Wu, Q.P.; Zhang, J.M.; Pang, R.; Zeng, H.Y.; Yang, X.Y.; Chen, M.T.; Wang, J.; et al. Prevalence and Characterization of Food-Related Methicillin-Resistant Staphylococcus aureus (MRSA) in China. Front. Microbiol. 2019, 10, 304. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Lee, I.; Kim, Y.O.; Park, S.C.; Chun, J. OrthoANI: An Improved Algorithm and Software for Calculating Average Nucleotide Identity. Int. J. Syst. Evol. Micr. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Goker, M. Genome Sequence-Based Species Delimitation with Confidence Intervals and Improved Distance Functions. BMC Bioinformatis 2013, 14, 60. [Google Scholar] [CrossRef] [Green Version]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Marnix, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary Metabolite Genome Mining Pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmadzadeh, M.; Tehrani, A.S. Evaluation of fluorescent Pseudomonads for Plant Growth Promotion, Antifungal Activity against Rhizoctonia solani on Common Bean, and Biocontrol Potential. Biol. Control 2009, 48, 101–107. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Abdel-Hamid, M.; Romeih, E.; Saporito, P.; Osman, A.; Mateiu, R.V.; Mojsoska, B.; Jenssen, H. Camel Milk Whey Hydrolysate Inhibits Growth and Biofilm Formation of Pseudomonas aeruginosa PAO1 and Methicillin-Resistant Staphylococcus aureus. Food Control. 2020, 11, 107056. [Google Scholar] [CrossRef]
- Kamei, Y.; Isnansetyo, A. Lysis of Methicillin-Resistant Staphylococcus aureus by 2,4-Diacetylphloroglucinol Produced by Pseudomonas sp. AMSN Isolated from a Marine Alga. Int. J. Antimicrob. A G 2003, 21, 71–74. [Google Scholar] [CrossRef]
- Zhao, H.; Liu, Y.P.; Zhang, L.Q. In silico and Genetic Analyses of Cyclic Lipopeptide Synthetic Gene Clusters in Pseudomonas sp. 11K1. Front. Microbiol. 2019, 10, 544. [Google Scholar] [CrossRef]
- Olorunleke, F.E.; Kieu, N.P.; De Waele, E.; Timmerman, M.; Ongena, M.; Hofte, M. Coregulation of the Cyclic Lipopeptides Orfamide and Sessilin in the Biocontrol Strain Pseudomonas sp. CMR12a. MicrobiologyOpen 2017, 6, e4995. [Google Scholar] [CrossRef] [Green Version]
- Powers, M.J.; Sanabria-Valentín, E.; Bowers, A.A.; Shank, E.A. Inhibition of Cell Differentiation in Bacillus subtilis by Pseudomonas protegens. J. Bacteriol. 2015, 197, 2129–2138. [Google Scholar] [CrossRef]
- Yarwood, J.M.; Douglas, J.; Bartels, D.J.; Volper, E.M.; Greenberg, E.P. Quorum Sensing in Staphylococcus aureus Biofilms. J. Bacteriol. 2004, 186, 1838–1850. [Google Scholar] [CrossRef] [Green Version]
- Gurney, R.; Thomas, C.M. Mupirocin: Biosynthesis, Special Features and Applications of an Antibiotic from a Gram-Negative Bacterium. Appl. Microbiol. Biot. 2011, 90, 11–21. [Google Scholar] [CrossRef]
- Cardozo, V.F.; Oliveira, A.G.; Nishio, E.K.; Perugini, M.R.E.; Andrade, C.; Silveira, W.D.; Durán, N.; Andrade, G.; Kobayashi, R.; Nakazato, G. Antibacterial Activity of Extracellular Compounds Produced by a Pseudomonas Strain against Methicillin-Resistant Staphylococcus aureus (MRSA) Strains. Ann. Clin. Microb. Anti. 2013, 12, 12. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.S.; Eom, S.H.; Jeong, S.Y.; Shin, H.J.; Je, J.Y.; Lee, E.W.; Chung, Y.H.; Kim, Y.M.; Kang, C.K.; Lee, M.S. Anti-Methicillin-Resistant Staphylococcus aureus (MRSA) Substance from the marine Bacterium Pseudomonas sp. UJ-6. Environ. Toxicol. Phar. 2013, 35, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Philmus, B.; Chang, J.; Loper, J.E. Novel Mechanism of Metabolic Co-Regulation Coordinates the Biosynthesis of Secondary Metabolites in Pseudomonas protegens. eLife 2017, 6, e22835. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.T.; von Tiedemann, A. 2,4-Diacetylphloroglucinol Suppresses Zoosporogenesis and Impairs Motility of Peronosporomycete Zoospores. World J. Microb. Biot. 2011, 27, 2071–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michelsen, C.F.; Watrous, J.; Glaring, M.A.; Kersten, R.; Koyama, N.; Dorrestein, P.C.; Stougaard, P. Nonribosomal Peptides, Key Biocontrol Components for Pseudomonas fluorescens In5, Isolated from a Greenlandic Suppressive Soil. mBio 2015, 6, e00079-15. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Qian, G.; Liu, F.; Li, Y.Z.; Shen, Y.; Du, L. Facile Method for Site-specific Gene Integration in Lysobacter enzymogenes for Yield Improvement of the Anti-MRSA Antibiotics WAP-8294A and the Antifungal Antibiotic HSAF. ACS Synth. Biol. 2013, 2, 670–678. [Google Scholar] [CrossRef] [Green Version]
- Chiellini, C.; Lombardo, K.; Mocali, S.; Miceli, E.; Fani, R. Pseudomonas Strains Isolated from Different Environmental Niches Exhibit Different Antagonistic Ability. Ethol. Ecol. Evol. 2019, 31, 399–420. [Google Scholar] [CrossRef]
- Hoffman, L.R.; D’Argenio, D.A.; MacCoss, M.J.; Zhang, Z.; Jones, R.A.; Miller, S.I. Aminoglycoside Antibiotics Induce Bacterial Biofilm Formation. Nature 2005, 436, 1171–1175. [Google Scholar] [CrossRef]
- Combes-Meynet, E.; Pothier, J.F.; Moënne-Loccoz, Y.; Prigent-Combaret, C. The Pseudomonas Secondary Metabolite 2,4-Diacetylphloroglucinol Is a Signal Inducing Rhizoplane Expression of Azospirillum Genes Involved in Plant-Growth Promotion. Mol. Plant Microbe. Interact. 2011, 24, 271–284. [Google Scholar] [CrossRef] [Green Version]
- Romero, D.; Traxler, M.F.; López, D.; Kolter, R. Antibiotics as Signal Molecules. Chem. Rev. 2011, 111, 5492–5505. [Google Scholar] [CrossRef] [Green Version]
- Lord, J.; Gikonyo, A.; Miwa, A.; Odoi, A. Antimicrobial Resistance among Enterobacteriaceae, Staphylococcus aureus, and Pseudomonas spp. Isolates from clinical Specimens from a Hospital in Nairobi, Kenya. PeerJ 2021, 9, e11958. [Google Scholar] [CrossRef]
- Isnansetyo, A.; Cui, L.Z.; Hiramatsu, K. Antibacterial Activity of 2,4-Diacetylphloroglucinol Produced by Pseudomonas sp. AMSN Isolated from a Marine Alga, against Vancomycin-Resistant Staphylococcus aureus. Int. J. Antimicrob. A G 2003, 22, 545–547. [Google Scholar] [CrossRef]

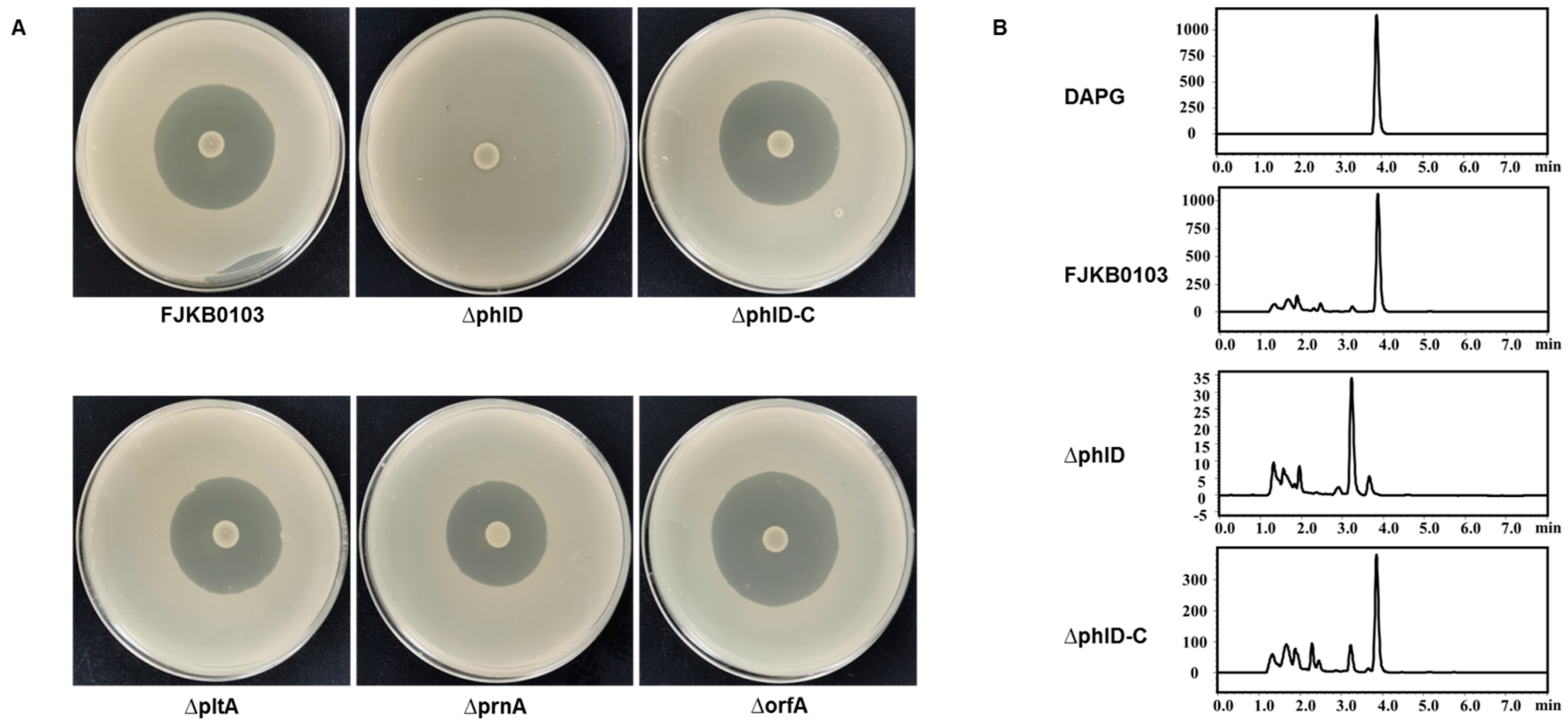
| Features | Chromosome |
|---|---|
| Number of contigs Size (bp) | 40 6,776,967 |
| G + C content | 63.4% |
| Number of genes | 6170 |
| Number of CDSs | 6096 |
| Number of rRNAs | 7 |
| Number of tRNAs | 66 |
| Number of tmRNAs | 1 |
| Species | ANIb (%) a | isDDH (%) b | |||
|---|---|---|---|---|---|
| ANIb | OrthoANI | Formula 1 | Formula 2 | Formula 3 | |
| P. protegens Cab57 (AP014522.1) | 98.44 | 98.51 | 96.80 | 86.90 | 97.10 |
| P. protegens CHA0 T (LS999205.1) | 98.13 | 98.27 | 95.10 | 84.00 | 95.60 |
| P. protegens H78 (CP013184.1) | 98.09 | 98.21 | 94.00 | 83.70 | 94.70 |
| P. protegens SN15-2 (CP043179.1) | 98.01 | 98.09 | 92.10 | 83.00 | 93.10 |
| P. protegens FDAARGOS_307 (CP022097.2) | 98.00 | 98.11 | 92.10 | 83.00 | 93.10 |
| P. protegens Pf-5 (NC_004129.6) | 97.98 | 98.11 | 92.10 | 83.00 | 93.10 |
| P. saponiphila DSM 9751 T (FNTJ00000000.1) | 89.30 | 89.71 | 62.90 | 38.60 | 57.50 |
| P. cerasi 58 T (LT222319.1) | 77.25 | 78.07 | 19.60 | 22.50 | 19.10 |
| P. meliae CFBP 3225 T (JYHE00000000.1) | 77.23 | 77.96 | 19.10 | 22.50 | 18.70 |
| P. congelans DSM 14939 T (NJH0000000) | 77.13 | 78.10 | 19.50 | 22.40 | 19.10 |
| P. tremae ICMP9151 T (LJRO00000000.1) | 76.67 | 77.50 | 18.10 | 22.30 | 17.80 |
| P. caspiana FBF102 T (LOHF00000000.1) | 76.45 | 77.20 | 17.80 | 22.60 | 17.60 |
| Strains | Total | DAPG | PLT | PRN | NRPS | Bacteriocin | Others |
|---|---|---|---|---|---|---|---|
| P. protegens FJKB0103 | 16 | 1 | 1 | 1 | 7 | 2 | 4 |
| P. protegens Cab57 | 14 | 1 | 1 | 1 | 5 | 2 | 4 |
| P. protegens CHA0 T | 14 | 1 | 1 | 1 | 5 | 3 | 3 |
| P. protegens H78 | 15 | 1 | 1 | 1 | 5 | 3 | 4 |
| P. protegens SN15-2 | 16 | 1 | 1 | 1 | 6 | 2 | 5 |
| P. protegens FDAARG | 15 | 1 | 1 | 1 | 5 | 3 | 4 |
| P. protegens Pf-5 | 15 | 1 | 1 | 1 | 5 | 3 | 4 |
| P. saponiphila DSM 9751 T | 14 | 1 | 0 | 0 | 4 | 3 | 6 |
| P. cerasi 58 T | 9 | 0 | 0 | 0 | 5 | 1 | 3 |
| P. meliae CFBP 3225 T | 9 | 0 | 0 | 0 | 5 | 0 | 4 |
| P. congelans DSM 14939 T | 9 | 0 | 0 | 0 | 7 | 0 | 2 |
| P. tremae ICMP9151 T | 15 | 0 | 0 | 0 | 9 | 0 | 6 |
| P. caspiana FBF102 T | 6 | 0 | 0 | 0 | 2 | 2 | 2 |
| Strains | MIC (μg/mL) | MBC (μg/mL) | ||||||
|---|---|---|---|---|---|---|---|---|
| DAPG | MAPG | PG | Vancomycin | DAPG | MAPG | PG | Vancomycin | |
| S. aureus ATCC25293 | 4 | >128 | >128 | 2 | 32 | >128 | >128 | 32 |
| S. aureus ATCC29213 | 4 | >128 | >128 | 2 | 32 | >128 | >128 | 16 |
| S. aureus Sta403 | 4 | >128 | >128 | 2 | 16 | >128 | >128 | 16 |
| S. aureus Sta24-1 | 4 | >128 | >128 | 1 | 32 | >128 | >128 | 32 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, H.; Liu, L.; Yang, L.; Gu, Q.; Li, Y.; Zhang, J.; Wu, S.; Chen, M.; Xie, X.; Wu, Q. Pseudomonas protegens FJKB0103 Isolated from Rhizosphere Exhibits Anti-Methicillin-Resistant Staphylococcus aureus Activity. Microorganisms 2022, 10, 315. https://doi.org/10.3390/microorganisms10020315
Zhao H, Liu L, Yang L, Gu Q, Li Y, Zhang J, Wu S, Chen M, Xie X, Wu Q. Pseudomonas protegens FJKB0103 Isolated from Rhizosphere Exhibits Anti-Methicillin-Resistant Staphylococcus aureus Activity. Microorganisms. 2022; 10(2):315. https://doi.org/10.3390/microorganisms10020315
Chicago/Turabian StyleZhao, Hui, Lu Liu, Lingshuang Yang, Qihui Gu, Ying Li, Jumei Zhang, Shi Wu, Moutong Chen, Xinqiang Xie, and Qingping Wu. 2022. "Pseudomonas protegens FJKB0103 Isolated from Rhizosphere Exhibits Anti-Methicillin-Resistant Staphylococcus aureus Activity" Microorganisms 10, no. 2: 315. https://doi.org/10.3390/microorganisms10020315
APA StyleZhao, H., Liu, L., Yang, L., Gu, Q., Li, Y., Zhang, J., Wu, S., Chen, M., Xie, X., & Wu, Q. (2022). Pseudomonas protegens FJKB0103 Isolated from Rhizosphere Exhibits Anti-Methicillin-Resistant Staphylococcus aureus Activity. Microorganisms, 10(2), 315. https://doi.org/10.3390/microorganisms10020315






