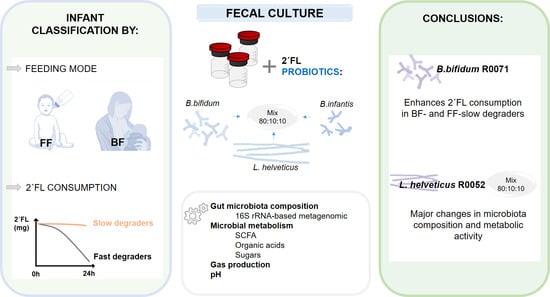
In Vitro Probiotic Modulation of the Intestinal Microbiota and 2′Fucosyllactose Consumption in Fecal Cultures from Infants at Two Months of Age
Abstract
:1. Introduction
2. Materials and Methods
2.1. 2′-Fucosyllactose Commercial Preparations and Probiotic Strains
2.2. Fecal Sample Collection and Batch Culture Fermentations
2.3. Microbiota Composition Analysis
2.4. Gas and pH Monitorization
2.5. Analysis of 2′Fucosyllactose, Lactose, Monosaccharides, and Organic Acids by HPLC
2.6. Analysis of Short-Chain Fatty Acids by Gas Chromatography
2.7. Bioinformatic Data Processing and Statistical Analyses of the Microbial Community
2.8. Statistical Analyses of Microbiota Composition and Microbial Metabolites
2.9. Nucleotide Sequence Accession Numbers
3. Results
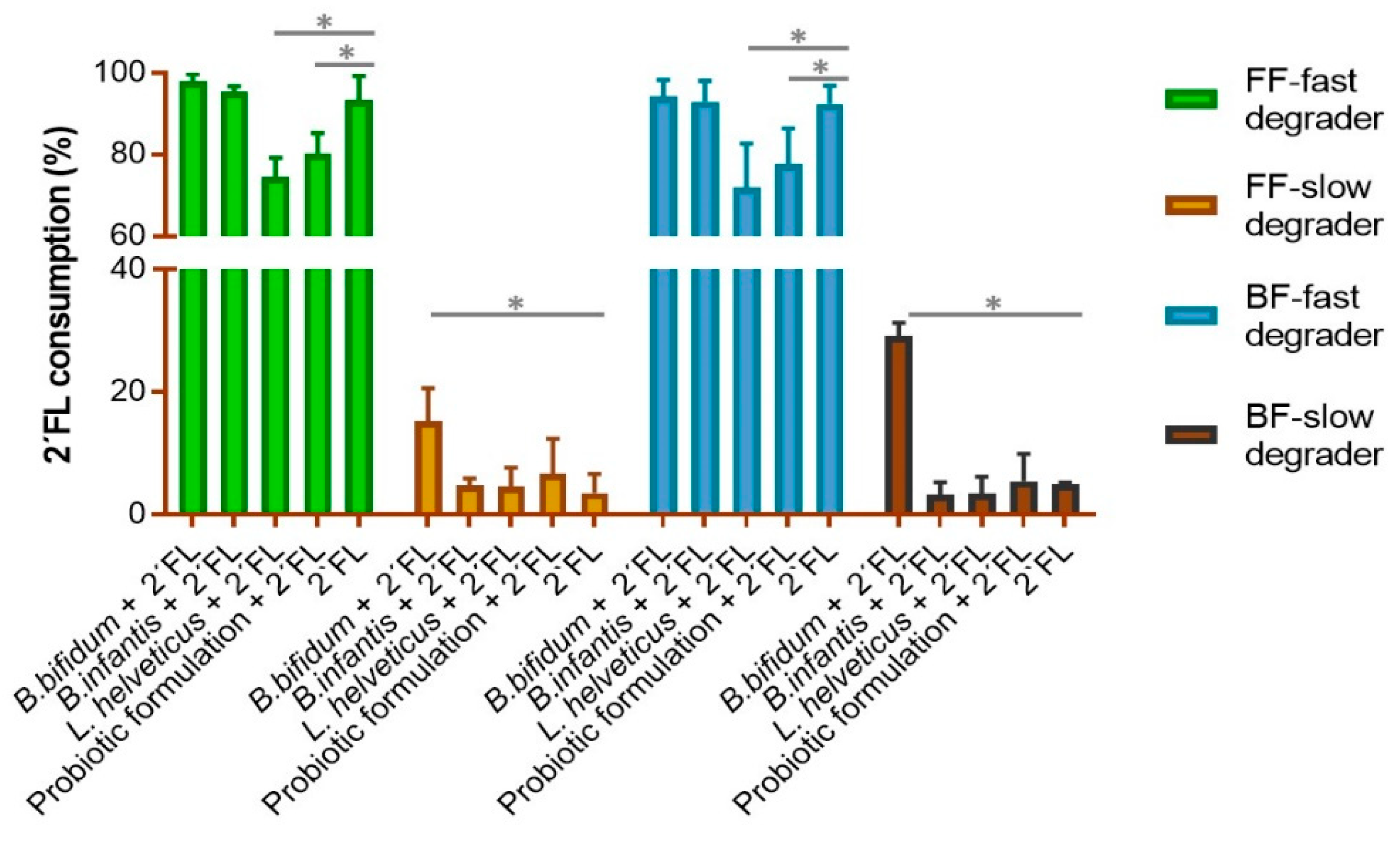
3.1. 2′-FL Degradation Profile in Fecal Cultures with Probiotics Added
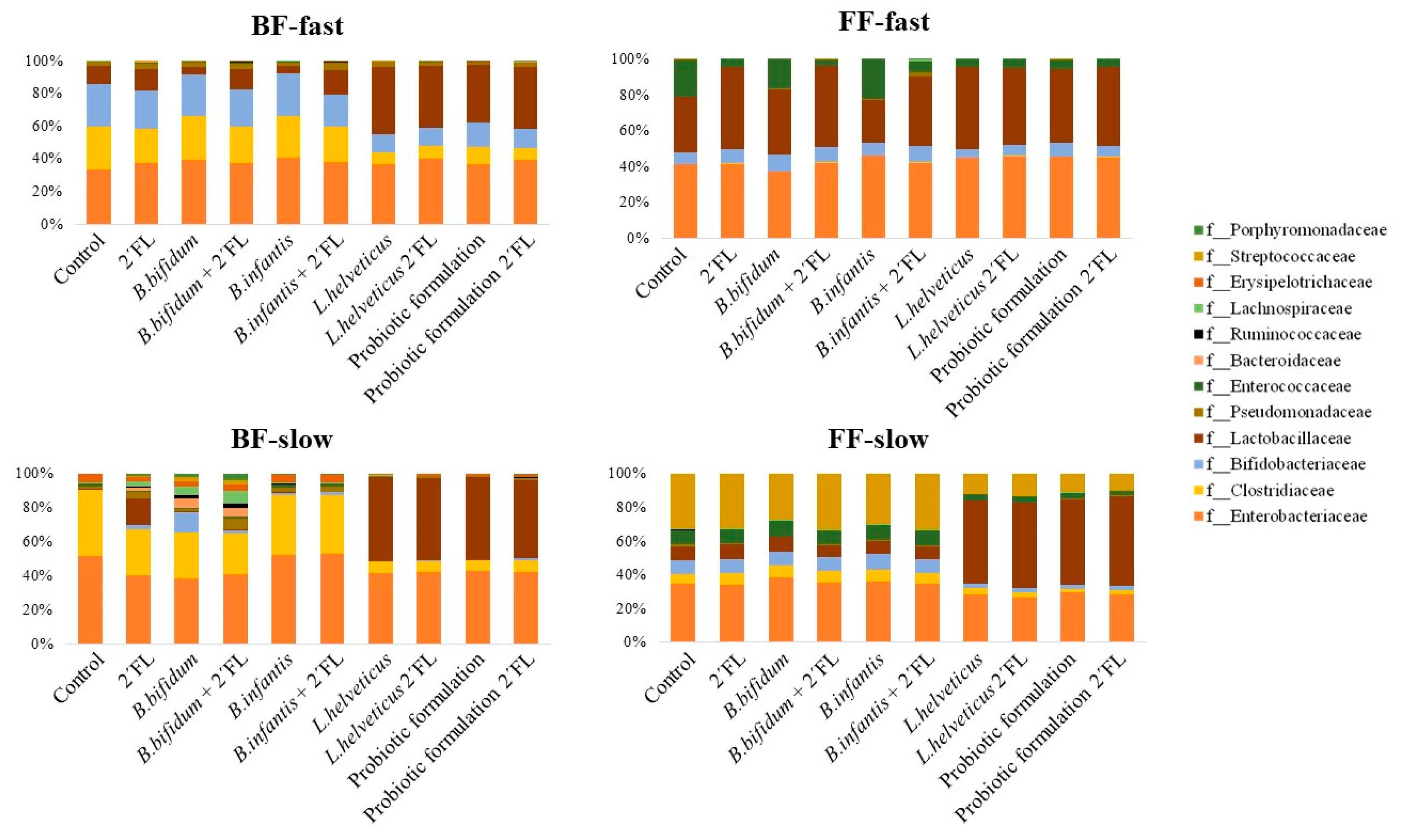
3.2. Effect of Probiotics and Their Combination with 2′FL on the Microbial Composition
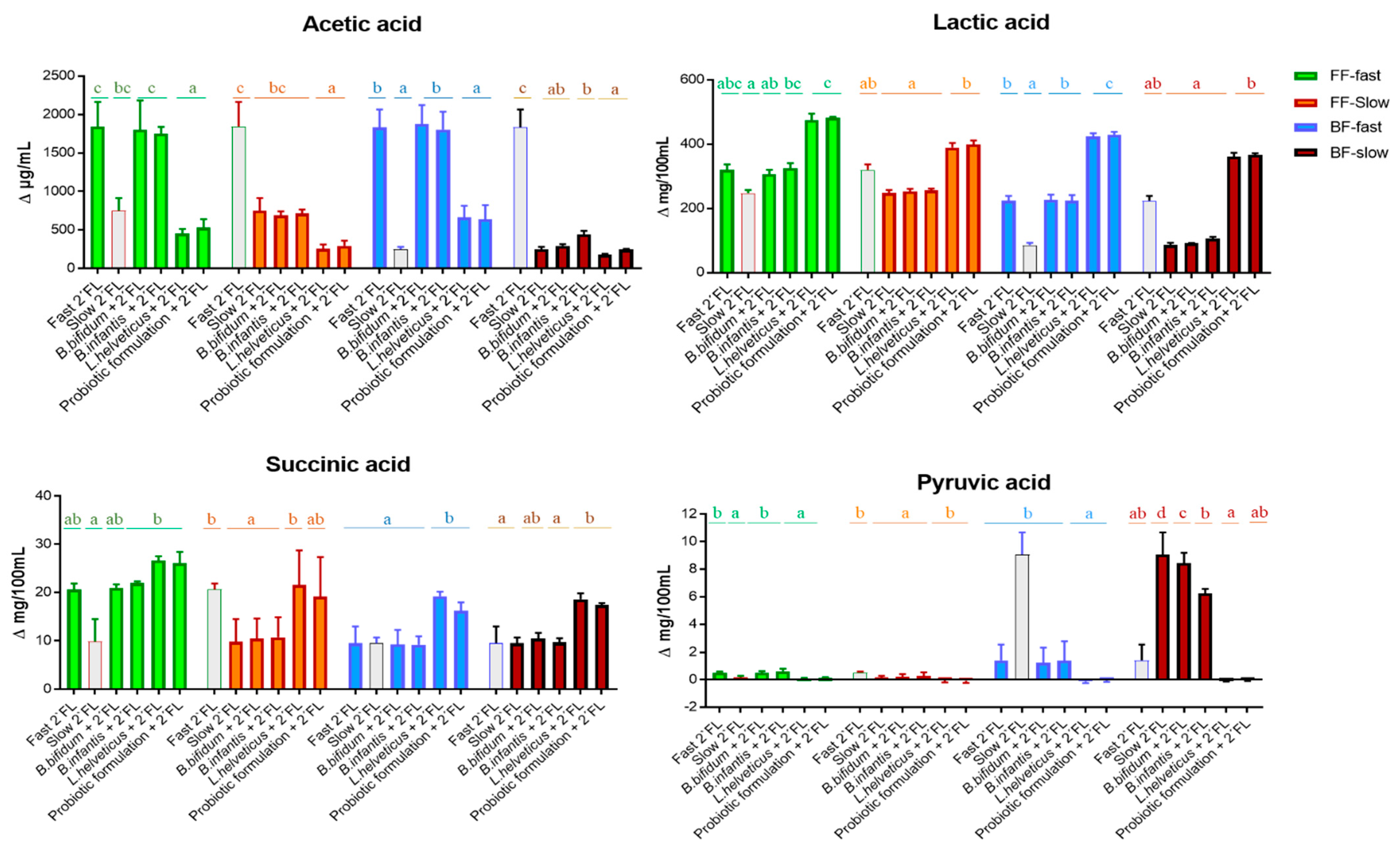
3.3. Effect of the Probiotic Treatment on the Microbial Metabolic Activity of Fecal Cultures with 2′FL Added
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Delgado Palacio, S.; Arboleya Montes, S.; Mancabelli, L.; et al. The first microbial colonizers of the human gut: Composition, activities, and health implications of the infant gut microbiota. Microbiol. Mol. Biol. Rev. 2017, 81, e00036-17. [Google Scholar] [CrossRef] [Green Version]
- Orczyk-Pawiłowicz, M.; Lis-Kuberka, J. The impact of dietary fucosylated oligosaccharides and glycoproteins of human milk on infant well-being. Nutrients 2020, 12, 1105. [Google Scholar] [CrossRef]
- Yu, Z.T.; Chen, C.; Newburg, D.S. Utilization of major fucosylated and sialylated human milk oligosaccharides by isolated human gut microbes. Glycobiology 2013, 23, 1281–1292. [Google Scholar] [CrossRef] [Green Version]
- Salli, K.; Hirvonen, J.; Siitonen, J.; Ahonen, I.; Anglenius, H.; Maukonen, J. Selective utilization of the human milk oligo-saccharides 2′-fucosyllactose and difucosyllactose by various probiotic and pathogenic bacteria. J. Agric. Food Chem. 2021, 69, 170–182. [Google Scholar] [CrossRef]
- Borewicz, K.; Gu, F.; Saccenti, E.; Arts, I.C.W.; Penders, J.; Thijs, C.; van Leeuwen, S.S.; Lindner, C.; Nauta, A.; van Leusen, E.; et al. Correlating infant fecal microbiota composition and human milk oligosaccharide consumption by microbiota of 1-month-old breastfed infants. Mol. Nutr. Food Res. 2019, 63, 1801214. [Google Scholar] [CrossRef]
- Borewicz, K.; Gu, F.; Saccenti, E.; Hechler, C.; Beijers, R.; Weerth, C.; van Leeuwen, S.S.; Schols, H.A.; Smidt, H. The associa-tion between breastmilk oligosaccharides and faecal microbiota in healthy breastfed infants at two, six, and twelve weeks of age. Sci. Rep. 2020, 10, 4270. [Google Scholar] [CrossRef]
- Nogacka, A.M.; Arboleya, S.; Nikpoor, N.; Auger, J.; Salazar, N.; Cuesta, I.; Mantecón, L.; Solís, G.; Gueimonde, M.; Tomp-kins, T.A.; et al. Influence of 2′-Fucosyllactose on the microbiota composition and metabolic activity of fecal cultures from breastfed and formula-fed infants at two months of age. Microorganisms 2021, 9, 1478. [Google Scholar] [CrossRef]
- Arboleya, S.; Saturio, S.; Suárez, M.; Fernández, N.; Mancabelli, L.; de los Reyes-Gavilán, C.G.; Ventura, M.; Solís, G.; Gueimonde, M. Donated human milk as a determinant factor for the gut bifidobacterial ecology in premature babies. Microorganisms 2020, 8, 760. [Google Scholar] [CrossRef]
- Song, M.W.; Kim, K.T.; Paik, H.D. Probiotics as a functional health supplement in infant formulas for the improvement of intestinal microflora and immunity. Food Rev. Int. 2021, 1–7. [Google Scholar] [CrossRef]
- Masum, A.K.M.; Chandrapala, J.; Huppertz, T.; Adhikari, B.; Zisu, B. Production and characterization of infant milk formula powders: A review. Dry Technol. 2021, 39, 1492–1512. [Google Scholar] [CrossRef]
- Salminen, S.; Stahl, B.; Vinderola, G.; Szajewska, H. Infant formula supplemented with biotics: Current knowledge and fu-ture perspectives. Nutrients 2020, 12, 1952. [Google Scholar] [CrossRef]
- Heeney, D.D.; Gareau, M.G.; Marco, M.L. Intestinal Lactobacillus in health and disease, a driver or just along for the ride? Curr. Opin. Biotechnol. 2018, 49, 140–147. [Google Scholar] [CrossRef] [Green Version]
- Arboleya, S.; Watkins, C.; Stanton, C.; Ross, R.P. Gut bifidobacteria populations in human health and aging. Front. Microbiol. 2016, 7, 1204. [Google Scholar] [CrossRef] [Green Version]
- Turroni, F.; Peano, C.; Pass, D.A.; Foroni, E.; Severgnini, M.; Claesson, M.J.; Kerr, C.; Hourihane, J.; Murray, D.; Fuligni, F.; et al. Diversity of bifidobacteria within the infant gut microbiota. PLoS ONE 2012, 7, e36957. [Google Scholar] [CrossRef] [Green Version]
- Arboleya, S.; Bottacini, F.; O’Connell-Motherway, M.; Ryan, C.A.; Ross, R.P.; van Sinderen, D.; Stanton, C. Gene-trait match-ing across the Bifidobacterium longum pan-genome reveals considerable diversity in carbohydrate catabolism among human infant strains. BMC Genom. 2018, 19, 33. [Google Scholar] [CrossRef]
- Ioannou, A.; Knol, J.; Belzer, C. Microbial glycoside hydrolases in the first year of life: An analysis review on their presence and importance in infant gut. Front. Microbiol. 2021, 12, 631282. [Google Scholar] [CrossRef]
- Tremblay, A.; Xu, X.; Colee, J.; Tompkins, T.A. Efficacy of a multi-strain probiotic formulation in pediatric populations: A comprehensive review of clinical studies. Nutrients 2021, 13, 1908. [Google Scholar] [CrossRef]
- Sherman, P.M.; Johnson-Henry, K.C.; Yeung, H.P.; Ngo, P.S.; Goulet, J.; Tompkins, T.A. Probiotics reduce enterohemorrhag-ic Escherichia coli O157:H7- and enteropathogenic E. coli O127:H6-induced changes in polarized T84 epithelial cell monolayers by reducing bacterial adhesion and cytoskeletal rearrangements. Infect. Immun. 2005, 73, 5183–5188. [Google Scholar] [CrossRef] [Green Version]
- Johnson-Henry, K.C.; Hagen, K.E.; Gordonpour, M.; Tompkins, T.A.; Sherman, P.M. Surface-layer protein extracts from Lactobacillus helveticus inhibit enterohaemorrhagic Escherichia coli O157:H7 adhesion to epithelial cells. Cell Microbiol. 2007, 9, 356–367. [Google Scholar] [CrossRef]
- MacPherson, C.W.; Shastri, P.; Mathieu, O.; Tompkins, T.A.; Burguière, P. Genome-wide immune modulation of TLR3-mediated inflammation in intestinal epithelial cells differs between single and multi-strain probiotic combination. PLoS ONE 2017, 12, e0169847. [Google Scholar] [CrossRef]
- Macpherson, C.; Audy, J.; Mathieu, O.; Tompkins, T.A. Multistrain probiotic modulation of intestinal epithelial cells’ im-mune response to a double-stranded RNA ligand, poly(i·c). Appl. Environ. Microbiol. 2014, 80, 1692–1700. [Google Scholar] [CrossRef] [Green Version]
- Arboleya, S.; Salazar, N.; Solís, G.; Fernández, N.; Hernández-Barranco, A.M.; Cuesta, I.; Gueimonde, M.; de los Reyes-Gavilán, C.G. Assessment of intestinal microbiota modulation ability of Bifidobacterium strains in in vitro fecal batch cultures from preterm neonates. Anaerobe 2013, 19, 9–16. [Google Scholar] [CrossRef]
- Nogacka, A.M.; de los Reyes-Gavilán, C.G.; Arboleya, S.; Ruas-Madiedo, P.; Martínez-Faedo, C.; Suarez, A.; He, F.; Harata, G.; Endo, A.; Salazar, N.; et al. In vitro selection of probiotics for microbiota modulation in normal-weight and severely obese individuals: Focus on gas production and interaction with intestinal epithelial cells. Front. Microbiol. 2021, 12, 630572. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Salazar, N.; Prieto, A.; Leal, J.A.; Mayo, B.; Bada-Gancedo, J.C.; de los Reyes-Gavilan, C.G.; Ruas-Madiedo, P. Production of exopolysaccharides by Lactobacillus and Bifidobacterium strains of human origin, and metabolic activity of the producing bac-teria in milk. J. Dairy Sci. 2009, 92, 4158–4168. [Google Scholar] [CrossRef] [Green Version]
- Amir, A.; McDonald, D.; Navas-Molina, J.A.; Kopylova, E.; Morton, J.T.; Xu, Z.Z.; Kightley, E.P.; Thompson, L.R.; Hyde, E.R.; Gonzalez, A.; et al. Deblur rapidly resolves single-nucleotide community sequence patterns. MSystems 2017, 2, e00191-16. [Google Scholar] [CrossRef] [Green Version]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing tax-onomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 17, 90. [Google Scholar] [CrossRef]
- Ríos-Covián, D.; Ruas-Madiedo, P.; Margolles, A.; Gueimonde, M.; de los Reyes-Gavilán, C.G.; Salazar, N. Intestinal short chain fatty acids and their link with diet and human health. Front. Microbiol. 2016, 7, 185. [Google Scholar] [CrossRef] [Green Version]
- Garrido, D.; Ruiz-Moyano, S.; Lemay, D.G.; Sela, D.A.; German, J.B.; Mills, D.A. Comparative transcriptomics reveals key differences in the response to milk oligosaccharides of infant gut-associated bifidobacteria. Sci. Rep. 2015, 5, 13517. [Google Scholar] [CrossRef]
- He, Z.; Yang, B.; Liu, X.; Ross, P.; Stanton, C.; Zhao, J.; Zhang, H.; Chen, W. Genotype-phenotype association analysis re-vealed different utilization ability of 2′-fucosyllactose in Bifidobacterium genus. J. Dairy Sci. 2021, 104, 1518–1523. [Google Scholar] [CrossRef]
- Turroni, F.; Milani, C.; Duranti, S.; Mancabelli, L.; Mangifesta, M.; Viappiani, A.; Lugli, G.A.; Ferrario, C.; Gioiosa, L.; Ferra-rini, A.; et al. Decipering bifidobacterial-mediated metabolic interactions and their impact on gut microbiota by a mul-ti-omics approach. ISME J. 2016, 10, 1656–1668. [Google Scholar] [CrossRef]
- Schwab, C.; Ruscheweyh, H.J.; Bunesova, V.; Pham, V.T.; Beerenwinkel, N.; Lacroix, C. Trophic interactions of infant bifidobacteria and Eubacterium hallii during L-fucose and fucosyllactose degradation. Front. Microbiol. 2017, 8, 95. [Google Scholar] [CrossRef]
- Centanni, M.; Ferguson, S.A.; Sims, I.M.; Biswas, A.; Tannock, G.W. Bifidobacterium bifidum ATCC 15696 and Bifidobacterium breve 24b metabolic interaction based on 2′-O-fucosyl- lactose studied in steady-state cultures in a Freter-style chemostat. Appl. Environ. Microbiol. 2019, 85, e02783-18. [Google Scholar] [CrossRef] [Green Version]
- Zúñiga, M.; Monedero, V.; Yebra, M.J. Utilization of host-derived glycans by intestinal Lactobacillus and Bifidobacterium spe-cies. Front. Microbiol. 2018, 9, 1917. [Google Scholar] [CrossRef] [Green Version]
- Mollet, B.; Pilloud, N. Galactose utilization in Lactobacillus helveticus: Isolation and characterization of the galactokinase (galK) and galactose-I-phosphate uridyl transferase (gaiT) genes. J. Bacteriol. 1991, 173, 4464–4473. [Google Scholar] [CrossRef] [Green Version]
- Grossiord, B.; Vaughan, E.E.; Luesink, E.; de Vos, W.M. Genetics of galactose utilization via the Leloir pathway in lactic acid bacteria. Lait 1998, 78, 77–84. [Google Scholar] [CrossRef] [Green Version]
- Pham, V.T.; Lacroix, C.; Braegger, C.P.; Chassard, C. Early colonization of functional groups of microbes in the infant gut. Environ. Microbiol. 2016, 18, 2246–2258. [Google Scholar] [CrossRef]
- Torino, M.I.; Taranto, M.P.; de Valdez, G.F. Citrate catabolism and production of acetate and succinate by Lactobacillus helvet-icus ATCC 15907. Appl. Microbiol. Biotechnol. 2005, 69, 79–85. [Google Scholar] [CrossRef]
- Tsukuda, N.; Yahagi, K.; Hara, T.; Watanabe, Y.; Matsumoto, H.; Mori, H.; Higashi, K.; Tsuji, H.; Matsumoto, S.; Kurokawa, K.; et al. Key bacterial taxa and metabolic pathways affecting gut short-chain fatty acid profiles in early life. ISME J. 2021, 15, 2574–2590. [Google Scholar] [CrossRef]
- Bittinger, K.; Zhao, C.; Li, Y.; Ford, E.; Friedman, E.S.; Ni, J.; Kulkarni, C.V.; Cai, J.; Tian, Y.; Liu, Q.; et al. Bacterial coloniza-tion reprograms the neonatal gut metabolome. Nat. Microbiol. 2020, 5, 838–847. [Google Scholar] [CrossRef]
- Morishita, T.; Yajima, M. Incomplete operation of biosynthetic and bioenergetic functions of the citric acid cycle in multiple auxotrophic lactobacilli. Biosci. Biotechnol. Biochem. 1995, 59, 251–255. [Google Scholar] [CrossRef] [Green Version]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef]
- Dogra, S.K.; Martin, F.P.; Donnicola, D.; Julita, M.; Berger, B.; Sprenger, N. Human milk oligosaccharide-stimulated Bifidobacterium species contribute to prevent later respiratory tract infections. Microorganisms 2021, 9, 1939. [Google Scholar] [CrossRef]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef]
- Kim, H.K.; Rutten, N.B.M.M.; van der Vaart, I.B.; Niers, L.E.M.; Choi, Y.H.; Rijkers, G.T.; van Hemert, S. Probiotic supple-mentation influences faecal short chain fatty acids in infants at high risk for eczema. Benef. Microbes 2015, 6, 783–790. [Google Scholar] [CrossRef]
- Kuugbee, E.D.; Shang, X.; Gamallat, Y.; Bamba, D.; Awadasseid, A.; Suliman, M.A.; Zang, S.; Ma, Y.; Chiwala, G.; Xin, Y.; et al. Structural change in microbiota by a probiotic cocktail enhances the gut barrier and reduces cancer via TLR2 signaling in a rat model of colon cancer. Dig. Dis. Sci. 2016, 61, 2908–2920. [Google Scholar] [CrossRef]
- Bajpai, P.; Darra, A.; Agrawal, A. Microbe-mitochondrion crosstalk and health: An emerging paradigm. Mitochondrion 2018, 39, 20–25. [Google Scholar] [CrossRef]
- Pham, V.T.; Chassard, C.; Rifa, E.; Braegger, C.; Geirnaert, A.; Rocha Martin, V.N.; Lacroix, C. Lactate metabolism is strongly modulated by fecal inoculum, pH, and retention time in PolyFermS continuous colonic fermentation models mimicking young infant proximal colon. MSystems 2019, 4, e00264-18. [Google Scholar] [CrossRef] [Green Version]
- Laursen, M.F.; Laursen, R.P.; Lamkjaer, A.; Michaelsen, K.F.; Bahl, M.I.; Licht, T.R. Administration of two probiotic strains during early childhood does not affect the endogenous gut microbiota composition despite probiotic proliferation. BMC Microbiol. 2017, 17, 175. [Google Scholar] [CrossRef] [Green Version]
| Group | Condition | Bifidobacteri- | Lactobacill- | Streptococc- | Lachnospir- | Ruminococc- | Erysipelotrich- |
|---|---|---|---|---|---|---|---|
| BF-Fast | Fast 2′FL | 23.77 ± 8.48 b | 13.08 ± 14.54 a | 0.18 ± 0.11 | 0.35 ± 0.25 | 0.07 ± 0.09 b | 0.01 ± 0.01 a |
| Slow 2′FL | 2.57 ± 4.23 a | 14.99 ± 25.63 a | 0.62 ± 0.83 | 2.88 ± 4.29 | 0.72 ± 0.72 c | 2.35 ± 1.35 b | |
| B.bifidum + 2′FL | 22.74 ± 10.83 b | 12.65 ± 13.81 a | 0.13 ± 0.04 | 0.44 ± 0.43 | 0.09 ± 0.08 b | 0.06 ± 0.07 a | |
| B.infantis + 2′FL | 19.56 ± 9.54 b | 15.43 ± 17.35 a | 0.10 ± 0.05 | 0.43 ± 0.34 | 0.12 ± 0.12 b | 0.13 ± 0.22 ab | |
| L.helveticus + 2′FL | 10.77 ± 2.76 a | 38.16 ± 1.3 b | 0.14 ± 0.05 | 0.14 ± 0.08 | 0.00 ± 0.01 a | 0.12 ± 0.28 a | |
| Combination + 2′FL | 11.81 ± 4.94 a | 37.39 ± 2.63 b | 0.10 ± 0.07 | 0.21 ± 0.16 | 0.07 ± 0.08 b | 0.10 ± 0.13 a | |
| BF-Slow | Fast 2′FL | 23.77 ± 8.48 b | 13.08 ± 14.54 a | 0.18 ± 0.11 | 0.35 ± 0.25 | 0.07 ± 0.09 a | 0.01 ± 0.01 a |
| Slow 2′FL | 2.57 ± 4.23 a | 14.99 ± 25.63 ab | 0.62 ± 0.83 | 2.88 ± 4.29 | 0.72 ± 0.72 b | 2.35 ± 1.35 b | |
| B.bifidum + 2′FL | 1.81 ± 1.30 ab | 0.61 ± 0.50 a | 2.45 ± 1.55 | 7.11 ± 10.35 | 2.38 ± 2.92 b | 3.63 ± 1.35 b | |
| B.infantis + 2′FL | 2.11 ± 0.43 ab | 0.18 ± 0.09 a | 0.18 ± 0.09 | 0.62 ± 0.34 | 0.18 ± 0.09 ab | 4.20 ± 1.19 b | |
| L.helveticus + 2′FL | 0.07 ± 0.02 a | 48.25 ± 5.83 b | 0.10 ± 0.09 | 0.16 ± 0.12 | 0.03 ± 0.02 a | 0.91 ± 0.24 ab | |
| Combination + 2′FL | 0.90 ± 1.21 a | 45.43 ± 5.95 b | 0.26 ± 0.3 | 0.48 ± 0.57 | 0.17 ± 0.18 ab | 1.04 ± 0.16 ab | |
| FF-Fast | Fast 2′FL | 7.78 ± 0.56 | 45.72 ± 5.14 b | 0.04 ± 0.01 a | 0.03 ± 0.03 a | 0.01 ± 0.01 | 0.18 ± 0.03 bc |
| Slow 2′FL | 8.02 ± 1.27 | 8.67 ± 9.09 a | 31.26 ± 2.43 b | 0.52 ± 0.20 b | 0.02 ± 0.02 | 0.03 ± 0.02 a | |
| B.bifidum + 2′FL | 8.50 ± 1.03 | 44.86 ± 0.83 b | 0.03 ± 0.03 a | 0.03 ± 0.02 a | 0.02 ± 0.01 | 0.22 ± 0.02 c | |
| B.infantis + 2′FL | 8.94 ± 1.32 | 38.67 ± 4.67 ab | 0.10 ± 0.09 a | 0.89 ± 1.46 ab | 0.22 ± 0.38 | 0.19 ± 0.05 bc | |
| L.helveticus + 2′FL | 6.18 ± 0.89 | 42.19 ± 2.15 b | 0.11 ± 0.05 ab | 0.06 ± 0.03 ab | 0.02 ± 0.03 | 0.15 ± 00 abc | |
| Combination + 2′FL | 5.95 ± 2.3 | 43.70 ± 4.81 ab | 0.12 ± 0.03 ab | 0.04 ± 0.04 a | 0.01 ± 0.01 | 0.09 ± 0.03 ab | |
| FF-Slow | Fast 2′FL | 7.78 ± 0.56 b | 45.72 ± 5.14 b | 0.04 ± 0.01 a | 0.03 ± 0.03 a | 0.01 ± 0.01 | 0.18 ± 0.03 |
| Slow 2′FL | 8.02 ± 1.27 b | 8.67 ± 9.09 a | 31.26 ± 2.4 b | 0.52 ± 0.20 b | 0.02 ± 0.02 | 0.03 ± 0.02 | |
| B.bifidum + 2′FL | 8.03 ± 1.82 b | 6.78 ± 6.86 a | 31.89 ± 3.06 b | 0.68 ± 0.3 c | 0.04 ± 0.05 | 0.02 ± 0.03 | |
| B.infantis + 2′FL | 8.17 ± 1.09 b | 7.31 ± 7.82 a | 32.09 ± 2.35 b | 0.54 ± 0.23 b | 0.02 ± 0.02 | 0.02 ± 0.04 | |
| L.helveticus + 2′FL | 2.86 ± 0.97 a | 49.58 ± 7.69 b | 12.76 ± 5.73 a | 0.42 ± 0.42 abc | 0.01 ± 0.02 | 0.02 ± 0.04 | |
| Combination + 2′FL | 2.13 ± 0.69 a | 52.34 ± 4.58 b | 9.87 ± 5.88 a | 0.27 ± 0.17 b | 0.01 ± 0.01 | 0.01 ± 0.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nogacka, A.M.; Arboleya, S.; Nikpoor, N.; Auger, J.; Salazar, N.; Cuesta, I.; Alvarez-Buylla, J.R.; Mantecón, L.; Solís, G.; Gueimonde, M.; et al. In Vitro Probiotic Modulation of the Intestinal Microbiota and 2′Fucosyllactose Consumption in Fecal Cultures from Infants at Two Months of Age. Microorganisms 2022, 10, 318. https://doi.org/10.3390/microorganisms10020318
Nogacka AM, Arboleya S, Nikpoor N, Auger J, Salazar N, Cuesta I, Alvarez-Buylla JR, Mantecón L, Solís G, Gueimonde M, et al. In Vitro Probiotic Modulation of the Intestinal Microbiota and 2′Fucosyllactose Consumption in Fecal Cultures from Infants at Two Months of Age. Microorganisms. 2022; 10(2):318. https://doi.org/10.3390/microorganisms10020318
Chicago/Turabian StyleNogacka, Alicja M., Silvia Arboleya, Naghmeh Nikpoor, Jeremie Auger, Nuria Salazar, Isabel Cuesta, Jorge R. Alvarez-Buylla, Laura Mantecón, Gonzalo Solís, Miguel Gueimonde, and et al. 2022. "In Vitro Probiotic Modulation of the Intestinal Microbiota and 2′Fucosyllactose Consumption in Fecal Cultures from Infants at Two Months of Age" Microorganisms 10, no. 2: 318. https://doi.org/10.3390/microorganisms10020318