Culture-Free Enumeration of Mycobacterium tuberculosis in Mouse Tissues Using the Molecular Bacterial Load Assay for Preclinical Drug Development
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Preparation of the Internal Control (IC)
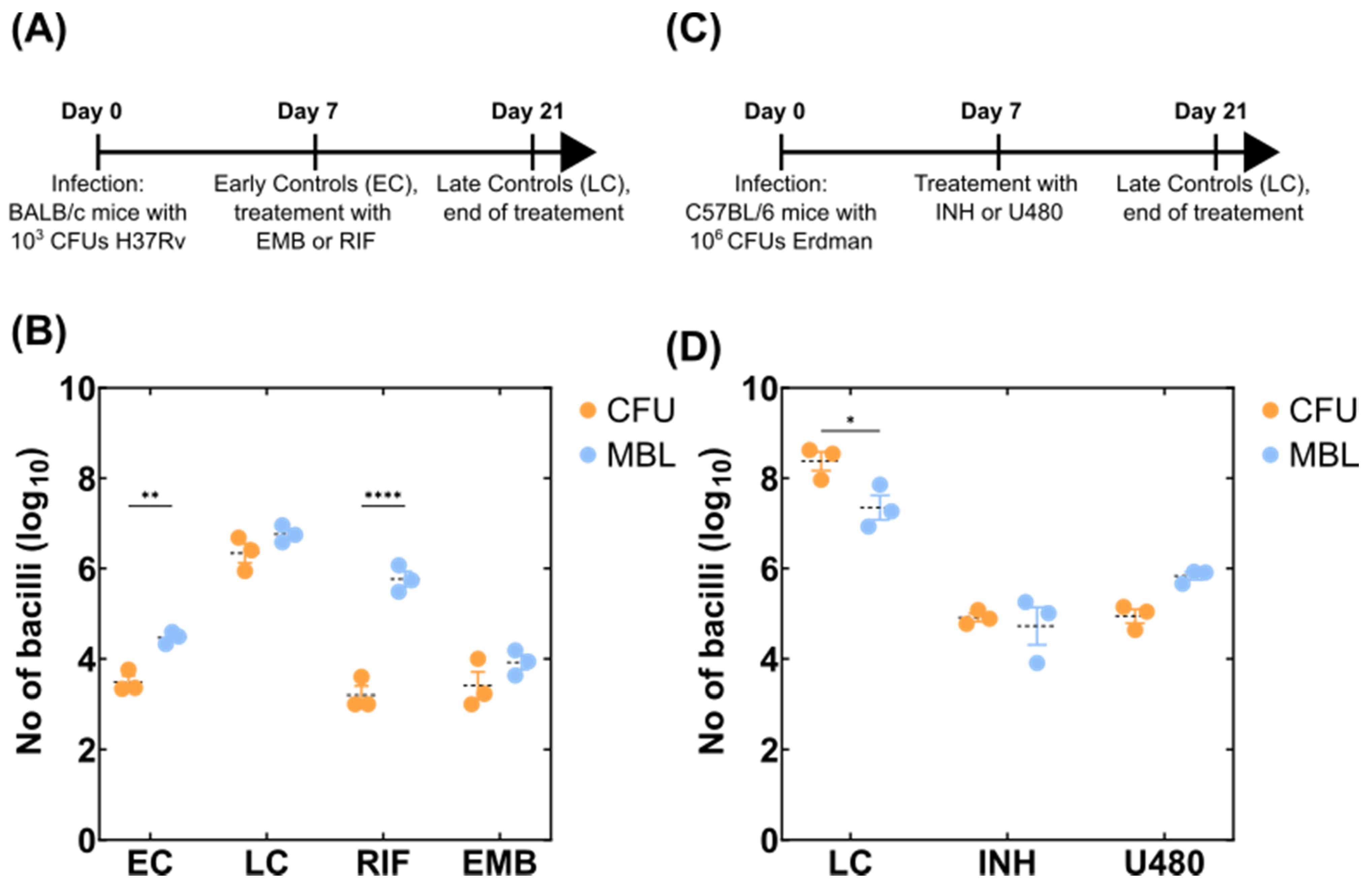
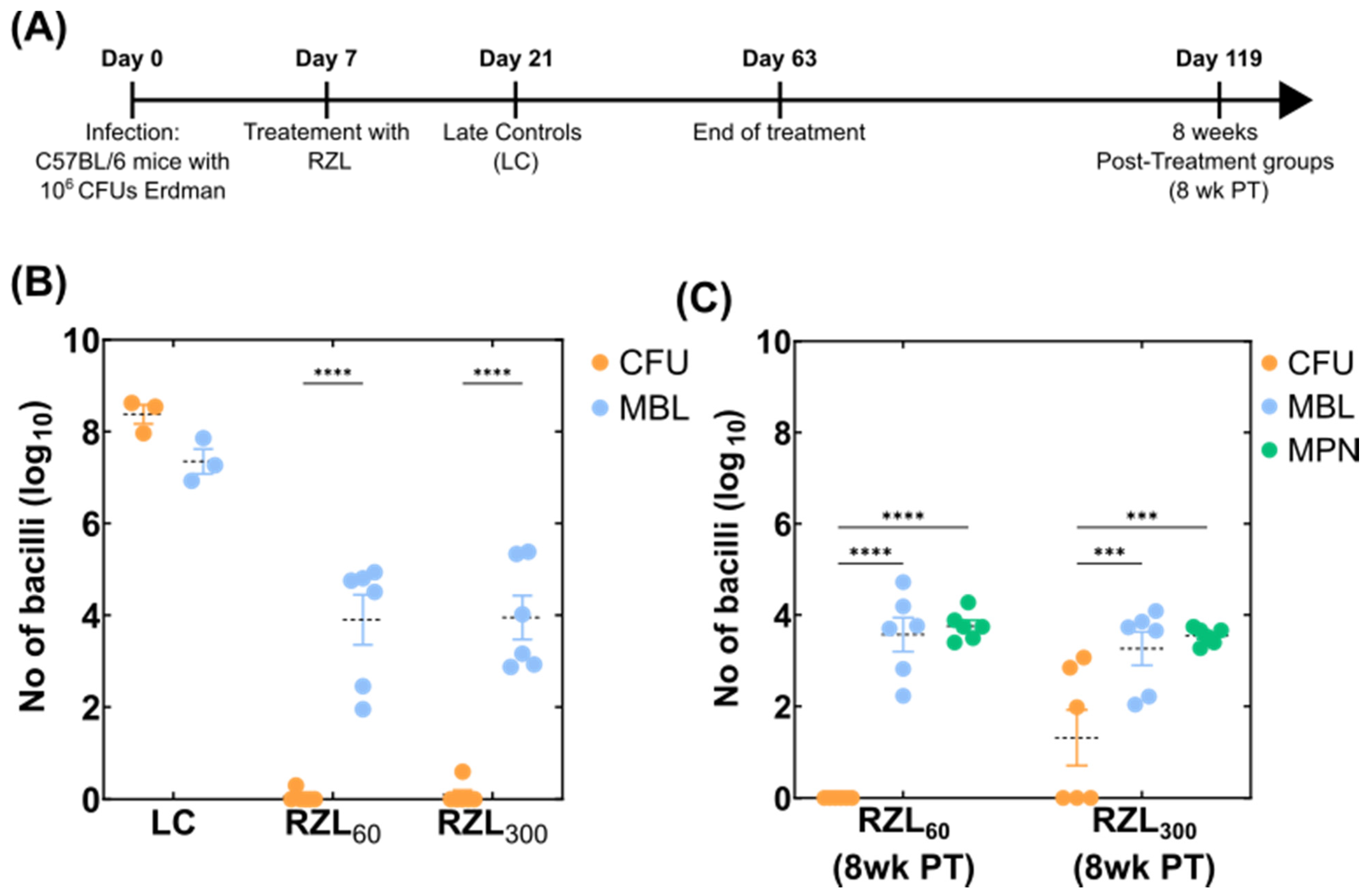
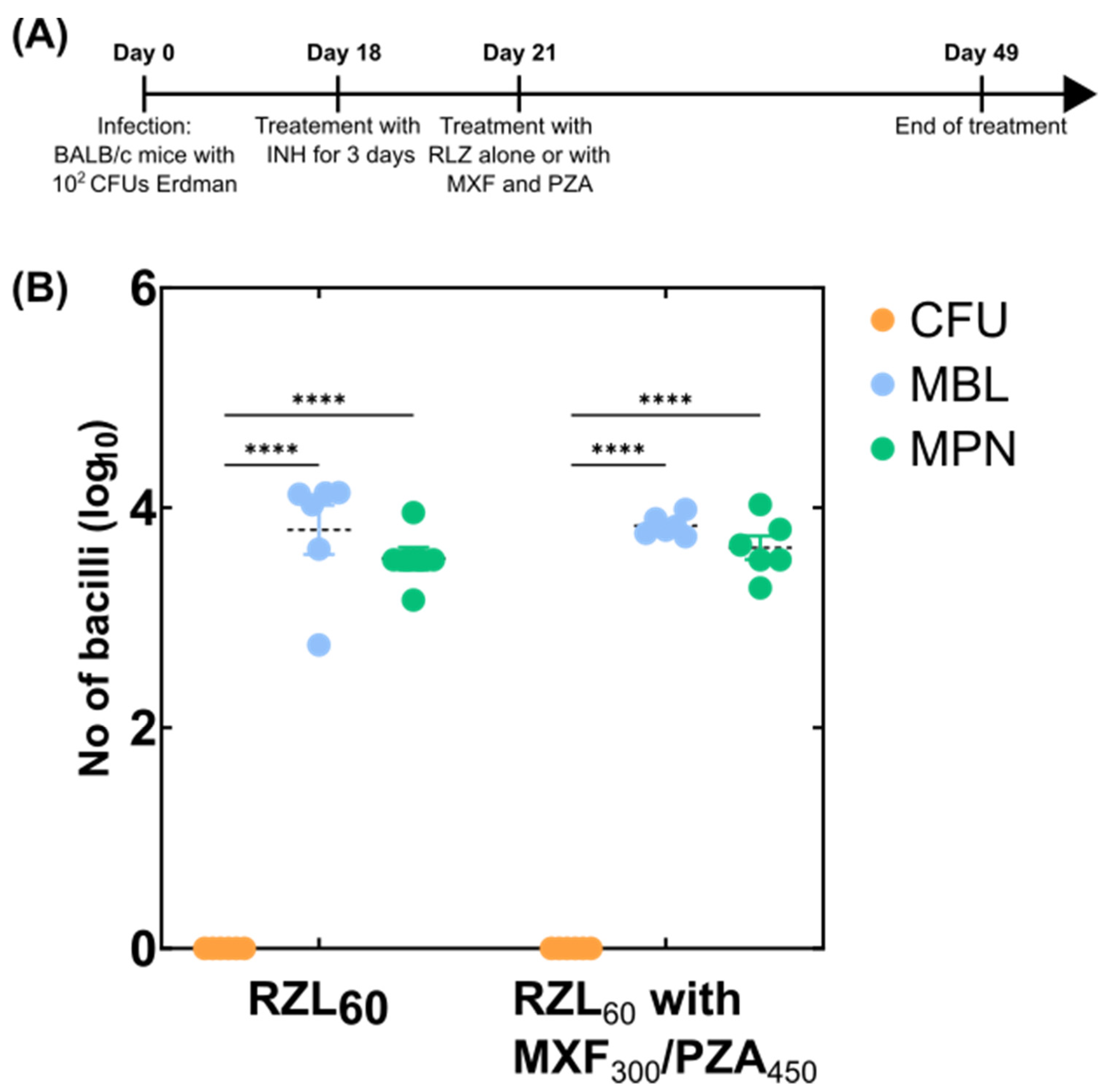
2.3. Mouse Experiments
2.4. Most Probable Number (MPN Assay)
2.5. MBL Assay
2.6. Statistical Analysis
3. Results
3.1. Adaptation of the MBL Assay to Tissue Homogenates
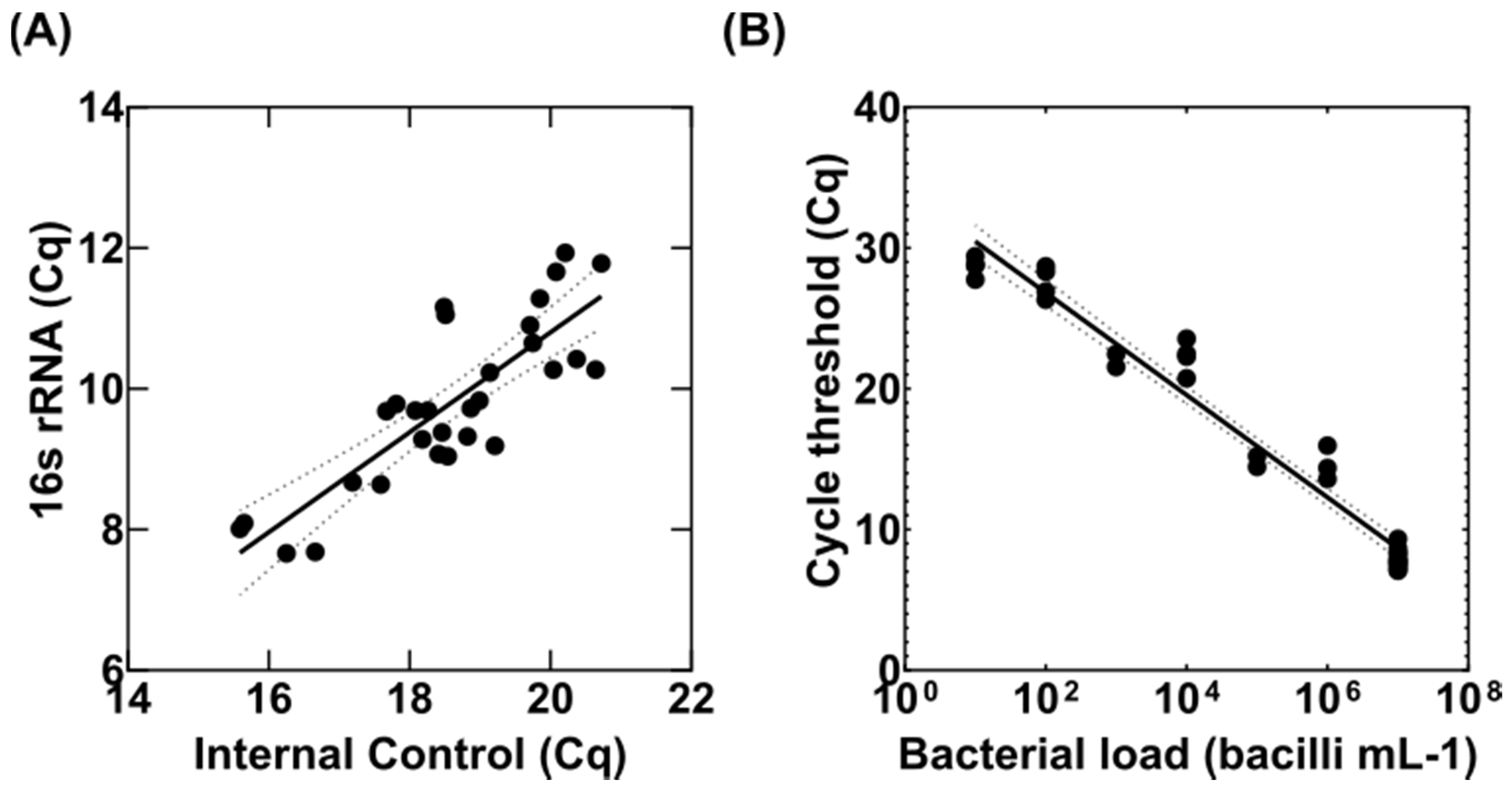
3.2. Detection of Variable Populations Using the MBL Assay
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO Global Tuberculosis Programme. Global Tuberculosis Report. 2021. Available online: https://www.who.int/publications/i/item/9789240037021 (accessed on 29 December 2021).
- Torres, N.M.C.; Rodríguez, J.J.Q.; Andrade, P.S.P.; Arriaga, M.B.; Netto, E.M. Factors predictive of the success of tuberculosis treatment: A systematic review with meta-analysis. PLoS ONE 2019, 14, e0226507. [Google Scholar] [CrossRef] [Green Version]
- Tiberi, S.; Vjecha, M.J.; Zumla Adam Galvin, J.; Migliori, G.B.; Zumla, A. Accelerating development of new shorter TB treatment regimens in anticipation of a resurgence of multi-drug resistant TB due to the COVID-19 pandemic. Int. J. Infect. Dis. 2021, 113, S96–S99. [Google Scholar] [CrossRef] [PubMed]
- Dorman, S.E.; Nahid, P.; Kurbatova, E.V.; Phillips, P.P.J.; Bryant, K.; Dooley, K.E.; Engle, M.; Goldberg, S.V.; Phan, H.T.T.; Hakim, J.; et al. Four-Month Rifapentine Regimens with or without Moxifloxacin for Tuberculosis. N. Engl. J. Med. 2021, 384, 1705–1718. [Google Scholar] [CrossRef] [PubMed]
- Kloprogge, F.; Hammond, R.; Kipper, K.; Gillespie, S.H.; Della Pasqua, O. Mimicking in-vivo exposures to drug combinations in-vitro: Anti-tuberculosis drugs in lung lesions and the hollow fiber model of infection. Sci. Rep. 2019, 9, 13228. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.K.; Gupta, U.D. Animal models of tuberculosis: Lesson learnt. Indian J. Med. Res. 2018, 147, 456–463. [Google Scholar] [CrossRef]
- Bowness, R.; Boeree, M.J.; Aarnoutse, R.; Dawson, R.; Diacon, A.; Mangu, C.; Heinrich, N.; Ntinginya, N.E.; Kohlenberg, A.; Mtafya, B.; et al. The relationship between Mycobacterium tuberculosis MGIT time to positivity and cfu in sputum samples demonstrates changing bacterial phenotypes potentially reflecting the impact of chemotherapy on critical sub-populations. J. Antimicrob. Chemother. 2015, 70, 448–455. [Google Scholar] [CrossRef]
- de Knegt, G.J.; Dickinson, L.; Pertinez, H.; Evangelopoulos, D.; McHugh, T.D.; Bakker-Woudenberg, I.A.J.M.; Davies, G.R.; de Steenwinkel, J.E.M. Assessment of treatment response by colony forming units, time to culture positivity and the molecular bacterial load assay compared in a mouse tuberculosis model. Tuberc. Edinb. Scotl. 2017, 105, 113–118. [Google Scholar] [CrossRef]
- Mukamolova, G.V.; Turapov, O.; Malkin, J.; Woltmann, G.; Barer, M.R. Resuscitation-promoting Factors Reveal an Occult Population of Tubercle Bacilli in Sputum. Am. J. Respir. Crit. Care Med. 2010, 181, 174–180. [Google Scholar] [CrossRef] [Green Version]
- Chengalroyen, M.D.; Beukes, G.M.; Gordhan, B.G.; Streicher, E.M.; Churchyard, G.; Hafner, R.; Warren, R.; Otwombe, K.; Martinson, N.; Kana, B.D. Detection and quantification of differentially culturable tubercle bacteria in sputum from patients with tuberculosis. Am. J. Respir. Crit. Care Med. 2016, 194, 1532–1540. [Google Scholar] [CrossRef] [Green Version]
- Turapov, O.; Glenn, S.; Kana, B.; Makarov, V.; Andrew, P.W.; Mukamolova, G.V. The In Vivo Environment Accelerates Generation of Resuscitation-Promoting Factor–Dependent Mycobacteria. Am. J. Respir. Crit. Care Med. 2014, 190, 1455–1457. [Google Scholar] [CrossRef] [Green Version]
- McAulay, K.; Saito, K.; Warrier, T.; Walsh, K.F.; Mathurin, L.D.; Royal-Mardi, G.; Lee, M.H.; Ocheretina, O.; Pape, J.W.; Fitzgerald, D.W.; et al. Differentially detectable Mycobacterium tuberculosis cells in sputum from treatment-naive subjects in Haiti and their proportionate increase after initiation of treatment. mBio 2018, 9, e02192-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Pertinez, H.; Ortega-Muro, F.; Alameda-Martin, L.; Liu, Y.; Schipani, A.; Davies, G.; Coates, A. Investigation of Elimination Rate, Persistent Subpopulation Removal, and Relapse Rates of Mycobacterium tuberculosis by Using Combinations of First-Line Drugs in a Modified Cornell Mouse Model. Antimicrob. Agents Chemother. 2016, 60, 4778–4785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boehme, C.C.; Nabeta, P.; Hillemann, D.; Nicol, M.P.; Shenai, S.; Krapp, F.; Allen, J.; Tahirli, R.; Blakemore, R.; Rustomjee, R.; et al. Rapid Molecular Detection of Tuberculosis and Rifampin Resistance. N. Engl. J. Med. 2010, 363, 1005–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedrich, S.O.; Rachow, A.; Saathoff, E.; Singh, K.; Mangu, C.D.; Dawson, R.; Phillips, P.P.; Venter, A.; Bateson, A.; Boehme, C.C.; et al. Assessment of the sensitivity and specificity of Xpert MTB/RIF assay as an early sputum biomarker of response to tuberculosis treatment. Lancet Respir. Med. 2013, 1, 462–470. [Google Scholar] [CrossRef] [Green Version]
- Kayigire, X.A.; Friedrich, S.O.; Venter, A.; Dawson, R.; Gillespie, S.H.; Boeree, M.J.; Heinrich, N.; Hoelscher, M.; Diacon, A.H. Direct Comparison of Xpert MTB/RIF Assay with Liquid and Solid Mycobacterial Culture for Quantification of Early Bactericidal Activity. J. Clin. Microbiol. 2013, 51, 1894–1898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devonshire, A.S.; O’Sullivan, D.M.; Honeyborne, I.; Jones, G.; Karczmarczyk, M.; Pavšič, J.; Gutteridge, A.; Milavec, M.; Mendoza, P.; Schimmel, H.; et al. The use of digital PCR to improve the application of quantitative molecular diagnostic methods for tuberculosis. BMC Infect. Dis. 2016, 16, 366. [Google Scholar] [CrossRef] [PubMed]
- Honeyborne, I.; McHugh, T.D.; Phillips, P.P.J.; Bannoo, S.; Bateson, A.; Carroll, N.; Perrin, F.M.; Ronacher, K.; Wright, L.; van Helden, P.D.; et al. Molecular bacterial load assay, a culture-free biomarker for rapid and accurate quantification of sputum Mycobacterium tuberculosis bacillary load during treatment. J. Clin. Microbiol. 2011, 49, 3905–3911. [Google Scholar] [CrossRef] [Green Version]
- Honeyborne, I.; Mtafya, B.; Phillips, P.P.J.; Hoelscher, M.; Ntinginya, E.N.; Kohlenberg, A.; Rachow, A.; Rojas-Ponce, G.; McHugh, T.D.; Heinrich, N. Pan African Consortium for the Evaluation of Anti-tuberculosis Antibiotics. The molecular bacterial load assay replaces solid culture for measuring early bactericidal response to antituberculosis treatment. J. Clin. Microbiol. 2014, 52, 3064–3067. [Google Scholar] [CrossRef] [Green Version]
- Evangelopoulos, D.; Whittaker, E.; Honeyborne, I.; McHugh, T.D.; Klein, N.; Shingadia, D. Pediatric tuberculosis-human immunodeficiency virus co-infection in the United Kingdom highlights the need for better therapy monitoring tools: A case report. J. Med. Case Rep. 2017, 11, 52. [Google Scholar] [CrossRef] [Green Version]
- Sabiiti, W.; Azam, K.; Farmer, E.C.W.; Kuchaka, D.; Mtafya, B.; Bowness, R.; Oravcova, K.; Honeyborne, I.; Evangelopoulos, D.; McHugh, T.D.; et al. Tuberculosis bacillary load, an early marker of disease severity: The utility of tuberculosis Molecular Bacterial Load Assay. Thorax 2020, 75, 606–608. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Bacteriological analytical manual, appendix 2: Most probable number from serial dilutions. Available online: https://www.fda.gov/food/laboratory-methods-food/bam-appendix-2-most-probable-number-serial-dilutions (accessed on 29 December 2021).
- Wallis, R.S.; Dawson, R.; Friedrich, S.O.; Venter, A.; Paige, D.; Zhu, T.; Silvia, A.; Gobey, J.; Ellery, C.; Zhang, Y.; et al. Mycobactericidal activity of sutezolid (PNU-100480) in sputum (EBA) and blood (WBA) of patients with pulmonary tuberculosis. PLoS ONE 2014, 9, e94462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mor, N.; Simon, B.; Heifets, L. Bacteriostatic and bactericidal activities of benzoxazinorifamycin KRM-1648 against Mycobacterium tuberculosis and Mycobacterium avium in human macrophages. Antimicrob. Agents Chemother. 1996, 40, 1482–1485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothstein, D.M.; Shalish, C.; Murphy, C.K.; Sternlicht, A.; Campbell, L.A. Development potential of rifalazil and other benzoxazinorifamycins. Expert. Opin. Investig. Drugs 2006, 15, 603–623. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, S.H.; Crook, A.M.; McHugh, T.D.; Mendel, C.M.; Meredith, S.K.; Murray, S.R.; Pappas, F.; Phillips, P.P.J.; Nunn, A.J. Four-Month Moxifloxacin-Based Regimens for Drug-Sensitive Tuberculosis. 2014, 371, 1577–1587. [CrossRef] [Green Version]
- Jawahar, M.S.; Banurekha, V.V.; Paramasivan, C.N.; Rahman, F.; Ramachandran, R.; Venkatesan, P.; Balasubramanian, R.; Selvakumar, N.; Ponnuraja, C.; Iliayas, A.S.; et al. Randomized Clinical Trial of Thrice-Weekly 4-Month Moxifloxacin or Gatifloxacin Containing Regimens in the Treatment of New Sputum Positive Pulmonary Tuberculosis Patients. PLoS ONE 2013, 8, e67030. [Google Scholar] [CrossRef]
- Merle, C.S.; Fielding, K.; Sow, O.B.; Gninafon, M.; Lo, M.B.; Mthiyane, T.; Odhiambo, J.; Amukoye, E.; Bah, B.; Kassa, F.; et al. A Four-Month Gatifloxacin-Containing Regimen for Treating Tuberculosis. New Engl. J. Med. 2014, 371, 1588–1598. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Liu, A.; Ortega-Muro, F.; Alameda-Martin, L.; Mitchison, D.; Coates, A. High-dose rifampicin kills persisters, shortens treatment duration, and reduces relapse rate in vitro and in vivo. Front. Microbiol. 2015, 6, 641. [Google Scholar] [CrossRef]
- Liu, Y.; Pertinez, H.; Ortega-Muro, F.; Alameda-Martin, L.; Harrison, T.; Davies, G.; Coates, A.; Hu, Y. Optimal doses of rifampicin in the standard drug regimen to shorten tuberculosis treatment duration and reduce relapse by eradicating persistent bacteria. J. Antimicrob. Chemother. 2018, 73, 724–731. [Google Scholar] [CrossRef]
- Walter, N.D.; Born, S.E.M.; Robertson, G.T.; Reichlen, M.; Dide-Agossou, C.; Ektnitphong, V.A.; Rossmassler, K.; Ramey, M.E.; Bauman, A.A.; Ozols, V.; et al. Mycobacterium tuberculosis precursor rRNA as a measure of treatment-shortening activity of drugs and regimens. Nat. Commun. 2021, 12, 2899. [Google Scholar] [CrossRef]
- Loraine, J.; Pu, F.; Turapov, O.; Mukamolova, G.V. Development of an in vitro assay for detection of drug-Induced Resuscitation-promoting-factor-dependent mycobacteria. Antimicrob. Agents Chemother. 2016, 60, 6227–6233. [Google Scholar] [CrossRef] [Green Version]
- Saito, K.; Warrier, T.; Somersan-Karakaya, S.; Kaminski, L.; Mi, J.; Jiang, X.; Park, S.; Shigyo, K.; Gold, B.; Roberts, J.; et al. Rifamycin action on RNA polymerase in antibiotic-tolerant Mycobacterium tuberculosis results in differentially detectable populations. Proc. Natl. Acad. Sci. USA 2017, 114, E4832–E4840. [Google Scholar] [CrossRef] [Green Version]
- Dusthackeer, A.; Balasubramanian, M.; Shanmugam, G.; Priya, S.; Nirmal, C.R.; Sam Ebenezer, R.; Balasubramanian, A.; Mondal, R.K.; Thiruvenkadam, K.; Hemanth Kumar, A.K.; et al. Differential Culturability of Mycobacterium tuberculosis in Culture-Negative Sputum of Patients With Pulmonary Tuberculosis and in a Simulated Model of Dormancy. Front. Microbiol. 2019, 10, 2381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordhan, B.G.; Peters, J.S.; McIvor, A.; Machowski, E.E.; Ealand, C.; Waja, Z.; Martinson, N.; Kana, B.D. Detection of differentially culturable tubercle bacteria in sputum using mycobacterial culture filtrates. Sci. Rep. 2021, 11, 6493. [Google Scholar] [CrossRef] [PubMed]
- Turapov, O.; O’Connor, B.D.; Sarybaeva, A.A.; Williams, C.; Patel, H.; Kadyrov, A.S.; Sarybaev, A.S.; Woltmann, G.; Barer, M.R.; Mukamolova, G.V. Phenotypically Adapted Mycobacterium tuberculosis Populations from Sputum Are Tolerant to First-Line Drugs. Antimicrob. Agents Chemother. 2016, 60, 2476–2483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Bacilli mL−1 Tissue | ||||||||
|---|---|---|---|---|---|---|---|---|
| 108 | 107 | 106 | 105 | 104 | 103 | 102 | <102 | |
| Mean (Cq) | 6.0 | 8.0 | 14.6 | 17.3 | 22.3 | 24.9 | 27.6 | 28.7 |
| ± SD | 0.9 | 0.6 | 1.0 | 0.9 | 1.2 | 0.9 | 1.1 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Evangelopoulos, D.; Shoen, C.M.; Honeyborne, I.; Clark, S.; Williams, A.; Mukamolova, G.V.; Cynamon, M.H.; McHugh, T.D. Culture-Free Enumeration of Mycobacterium tuberculosis in Mouse Tissues Using the Molecular Bacterial Load Assay for Preclinical Drug Development. Microorganisms 2022, 10, 460. https://doi.org/10.3390/microorganisms10020460
Evangelopoulos D, Shoen CM, Honeyborne I, Clark S, Williams A, Mukamolova GV, Cynamon MH, McHugh TD. Culture-Free Enumeration of Mycobacterium tuberculosis in Mouse Tissues Using the Molecular Bacterial Load Assay for Preclinical Drug Development. Microorganisms. 2022; 10(2):460. https://doi.org/10.3390/microorganisms10020460
Chicago/Turabian StyleEvangelopoulos, Dimitrios, Carolyn M. Shoen, Isobella Honeyborne, Simon Clark, Ann Williams, Galina V. Mukamolova, Michael H. Cynamon, and Timothy D. McHugh. 2022. "Culture-Free Enumeration of Mycobacterium tuberculosis in Mouse Tissues Using the Molecular Bacterial Load Assay for Preclinical Drug Development" Microorganisms 10, no. 2: 460. https://doi.org/10.3390/microorganisms10020460
APA StyleEvangelopoulos, D., Shoen, C. M., Honeyborne, I., Clark, S., Williams, A., Mukamolova, G. V., Cynamon, M. H., & McHugh, T. D. (2022). Culture-Free Enumeration of Mycobacterium tuberculosis in Mouse Tissues Using the Molecular Bacterial Load Assay for Preclinical Drug Development. Microorganisms, 10(2), 460. https://doi.org/10.3390/microorganisms10020460






