Pre-Clinical Tools for Predicting Drug Efficacy in Treatment of Tuberculosis
Abstract
:1. Introduction
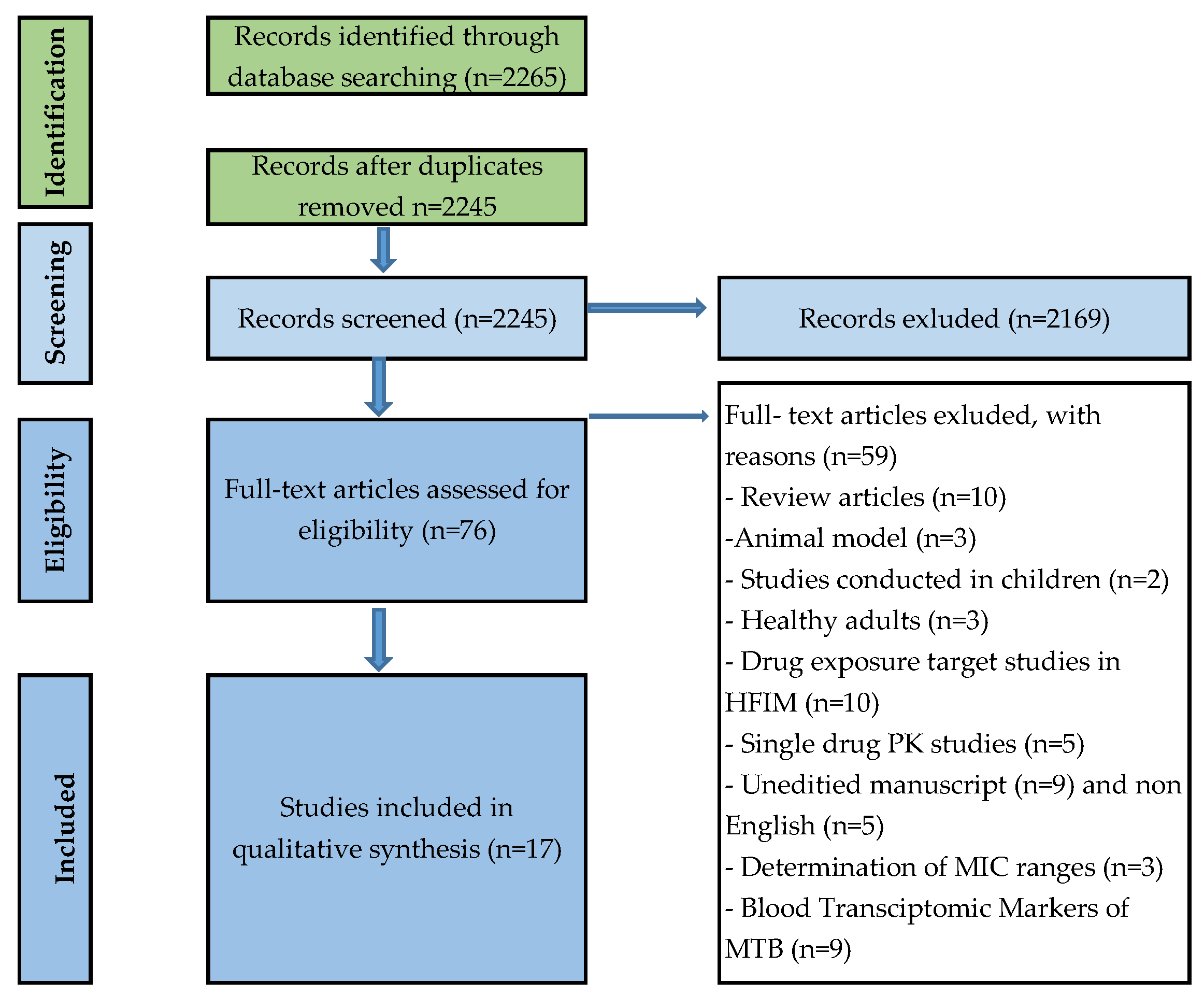
2. Methods
Study Selection and Search Strategy
3. Results
3.1. In Vitro Microbiological Based Assays Using In Vitro Checkerboard Models
3.2. In Vitro Time-Kill Kinetic Assay
3.3. In Vitro Models: Use of the Hollow Fibre Infection Model
3.4. Theoretical/Mathematical Models Used to Identify Potential Regimens
3.5. High-Throughput Combinatorial Screening
4. Discussion
5. Conclusions
- Studies including three or more drug combinations should test the drug concentration range in separate and combined assays.
- Testing should be performed on bacteria in different metabolic states.
- The use of in vitro methods such as the checkerboard assay is a useful first step; however, a standardised method of interpretation must be validated in all laboratories involved in the studies.
- Drug concentrations used should be pharmacologically relevant.
- Standardised approaches are needed in evaluating all drug combinations in an in vitro HFIM, where drug exposures and human pharmacokinetic profiles of the drug in the target site are simulated to evaluate the impact of these combinations for cell killing and the suppression of resistance [41].
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- WHO. Global Tuberculosis Report; WHO: Geneve, Switzerland, 2021. [Google Scholar]
- WHO. Global Tuberculosis Report; WHO: Geneve, Switzerland, 2019. [Google Scholar]
- WHO. Module 4: Treatment—Drug-Resistant Tuberculosis Treatment; WHO: Geneve, Switzerland, 2020. [Google Scholar]
- WHO. Consolidated Guidelines on Tuberculosis: Module 4: Treatment: Drug-Resistant Tuberculosis Treatment; World Health Organization: Geneve, Switzerland, 2020. [Google Scholar]
- Shetye, G.S.; Franzblau, S.G.; Cho, S. New tuberculosis drug targets, their inhibitors, and potential therapeutic impact. Transl. Res. 2020, 220, 68–97. [Google Scholar] [CrossRef]
- Zumla, A.; Nahid, P.; Cole, S.T. Advances in the development of new tuberculosis drugs and treatment regimens. Nat. Rev. Drug Discov. 2013, 12, 388–404. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.R.; Dalcolmo, M.; Tiberi, S.; Arbex, M.A.; Munoz-Torrico, M.; Duarte, R.; D’Ambrosio, L.; Visca, D.; Rendon, A.; Gaga, M.; et al. New and repurposed drugs to treat multidrug- and extensively drug-resistant tuberculosis. J. Bras. Pneumol. 2018, 44, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Conradie, F.; Diacon, A.H.; Ngubane, N.; Howell, P.; Everitt, D.; Crook, A.M.; Mendel, C.M.; Egizi, E.; Moreira, J.; Timm, J. Bedaquiline, pretomanid and linezolid for treatment of extensively drug resistant, intolerant or non-responsive multidrug resistant pulmonary tuberculosis. N. Engl. J. Med. 2020, 382, 893–902. [Google Scholar] [CrossRef] [PubMed]
- WHO. Global Tuberculosis Report 2018; World Health Organization: Geneve, Switzerland, 2018. [Google Scholar]
- Nyang’wa, B.; Motta, I.; Kazounis, E.; Berry, C. Early termination of randomisation into TB-PRACTECAL, a novel six months all-oral regimen Drug Resistant TB study. J. Int. AIDS Soc. 2021, 24, 70–71. [Google Scholar]
- Nunn, A.J.; Phillips, P.P.J.; Meredith, S.K.; Chiang, C.Y.; Conradie, F.; Dalai, D.; van Deun, A.; Dat, P.T.; Lan, N.; Master, I.; et al. A Trial of a Shorter Regimen for Rifampin-Resistant Tuberculosis. N. Engl. J. Med. 2019, 380, 1201–1213. [Google Scholar] [CrossRef] [PubMed]
- Sotgiu, G.; Centis, R.; D’Ambrosio, L.; Alffenaar, J.W.; Anger, H.A.; Caminero, J.A.; Castiglia, P.; De Lorenzo, S.; Ferrara, G.; Koh, W.J.; et al. Efficacy, safety and tolerability of linezolid containing regimens in treating MDR-TB and XDR-TB: Systematic review and meta-analysis. Eur. Respir. J. 2012, 40, 1430–1442. [Google Scholar] [CrossRef] [Green Version]
- Berry, C.; Yates, T.A.; Seddon, J.A.; Phillips, P.P.; du Cros, P.; North London, T.B.J.C. Efficacy, safety and tolerability of linezolid for the treatment of XDR-TB: A study in China. Eur. Respir. J. 2016, 47, 1591–1592. [Google Scholar] [CrossRef] [Green Version]
- D’Ambrosio, L.; Tadolini, M.; Centis, R.; Duarte, R.; Sotgiu, G.; Aliberti, S.; Dara, M.; Migliori, G.B. Supporting clinical management of the difficult-to-treat TB cases: The ERS-WHO TB Consilium. Int. J. Infect. Dis. 2015, 32, 156–160. [Google Scholar] [CrossRef] [Green Version]
- Lienhardt, C.; Weyer, K.; Falzon, D. The Use of Bedaquiline in the Treatment of Multi-Drug Resistant Tuberculosis: Interim Policy Guidance; World Health Organization: Geneve, Switzerland, 2013. [Google Scholar]
- Deb, U.; Biswas, S. Pretomanid: The latest USFDA-approved anti-tuberculosis drug. Indian J. Tuberc. 2021, 68, 287–291. [Google Scholar] [CrossRef]
- Yepes-Nunez, J.J.; Urrutia, G.; Romero-Garcia, M.; Alonso-Fernandez, S. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Rev. Esp. Cardiol. 2021, 74, 790–799. [Google Scholar] [CrossRef]
- Maltempe, F.G.; Caleffi-Ferracioli, K.R.; do Amaral, R.C.R.; de Oliveira Demitto, F.; Siqueira, V.L.D.; de Lima Scodro, R.B.; Hirata, M.H.; Pavan, F.R.; Cardoso, R.F. Activity of rifampicin and linezolid combination in Mycobacterium tuberculosis. Tuberculosis 2017, 104, 24–29. [Google Scholar] [CrossRef] [Green Version]
- Drusano, G.L.; Neely, M.; Van Guilder, M.; Schumitzky, A.; Brown, D.; Fikes, S.; Peloquin, C.; Louie, A. Analysis of combination drug therapy to develop regimens with shortened duration of treatment for tuberculosis. PLoS ONE 2014, 9, e101311. [Google Scholar] [CrossRef] [PubMed]
- Caleffi-Ferracioli, K.R.; Maltempe, F.G.; Siqueira, V.L.; Cardoso, R.F. Fast detection of drug interaction in Mycobacterium tuberculosis by a checkerboard resazurin method. Tuberculosis 2013, 93, 660–663. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Gavin, A.; Tudo, G.; Vergara, A.; Hurtado, J.C.; Gonzalez-Martin, J. In vitro activity against Mycobacterium tuberculosis of levofloxacin, moxifloxacin and UB-8902 in combination with clofazimine and pretomanid. Int. J. Antimicrob. Agents 2015, 46, 582–585. [Google Scholar] [CrossRef]
- De Miranda Silva, C.; Hajihosseini, A.; Myrick, J.; Nole, J.; Louie, A.; Schmidt, S.; Drusano, G.L. Effect of Moxifloxacin plus Pretomanid against Mycobacterium tuberculosis in Log Phase, Acid Phase, and Nonreplicating-Persister Phase in an In Vitro Assay. Antimicrob. Agents Chemother. 2019, 63, e01695-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Miranda Silva, C.; Hajihosseini, A.; Myrick, J.; Nole, J.; Louie, A.; Schmidt, S.; Drusano, G.L. Effect of Linezolid plus Bedaquiline against Mycobacterium tuberculosis in Log Phase, Acid Phase, and Nonreplicating-Persister Phase in an In Vitro Assay. Antimicrob. Agents Chemother. 2018, 62, e00856-18. [Google Scholar] [CrossRef] [Green Version]
- Pang, Y.; Jing, W.; Lu, J.; Zong, Z.; Huo, F.; Dong, L.; Dai, G.; Li, Y.; Huang, H.; Chu, N. No in vitro synergistic effect of bedaquiline combined with fluoroquinolones, linezolid, and clofazimine against extensively drug-resistant tuberculosis. Diagn. Microbiol. Infect. Dis. 2019, 94, 361–364. [Google Scholar] [CrossRef]
- Santos, N.C.S.; Scodro, R.B.L.; de Almeida, A.L.; Baldin, V.P.; Nakamura de Vasconcelos, S.S.; Siqueira, V.L.D.; Caleffi-Ferracioli, K.R.; Campanerut-Sa, P.A.Z.; Cardoso, R.F. Combinatory activity of linezolid and levofloxacin with antituberculosis drugs in Mycobacterium tuberculosis. Tuberculosis 2018, 111, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Zheng, M.; Wang, B.; Mu, X.; Li, P.; Fu, L.; Liu, S.; Guo, Z. Interactions of linezolid and second-line anti-tuberculosis agents against multidrug-resistant Mycobacterium tuberculosis in vitro and in vivo. Int. J. Infect. Dis. 2016, 52, 23–28. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Xu, Z.; Jiang, Y.; Liu, H.; Zhao, L.L.; Li, M.; Xu, D.; Zhao, X.; Liu, Z.; Wang, R.; et al. Synergistic activities of clofazimine with moxifloxacin or capreomycin against Mycobacterium tuberculosis in China. Int. J. Antimicrob. Agents 2019, 54, 642–646. [Google Scholar] [CrossRef]
- Bax, H.I.; Bakker-Woudenberg, I.; de Vogel, C.P.; van der Meijden, A.; Verbon, A.; de Steenwinkel, J.E.M. The role of the time-kill kinetics assay as part of a preclinical modeling framework for assessing the activity of anti-tuberculosis drugs. Tuberculosis 2017, 105, 80–85. [Google Scholar] [CrossRef]
- Rey-Jurado, E.; Tudo, G.; Martinez, J.A.; Gonzalez-Martin, J. Synergistic effect of two combinations of antituberculous drugs against Mycobacterium tuberculosis. Tuberculosis 2012, 92, 260–263. [Google Scholar] [CrossRef]
- Louie, A.; Duncanson, B.; Myrick, J.; Maynard, M.; Nole, J.; Brown, D.; Schmidt, S.; Neely, M.; Scanga, C.A.; Peloquin, C.; et al. Activity of Moxifloxacin against Mycobacterium tuberculosis in Acid Phase and Nonreplicative-Persister Phenotype Phase in a Hollow-Fiber Infection Model. Antimicrob. Agents Chemother. 2018, 62, e01470-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cokol, M.; Kuru, N.; Bicak, E.; Larkins-Ford, J.; Aldridge, B.B. Efficient measurement and factorization of high-order drug interactions in Mycobacterium tuberculosis. Sci. Adv. 2017, 3, e1701881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yilancioglu, K.; Cokol, M. Design of high-order antibiotic combinations against M. tuberculosis by ranking and exclusion. Sci. Rep. 2019, 9, 11876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, S.; Jaipalli, S.; Larkins-Ford, J.; Lohmiller, J.; Aldridge, B.B.; Sherman, D.R.; Chandrasekaran, S. Transcriptomic Signatures Predict Regulators of Drug Synergy and Clinical Regimen Efficacy against Tuberculosis. mBio 2019, 10, e02627-19. [Google Scholar] [CrossRef] [Green Version]
- Peterson, E.J.R.; Ma, S.; Sherman, D.R.; Baliga, N.S. Network analysis identifies Rv0324 and Rv0880 as regulators of bedaquiline tolerance in Mycobacterium tuberculosis. Nat. Microbiol. 2016, 1, 16078. [Google Scholar] [CrossRef] [Green Version]
- Den Hollander, J.G.; Mouton, J.W.; Verbrugh, H.A. Use of pharmacodynamic parameters to predict efficacy of combination therapy by using fractional inhibitory concentration kinetics. Antimicrob. Agents Chemother. 1998, 42, 744–748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergmann, J.S.; Woods, G.L. In vitro activity of antimicrobial combinations against clinical isolates of susceptible and resistant Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 1998, 2, 621–626. [Google Scholar]
- Zhu, C.; Liu, Y.; Hu, L.; Yang, M.; He, Z.G. Molecular mechanism of the synergistic activity of ethambutol and isoniazid against Mycobacterium tuberculosis. J. Biol. Chem. 2018, 293, 16741–16750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillespie, S.H.; Crook, A.M.; McHugh, T.D.; Mendel, C.M.; Meredith, S.K.; Murray, S.R.; Pappas, F.; Phillips, P.P.; Nunn, A.J.; Consortium, R.E. Four-month moxifloxacin-based regimens for drug-sensitive tuberculosis. N. Engl. J. Med. 2014, 371, 1577–1587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Li, T.; Qu, G.; Pang, Y.; Zhao, Y. In vitro synergistic activity of clofazimine and other antituberculous drugs against multidrug-resistant Mycobacterium tuberculosis isolates. Int. J. Antimicrob. Agents 2015, 45, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Pullan, S.T.; Allnutt, J.C.; Devine, R.; Hatch, K.A.; Jeeves, R.E.; Hendon-Dunn, C.L.; Marsh, P.D.; Bacon, J. The effect of growth rate on pyrazinamide activity in Mycobacterium tuberculosis - insights for early bactericidal activity? BMC Infect. Dis. 2016, 16, 205. [Google Scholar] [CrossRef] [Green Version]
- Sadouki, Z.; McHugh, T.D.; Aarnoutse, R.; Ortiz Canseco, J.; Darlow, C.; Hope, W.; van Ingen, J.; Longshaw, C.; Manissero, D.; Mead, A.; et al. Application of the hollow fibre infection model (HFIM) in antimicrobial development: A systematic review and recommendations of reporting. J. Antimicrob. Chemother. 2021, 76, 2252–2259. [Google Scholar] [CrossRef]
- Peterson, E.J.; Reiss, D.J.; Turkarslan, S.; Minch, K.J.; Rustad, T.; Plaisier, C.L.; Longabaugh, W.J.; Sherman, D.R.; Baliga, N.S. A high-resolution network model for global gene regulation in Mycobacterium tuberculosis. Nucleic Acids Res. 2014, 42, 11291–11303. [Google Scholar] [CrossRef] [Green Version]
- Cokol-Cakmak, M.; Bakan, F.; Cetiner, S.; Cokol, M. Diagonal Method to Measure Synergy Among Any Number of Drugs. J. Vis. Exp. 2018, 136, e57713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez, J.E.; McKinney, J.D.M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis 2004, 84, 29–44. [Google Scholar] [CrossRef]
- Dhar, N.; McKinney, J.D. Microbial phenotypic heterogeneity and antibiotic tolerance. Curr. Opin. Microbiol. 2007, 10, 30–38. [Google Scholar] [CrossRef]
- Chandrasekaran, S.; Price, N.D. Probabilistic integrative modeling of genome-scale metabolic and regulatory networks in Escherichia coli and Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2010, 107, 17845–17850. [Google Scholar] [CrossRef] [Green Version]
- Ekins, S.; Pottorf, R.; Reynolds, R.C.; Williams, A.J.; Clark, A.M.; Freundlich, J.S. Looking back to the future: Predicting in vivo efficacy of small molecules versus Mycobacterium tuberculosis. J. Chem. Inf. Model. 2014, 54, 1070–1082. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Minch, K.J.; Rustad, T.R.; Hobbs, S.; Zhou, S.L.; Sherman, D.R.; Price, N.D. Integrated Modeling of Gene Regulatory and Metabolic Networks in Mycobacterium tuberculosis. PLoS Comput. Biol. 2015, 11, e1004543. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.Y.; Clemens, D.L.; Silva, A.; Dillon, B.J.; Maslesa-Galic, S.; Nava, S.; Ding, X.; Ho, C.M.; Horwitz, M.A. Drug regimens identified and optimized by output-driven platform markedly reduce tuberculosis treatment time. Nat. Commun. 2017, 8, 14183. [Google Scholar] [CrossRef] [Green Version]
- Andries, K.; Verhasselt, P.; Guillemont, J.; Gohlmann, H.W.; Neefs, J.M.; Winkler, H.; Van Gestel, J.; Timmerman, P.; Zhu, M.; Lee, E.; et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 2005, 307, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Borrell, S.; Trauner, A.; Brites, D.; Rigouts, L.; Loiseau, C.; Coscolla, M.; Niemann, S.; De Jong, B.; Yeboah-Manu, D.; Kato-Maeda, M.; et al. Reference set of Mycobacterium tuberculosis clinical strains: A tool for research and product development. PLoS ONE 2019, 14, e0214088. [Google Scholar] [CrossRef] [Green Version]
- Bonapace, C.R.; Bosso, J.A.; Friedrich, L.V.; White, R.L. Comparison of methods of interpretation of checkerboard synergy testing. Diagn. Microbiol. Infect. Dis. 2002, 44, 363–366. [Google Scholar] [CrossRef]
- Zimmer, A.; Katzir, I.; Dekel, E.; Mayo, A.E.; Alon, U. Prediction of multidimensional drug dose responses based on measurements of drug pairs. Proc. Natl. Acad. Sci. USA 2016, 113, 10442–10447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haver, H.L.; Chua, A.; Ghode, P.; Lakshminarayana, S.B.; Singhal, A.; Mathema, B.; Wintjens, R.; Bifani, P. Mutations in genes for the F420 biosynthetic pathway and a nitroreductase enzyme are the primary resistance determinants in spontaneous in vitro-selected PA-824-resistant mutants of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2015, 59, 5316–5323. [Google Scholar] [CrossRef] [Green Version]
- Manjunatha, U.; Boshoff, H.I.; Barry, C.E. The mechanism of action of PA-824: Novel insights from transcriptional profiling. Commun. Integr. Biol. 2009, 2, 215–218. [Google Scholar] [CrossRef] [Green Version]
- Swaney, S.M.; Aoki, H.; Ganoza, M.C.; Shinabarger, D.L. The oxazolidinone linezolid inhibits initiation of protein synthesis in bacteria. Antimicrob. Agents Chemother. 1998, 42, 3251–3255. [Google Scholar] [CrossRef] [Green Version]
- Beckert, P.; Hillemann, D.; Kohl, T.A.; Kalinowski, J.; Richter, E.; Niemann, S.; Feuerriegel, S. rplC T460C identified as a dominant mutation in linezolid-resistant Mycobacterium tuberculosis strains. Antimicrob. Agents Chemother. 2012, 56, 2743–2745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balasubramanian, V.; Solapure, S.; Iyer, H.; Ghosh, A.; Sharma, S.; Kaur, P.; Deepthi, R.; Subbulakshmi, V.; Ramya, V.; Ramachandran, V.; et al. Bactericidal activity and mechanism of action of AZD5847, a novel oxazolidinone for treatment of tuberculosis. Antimicrob. Agents Chemother. 2014, 58, 495–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cholo, M.C.; Mothiba, M.T.; Fourie, B.; Anderson, R. Mechanisms of action and therapeutic efficacies of the lipophilic antimycobacterial agents clofazimine and bedaquiline. J. Antimicrob. Chemother. 2017, 72, 338–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartkoorn, R.C.; Uplekar, S.; Cole, S.T. Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2014, 58, 2979–2981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koul, A.; Dendouga, N.; Vergauwen, K.; Molenberghs, B.; Vranckx, L.; Willebrords, R.; Ristic, Z.; Lill, H.; Dorange, I.; Guillemont, J.; et al. Diarylquinolines target subunit c of mycobacterial ATP synthase. Nat. Chem. Biol. 2007, 3, 323–324. [Google Scholar] [CrossRef]
- Huitric, E.; Verhasselt, P.; Koul, A.; Andries, K.; Hoffner, S.; Andersson, D.I. Rates and mechanisms of resistance development in Mycobacterium tuberculosis to a novel diarylquinoline ATP synthase inhibitor. Antimicrob. Agents Chemother. 2010, 54, 1022–1028. [Google Scholar] [CrossRef] [Green Version]
- Ramaswamy, S.V.; Reich, R.; Dou, S.J.; Jasperse, L.; Pan, X.; Wanger, A.; Quitugua, T.; Graviss, E.A. Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2003, 47, 1241–1250. [Google Scholar] [CrossRef] [Green Version]
- Vilcheze, C.; Wang, F.; Arai, M.; Hazbon, M.H.; Colangeli, R.; Kremer, L.; Weisbrod, T.R.; Alland, D.; Sacchettini, J.C.; Jacobs, W.R., Jr. Transfer of a point mutation in Mycobacterium tuberculosis inhA resolves the target of isoniazid. Nat. Med. 2006, 12, 1027–1029. [Google Scholar] [CrossRef]
- Massengo-Tiasse, R.P.; Cronan, J.E. Diversity in enoyl-acyl carrier protein reductases. Cell. Mol. Life Sci. 2009, 66, 1507–1517. [Google Scholar] [CrossRef] [Green Version]
- Thirumurugan, R.; Kathirvel, M.; Vallayyachari, K.; Surendar, K.; Samrot, A.V.; Muthaiah, M. Molecular analysis of rpoB gene mutations in rifampicin resistant Mycobacterium tuberculosis isolates by multiple allele specific polymerase chain reaction in Puducherry, South India. J. Infect. Public Health 2015, 8, 619–625. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; An, X.; Liu, H.; Wang, S.; Xiao, T.; Liu, H. Uncovering the resistance mechanism of Mycobacterium tuberculosis to rifampicin due to RNA polymerase H451D/Y/R mutations from computational perspective. Front. Chem. 2019, 7, 819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safi, H.; Lingaraju, S.; Amin, A.; Kim, S.; Jones, M.; Holmes, M.; McNeil, M.; Peterson, S.N.; Chatterjee, D.; Fleischmann, R.; et al. Evolution of high-level ethambutol-resistant tuberculosis through interacting mutations in decaprenylphosphoryl-beta-D-arabinose biosynthetic and utilization pathway genes. Nat. Genet. 2013, 45, 1190–1197. [Google Scholar] [CrossRef] [PubMed]
- Jureen, P.; Werngren, J.; Toro, J.C.; Hoffner, S. Pyrazinamide resistance and pncA gene mutations in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2008, 52, 1852–1854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Njire, M.; Tan, Y.; Mugweru, J.; Wang, C.; Guo, J.; Yew, W.; Tan, S.; Zhang, T. Pyrazinamide resistance in Mycobacterium tuberculosis: Review and update. Adv. Med. Sci. 2016, 61, 63–71. [Google Scholar] [CrossRef]
- Sharma, D.; Cukras, A.R.; Rogers, E.J.; Southworth, D.R.; Green, R. Mutational analysis of S12 protein and implications for the accuracy of decoding by the ribosome. J. Mol. Biol. 2007, 374, 1065–1076. [Google Scholar] [CrossRef] [Green Version]
- Springer, B.; Kidan, Y.G.; Prammananan, T.; Ellrott, K.; Bottger, E.C.; Sander, P. Mechanisms of streptomycin resistance: Selection of mutations in the 16S rRNA gene conferring resistance. Antimicrob. Agents Chemother. 2001, 45, 2877–2884. [Google Scholar] [CrossRef] [Green Version]
- Nosova, E.Y.; Bukatina, A.A.; Isaeva, Y.D.; Makarova, M.V.; Galkina, K.Y.; Moroz, A.M. Analysis of mutations in the gyrA and gyrB genes and their association with the resistance of Mycobacterium tuberculosis to levofloxacin, moxifloxacin and gatifloxacin. J. Med. Microbiol. 2013, 62, 108–113. [Google Scholar] [CrossRef] [Green Version]
- Aubry, A.; Pan, X.S.; Fisher, L.M.; Jarlier, V.; Cambau, E. Mycobacterium tuberculosis DNA gyrase: Interaction with quinolones and correlation with antimycobacterial drug activity. Antimicrob. Agents Chemother. 2004, 48, 1281–1288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reeves, A.Z.; Campbell, P.J.; Sultana, R.; Malik, S.; Murray, M.; Plikaytis, B.B.; Shinnick, T.M.; Posey, J.E. Aminoglycoside cross-resistance in Mycobacterium tuberculosis due to mutations in the 5′ untranslated region of whiB7. Antimicrob. Agents Chemother. 2013, 57, 1857–1865. [Google Scholar] [CrossRef] [Green Version]
- Sowajassatakul, A.; Prammananan, T.; Chaiprasert, A.; Phunpruch, S. Molecular characterization of amikacin, kanamycin and capreomycin resistance in M/XDR-TB strains isolated in Thailand. BMC Microbiol. 2014, 14, 165. [Google Scholar] [CrossRef] [Green Version]
- Smith, T.; Wolff, K.A.; Nguyen, L. Molecular biology of drug resistance in Mycobacterium tuberculosis. Pathog. Mycobacterium Tuberc. Its Interact. Host Org. 2012, 53–80. [Google Scholar]
- Zhao, F.; Wang, X.D.; Erber, L.N.; Luo, M.; Guo, A.Z.; Yang, S.S.; Gu, J.; Turman, B.J.; Gao, Y.R.; Li, D.F.; et al. Binding pocket alterations in dihydrofolate synthase confer resistance to para-aminosalicylic acid in clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2014, 58, 1479–1487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rengarajan, J.; Sassetti, C.M.; Naroditskaya, V.; Sloutsky, A.; Bloom, B.R.; Rubin, E.J. The folate pathway is a target for resistance to the drug para-aminosalicylic acid (PAS) in mycobacteria. Mol. Microbiol. 2004, 53, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Mathys, V.; Wintjens, R.; Lefevre, P.; Bertout, J.; Singhal, A.; Kiass, M.; Kurepina, N.; Wang, X.M.; Mathema, B.; Baulard, A.; et al. Molecular genetics of para-aminosalicylic acid resistance in clinical isolates and spontaneous mutants of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2009, 53, 2100–2109. [Google Scholar] [CrossRef] [Green Version]
- Grant, S.S.; Wellington, S.; Kawate, T.; Desjardins, C.A.; Silvis, M.R.; Wivagg, C.; Thompson, M.; Gordon, K.; Kazyanskaya, E.; Nietupski, R.; et al. Baeyer-Villiger Monooxygenases EthA and MymA Are Required for Activation of Replicating and Non-replicating Mycobacterium tuberculosis Inhibitors. Cell Chem. Biol. 2016, 23, 666–677. [Google Scholar] [CrossRef] [Green Version]
- Carette, X.; Blondiaux, N.; Willery, E.; Hoos, S.; Lecat-Guillet, N.; Lens, Z.; Wohlkonig, A.; Wintjens, R.; Soror, S.H.; Frenois, F.; et al. Structural activation of the transcriptional repressor EthR from Mycobacterium tuberculosis by single amino acid change mimicking natural and synthetic ligands. Nucleic Acids Res. 2012, 40, 3018–3030. [Google Scholar] [CrossRef] [Green Version]
- Rueda, J.; Realpe, T.; Mejia, G.I.; Zapata, E.; Rozo, J.C.; Ferro, B.E.; Robledo, J. Genotypic analysis of genes associated with independent resistance and cross-resistance to isoniazid and ethionamide in Mycobacterium tuberculosis clinical isolates. Antimicrob. Agents Chemother. 2015, 59, 7805–7810. [Google Scholar] [CrossRef] [Green Version]
| First Author, Year | Source of Patients Data | Total Number of Samples Used | TB/DST/MIC Test Results | Material | MIC Value | Validated Analytical Determination/Methodology | Drug Interaction | Sample Handling Described | Endpoints Method AUC Calculation | Endpoints Method FICI Calculation | Endpoints Method EBA Calculation Cmax | Grading Risk of Bias (High, Medium, Low) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Maltempe 2017 [18] | (14 susceptible, 9 INH mono-resistant and 14 MDR and laboratory strains (H37Rv) | 37 | +, +, + | Culture | RIF (0.004 to 0.25 μg/mL and 4–250 μg/mL). LZD (0.125 to 0.5 μg/mL for susceptible and 0.125–2.5 μg/mL for RIF) | Checkerboard, REDCA assay. Time-kill curve assay | LZD and RIF | + | − | + | − | Low |
| Drusano 2014 [19] | H37Rv | Not specified | +, −, +, | Culture | LZD (1.0 mg/L) RIF (0.25 mg/L) | HFIM | LZD and RIF | + | − | − | − | High |
| Calefi- Ferracioli 2013 [20] | H37Rv, 9 susceptible and 10 resistant clinical isolates | 19 | +, + 1, + | Culture | INH, EMB and LFX (0.03–32 mg/L, 0.5–032 mg/L and 0.06–4 mg/L). | REDCA, classical checkerboard assay | INH/ LFX EMB | + | − | + | − | Low |
| Lopez-Gavin 2015 [21] | 7 MDR and 11 DS clinical isolates | 17 | +, +, + | Culture | CFX, LFX, MFX and UB-8902 (0.0625–1 mg/L); Pa (0.0313–1 mg/L) | Checkerboard | CFZ/Pa/LFX CFZ/Pa/MFX CFX/Pa/Ub-8902 | + | − | + | − | Low |
| Miranda Silva, 2019 [22] | M. tuberculosis 18b, H37Rv | Not specified | +, +, + | Culture | MFX (0.25 mg/L and 0.5 mg/L). Pa (0.125 mg/L) | Checkerboard, URSA | MFX and Pa Log, acid, NRP phases | + | − | − | − | High |
| Miranda Silva, 2018 [23] | M. tuberculosis 18b, H37Rv | Not specified | +, +, + | Culture | LZD (1 mg/L) BDQ (0.25–0.5 mg/L), 0.5) | Checkerboard, URSA | LZD and BDQ | + | − | − | − | High |
| Pang, 2019 [24] | XDR-TB | 191 2 | +, +, + | Culture | BDQ ≥ 0.063 mg/L, MFXx and GFX (0.125 mg/L), LZD (0.5 mg/L), Cfz (0.25 mg/L) | Checkerboard | BDQ/MFX/GFX/ CFZ, LZD | + | − | + | − | Low |
| Santos, 2018 [25] | M. tuberculosis H37Rv, 2 susceptible and 10 resistant clinical isolate | 12 | +, +, + | Culture | INH (0.03–6.25 μg/mL) RIF (0.008–100 μg/mL), LFX (0.12–0.25 μg/mL) LZD (0.25–0.5 μg/mL) | Three-dimensional checkerboard | LZD and LFX | + | − | + | − | low |
| Zhao, 2016 [26] | M. tuberculosis H37Rv, 3 MDR-TB clinical isolate | 3 | +, +, + | Culture | LZD (0.06 to 1 mg/mL) and MFX, LFX, PAS, KAN, CAP, AMK, and CFZ (0.125 mg/Land 8 mg/L). | Checkerboard 2 | CAP, AMK KAN, LFX, MFX PAS and CFZ | + | − | + | − | High |
| Li 2019 [27] | M. tuberculosis H37Rv, 3 MDR-TB, 2 XDR-TB, 3 Pan- susceptible clinical isolate, and 12 resistant strains to other drugs | 30 | +, +, + | Culture | CFZ (0.016–2 μg/mL), CAP (0.25–4 μg/mL), MFX (0.016–1 μg/mL). | Checkerboard | CFZ and MFX or CAP | + | − | + | − | Low |
| Bax 2017 [28] | M. tuberculosis Beijing VN 2002-1585 (BE 1585), R-TB | 2 | +, +, + | Culture | INH (0.125 mg/L), RIF(0.25 mg/L), STR (2 mg/L), EMB (5 mg/L), PAS (0.125 mg/L). | Time-kill kinetics assay | STR, INH, RIF, EMB, PAS and PZA | + | − | − | + | High |
| Rey-Jurado, 2012 [29] | 12 H mono-res or H/S –res, 11 DS clinical isolates | 32 | +, +, + | Culture | EMB (0.31–5 mg/mL), RIF (0.125–2 mg/mL), OFX (0.125–2 mg/mL) INH (0.025–102.4 mg/mL) | Two-dimensional checkerboard | INH/RIF, and EMB/OFX, RIF and EMB | + | − | + | − | Low |
| Louie, 2018 [30] | M. tuberculosis strain H37Rv and strain 18 b | 2 | Mutational frequency determination, MIC | Culture | N/A | HFIM | MFX activity Acid, NRP phases | + | + | − | − | High |
| Cokol, 2017 [31] | Panthotenate and leucine auxothrophic strain of M. tuberculosis | Not specified | +, +, + | Culture | N/A | Three-dimensional checkerboard DiaMOND | BDQ + CFZ+ RIF and BDQ + Pa + RIF and BDQ + CFZ+ INH + RIF and CFZ + INH + Pa+ RIF | + | − | + | − | High |
| Cokol, 2019 [32] | M. tuberculosis strain | Not specified | +, +, + | - | N/A | R/ED checkerboard | Pa + ETO and BDQ + CFZ | + | − | + 3 | − | High |
| (Ma, 2019 [33]) | Genetic wild-type strain, H37Rv and the TFI strain | 14 | +, +, + | Culture | N/A | INDIGO-MTB checkerboard assays and high-throughput DiaMOND method | BDQ/ CFZ alone or in a three-drug combination with PZA, EMB, RIF, or INH. INH-RIF-STR | + | + | + | − | High |
| (Peterson, 2016 [34]). | MTB wild-type H37Rv, ΔRv0324 and ΔRv0880 strains | Not specified | +, −, + | Culture | N/A | INDIGO model, EGRIN and PROM computational models | BDQ and Pa | + | − | + | − | Low |
| Drug Combination | Synergism/Additive | Antagonism |
|---|---|---|
| Computational model INDIGO-MTB, checkerboard assays, and the high-throughput DiaMOND method (Ma, 2019 [33]) | BDQ/CFZ alone or in a three-drug combination with PZA, EMB, RIF, or INH. INH-RIF-STR. When Rv1353c is induced, BDQ-STR and CAP-STR shift toward synergy | INH-STR and INH-RIF RIF-MFX. BDQ-STR and CAP-STM shift toward antagonism |
| BDQ and Pa, INDIGO model, EGRIN, and PROM computational models (Peterson, 2016 [34]) | BDQ and Pa Un-induced overexpression of Rv0880 (additive to moderately synergistic BDQ and Pa) Downregulation of the expression of Rv0324 and Rv0880 (considerable synergism) | Induced overexpression of Rv0880 (BDQ and Pa) Increased expression of Rv0324 (BDQ and Pa) |
| INH and EMB, DNA footprinting, and isothermal titration calorimetry and surface plasmon resonance assays (Zhu, 2018 [37]) | INH and EMB | N/A |
| LZD and RIF, modified checkerboard-REDCA model (Maltempe, 2017 [18]) | LZD and RIF (M. tuberculosis H37Rv) and 8 (20.5%) clinical isolates. Out of eight, three DS, two INH mono-resistant, and three MDR isolates. | N/A |
| LZD and RIF (Drusano, 2014 [19]) | LZD and RIF interact in a non-significant tendency towards antagonism for killing the wild-type (WT) population. | N/A |
| INH or EMB interaction with LFX, modified checkerboard assay, REDCA (Calefi-Ferraciol, 2013 [20]) | M. tuberculosis H37Rv and resistant isolates (EMB and LFX) | INH vs. LFX no synergism |
| CFZ/Pa/LFX and CFX/Pa/MFX and CFZ/Pa/Ub-8902 Checkerboard assay (López-Gavín, 2015 [21]) | CFZ/Pa/LFX, CFZ/Pa/MFX, and CFZ/Pa/Ub-8902 combination (MDR and drug-susceptible isolates) | N/A |
| MFX/Pa interaction in Log, Acid and NRP phases using a 9 by 8 well checkerboard assay (Miranda Silva, 2019 [22]) | MFX and Pa additive for all metabolic state | N/A |
| LZD/BDQ in Log, Acid, and NRP Phases,9 by 8 well Checkerboard assay (Miranda Silva, 2018 [23]), | LZD and BDQ is additive for bacterial killing in both strains for all metabolic states. | N/A |
| BDQ/MFX/GFX/CFZ, and LZD, checkerboard assay (Pang, 2019 [24]) | BDQ combination with MFX, GFX, CFZ, and LZD for treatment XDR-TB | XDR-TB isolates for BDQ-MFX, BDQ-GFX, BDQ-LZD, and BDQ-CFZ |
| LZD and LFX three-dimensional checkerboard (Santos, 2018 [25]) | 40% of resistant clinical isolates INH/RIF/LFX and 50% resistant clinical isolates INH/RIF/LZD, with a better synergism observed for INH and RIF combined to LVX or LZD at 1/4 MIC | N/A |
| LZD and CAP, AMK KAM, LFX, MFX, PAS, and CFZ, checkerboard assay (Zhao, 2016 [26]) | LZD/CAP/ LZD/PAS, LZD/LFX and LZD/AMK showed partial synergism in 3/4, 2/4, 1/4 isolates, respectively (REDCA) | N/A |
| CFZ with MFX or CAP checkerboard assay (Li, 2019 [27]) | CFZ/CAP CFZ/MFX. | M/XDR strains in increased concentration of CFZ in CFZ/CAP and CFZ/MFX combination |
| STR, INH, RIF, EMB, Pas and PZA time-kill kinetics (Bax, 2017 [28]) | INH/RIF at clinically used concentrations | N/A |
| INH/RIF, EMB/OFX RIF/EMB, two-dimensional checkerboard assay (Rey Jurado, 2012 [29]) | INH, RIF and EMB synergism in the INH drug res isolates OFX, RIF and EMB in the res and DS isolates | N/A |
| High-throughput combinational screening, checkerboard and DiAMOND (Cokol, 2017 [31]) | BDQ + CFZ + INH, BDQ + CFZ + RIF and BDQ + Pa + RIF and four-way combinations BDQ + CFZ + INH + RIF and CFZ + INH+ Pa+ RIF | N/A |
| Pa + ETO and BDQ + CFZ, R/ED checkerboard assay (Cokol, 2019 [32]) | Pa + ETO and BDQ + CFZ is against RIF-resistant M. tuberculosis. Pa + VAN and FUS + CFZ CFZ + FUS and (LAS) + Pa against MDR isolates CFZ + INH and ETO + RIF | N/A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Margaryan, H.; Evangelopoulos, D.D.; Muraro Wildner, L.; McHugh, T.D. Pre-Clinical Tools for Predicting Drug Efficacy in Treatment of Tuberculosis. Microorganisms 2022, 10, 514. https://doi.org/10.3390/microorganisms10030514
Margaryan H, Evangelopoulos DD, Muraro Wildner L, McHugh TD. Pre-Clinical Tools for Predicting Drug Efficacy in Treatment of Tuberculosis. Microorganisms. 2022; 10(3):514. https://doi.org/10.3390/microorganisms10030514
Chicago/Turabian StyleMargaryan, Hasmik, Dimitrios D. Evangelopoulos, Leticia Muraro Wildner, and Timothy D. McHugh. 2022. "Pre-Clinical Tools for Predicting Drug Efficacy in Treatment of Tuberculosis" Microorganisms 10, no. 3: 514. https://doi.org/10.3390/microorganisms10030514






