Norepinephrine and Serotonin Can Modulate the Behavior of the Probiotic Enterococcus faecium NCIMB10415 towards the Host: Is a Putative Surface Sensor Involved?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Media and Growth Conditions
2.2. Cell Lines
2.3. Confocal Laser Scanning Microscopy (CLSM) Biofilm Formation
2.4. Adhesion
2.5. Transepithelial Electrical Resistance (TER)
2.6. DCs Stimulation
2.7. Flow Cytometric Analysis and Cytokine Quantification in Culture Supernatants
2.8. Genome Sequencing and Annotation
2.9. Putative Sensor Sequence Screening and Alignment
2.10. Structure Modeling and Docking
2.11. VicK (WalK) Expression and Purification
2.12. Micro-Scale Thermophoresis (MST)
2.13. Statistical Analysis
3. Results
3.1. Biofilm
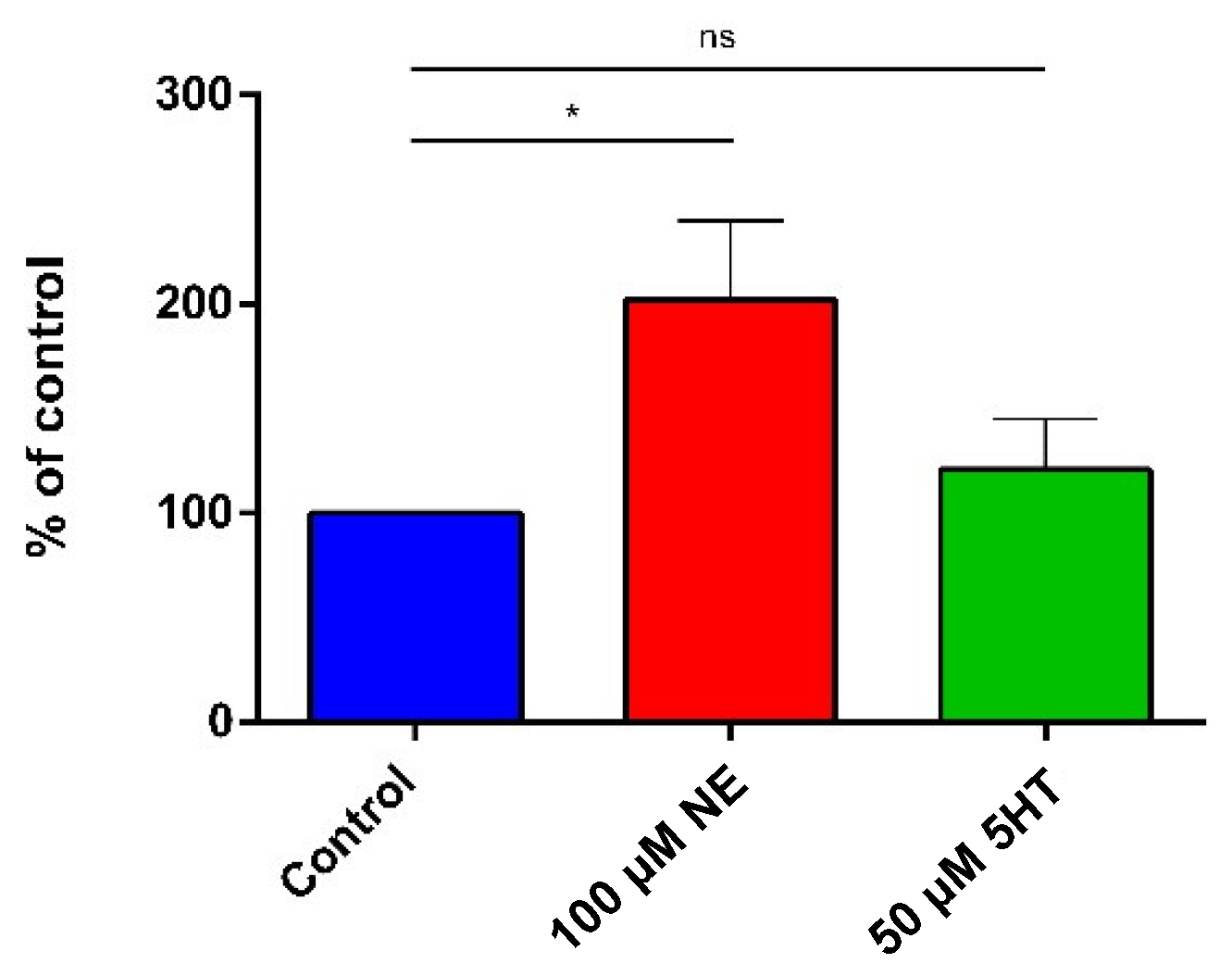
3.2. Adhesion on Caco-2/TC7 Cells
3.3. Effect on the Epithelial Barrier Function (TER on Differentiated Caco-2/TC7 Cells)
3.4. Immune Modulation
3.4.1. DC Differentiation and Maturation
3.4.2. Cytokine Quantification in Culture Supernatants
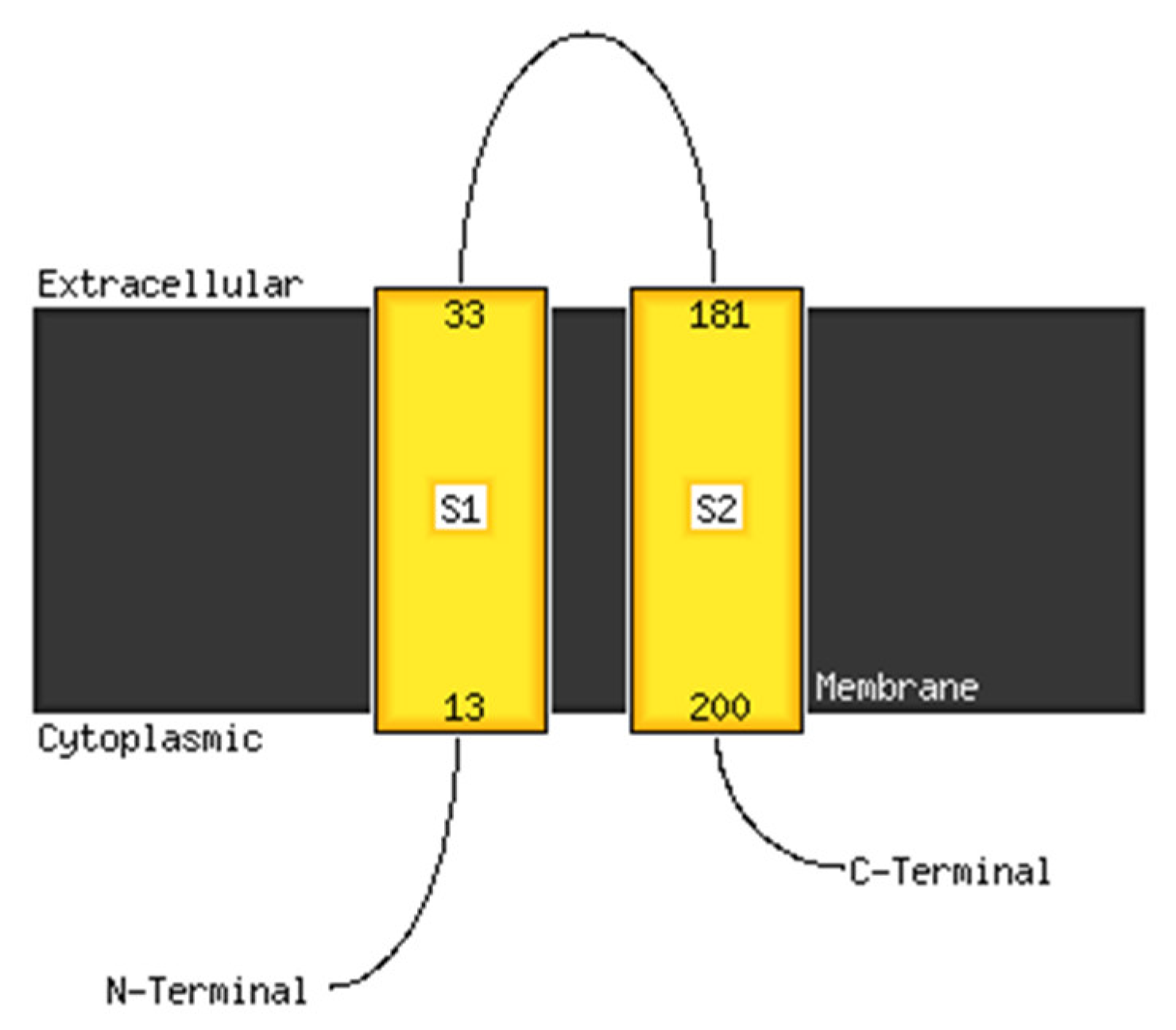
3.5. In Silico Analyses of a Putative Sensor
3.5.1. Structure Modeling
3.5.2. Molecular Docking
3.6. Binding Studies—Micro-Scale Thermophoresis
4. Discussion
4.1. Biofilm, Adhesion and TER
4.2. Immune-Targeted Activity
4.3. Sensor
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Franz, C. Enterococci in Foods—A Conundrum for Food Safety. Int. J. Food Microbiol. 2003, 88, 105–122. [Google Scholar] [CrossRef]
- Hanchi, H.; Mottawea, W.; Sebei, K.; Hammami, R. The Genus Enterococcus: Between Probiotic Potential and Safety Concerns—An Update. Front. Microbiol. 2018, 9, 1791. [Google Scholar] [CrossRef]
- Bondi, M.; Laukova, A.; de Niederhausern, S.; Messi, P.; Papadopoulou, C.; Economou, V. Controversial Aspects Displayed by Enterococci: Probiotics or Pathogens? BioMed Res. Int. 2020, 2020, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Giraffa, G. Functionality of Enterococci in Dairy Products. Int. J. Food Microbiol. 2003, 88, 215–222. [Google Scholar] [CrossRef]
- Hosseini, S.V.; Arlindo, S.; Böhme, K.; Fernández-No, C.; Calo-Mata, P.; Barros-Velázquez, J. Molecular and Probiotic Characterization of Bacteriocin-Producing Enterococcus Faecium Strains Isolated from Nonfermented Animal Foods: Probiotic E. Faecium Strains. J. Appl. Microbiol. 2009, 107, 1392–1403. [Google Scholar] [CrossRef] [PubMed]
- Ziadi, M.; Mhir, S.; Dubois-Dauphin, R.; Chambellon, E.; Yvon, M.; Thonart, P.; Hamdi, M. Analysis of Volatile Compounds, Amino Acid Catabolism and Some Technological Properties of Enterococcus Faecalis Strain SLT13 Isolated from Artisanal Tunisian Fermented Milk. BMR J. 2016, 14, 1–12. [Google Scholar] [CrossRef]
- Zommiti, M.; Cambronel, M.; Maillot, O.; Barreau, M.; Sebei, K.; Feuilloley, M.; Ferchichi, M.; Connil, N. Evaluation of Probiotic Properties and Safety of Enterococcus Faecium Isolated From Artisanal Tunisian Meat “Dried Ossban”. Front. Microbiol. 2018, 9, 1685. [Google Scholar] [CrossRef] [Green Version]
- Baccouri, O.; Boukerb, A.M.; Farhat, L.B.; Zébré, A.; Zimmermann, K.; Domann, E.; Cambronel, M.; Barreau, M.; Maillot, O.; Rincé, I.; et al. Probiotic Potential and Safety Evaluation of Enterococcus Faecalis OB14 and OB15, Isolated From Traditional Tunisian Testouri Cheese and Rigouta, Using Physiological and Genomic Analysis. Front. Microbiol. 2019, 10, 881. [Google Scholar] [CrossRef]
- Franz, C.M.A.P.; Huch, M.; Abriouel, H.; Holzapfel, W.; Gálvez, A. Enterococci as Probiotics and Their Implications in Food Safety. Int. J. Food Microbiol. 2011, 151, 125–140. [Google Scholar] [CrossRef] [Green Version]
- Pessione, A.; Lamberti, C.; Cocolin, L.; Campolongo, S.; Grunau, A.; Giubergia, S.; Eberl, L.; Riedel, K.; Pessione, E. Different Protein Expression Profiles in Cheese and Clinical Isolates of Enterococcus Faecalis Revealed by Proteomic Analysis. Proteomics 2012, 12, 431–447. [Google Scholar] [CrossRef]
- Pillar, C.M.; Gilmore, M.S. Enterococcal virulence—Pathogenicity island of E. faecalis. Front. Biosci. 2004, 9, 2335–2346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crank, C.; O’Driscoll, T. Vancomycin-Resistant Enterococcal Infections: Epidemiology, Clinical Manifestations, and Optimal Management. IDR 2015, 8, 217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben Braïek, O.; Smaoui, S. Enterococci: Between Emerging Pathogens and Potential Probiotics. BioMed Res. Int. 2019, 2019, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Palmer, K.L.; Gilmore, M.S. Multidrug-Resistant Enterococci Lack CRISPR-Cas. mBio 2010, 1, 4. [Google Scholar] [CrossRef] [Green Version]
- Laverde Gomez, J.A.; Hendrickx, A.P.A.; Willems, R.J.; Top, J.; Sava, I.; Huebner, J.; Witte, W.; Werner, G. Intra- and Interspecies Genomic Transfer of the Enterococcus Faecalis Pathogenicity Island. PLoS ONE 2011, 6, e16720. [Google Scholar] [CrossRef] [Green Version]
- Manson, J.M.; Hancock, L.E.; Gilmore, M.S. Mechanism of Chromosomal Transfer of Enterococcus Faecalis Pathogenicity Island, Capsule, Antimicrobial Resistance, and Other Traits. Proc. Natl. Acad. Sci. USA 2010, 107, 12269–12274. [Google Scholar] [CrossRef] [Green Version]
- Cirrincione, S.; Neumann, B.; Zühlke, D.; Riedel, K.; Pessione, E. Detailed Soluble Proteome Analyses of a Dairy-Isolated Enterococcus Faecalis: A Possible Approach to Assess Food Safety and Potential Probiotic Value. Front. Nutr. 2019, 6, 71. [Google Scholar] [CrossRef]
- Hew, C.M.; Korakli, M.; Vogel, R.F. Expression of Virulence-Related Genes by Enterococcus Faecalis in Response to Different Environments. Syst. Appl. Microbiol. 2007, 30, 257–267. [Google Scholar] [CrossRef]
- Biaggini, K.; Barbey, C.; Borrel, V.; Feuilloley, M.; Déchelotte, P.; Connil, N. The Pathogenic Potential of Pseudomonas Fluorescens MFN1032 on Enterocytes Can Be Modulated by Serotonin, Substance P and Epinephrine. Arch. Microbiol. 2015, 197, 983–990. [Google Scholar] [CrossRef]
- Cambronel, M.; Tortuel, D.; Biaggini, K.; Maillot, O.; Taupin, L.; Réhel, K.; Rincé, I.; Muller, C.; Hardouin, J.; Feuilloley, M.; et al. Epinephrine Affects Motility, and Increases Adhesion, Biofilm and Virulence of Pseudomonas Aeruginosa H103. Sci. Rep. 2019, 9, 2021. [Google Scholar] [CrossRef]
- Cambronel, M.; Nilly, F.; Mesguida, O.; Boukerb, A.M.; Racine, P.-J.; Baccouri, O.; Borrel, V.; Martel, J.; Fécamp, F.; Knowlton, R.; et al. Influence of Catecholamines (Epinephrine/Norepinephrine) on Biofilm Formation and Adhesion in Pathogenic and Probiotic Strains of Enterococcus Faecalis. Front. Microbiol. 2020, 11, 1501. [Google Scholar] [CrossRef] [PubMed]
- Scardaci, R.; Varese, F.; Manfredi, M.; Marengo, E.; Mazzoli, R.; Pessione, E. Enterococcus Faecium NCIMB10415 Responds to Norepinephrine by Altering Protein Profiles and Phenotypic Characters. J. Proteom. 2021, 231, 104003. [Google Scholar] [CrossRef] [PubMed]
- Scardaci, R.; Manfredi, M.; Barberis, E.; Scutera, S.; Marengo, E.; Pessione, E. Serotonin Exposure Improves Stress Resistance, Aggregation, and Biofilm Formation in the Probiotic Enterococcus Faecium NCIMB10415. Microbiol. Res. 2021, 12, 606–625. [Google Scholar] [CrossRef]
- Lesouhaitier, O.; Veron, W.; Chapalain, A.; Madi, A.; Blier, A.-S.; Dagorn, A.; Connil, N.; Chevalier, S.; Orange, N.; Feuilloley, M. Gram-Negative Bacterial Sensors for Eukaryotic Signal Molecules. Sensors 2009, 9, 6967–6990. [Google Scholar] [CrossRef] [Green Version]
- Clarke, M.B.; Hughes, D.T.; Zhu, C.; Boedeker, E.C.; Sperandio, V. The QseC Sensor Kinase: A Bacterial Adrenergic Receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 10420–10425. [Google Scholar] [CrossRef] [Green Version]
- Rasko, D.A.; Moreira, C.G.; Li, D.R.; Reading, N.C.; Ritchie, J.M.; Waldor, M.K.; Williams, N.; Taussig, R.; Wei, S.; Roth, M.; et al. Targeting QseC Signaling and Virulence for Antibiotic Development. Science 2008, 321, 1078–1080. [Google Scholar] [CrossRef] [Green Version]
- Mayer, E.A. Gut Feelings: The Emerging Biology of Gut–Brain Communication. Nat. Rev. Neurosci. 2011, 12, 453–466. [Google Scholar] [CrossRef]
- Yan, F.; Polk, D.B. Probiotics and Immune Health. Curr. Opin. Gastroenterol. 2011, 27, 496–501. [Google Scholar] [CrossRef] [Green Version]
- Molina, M.A.; Díaz, A.M.; Hesse, C.; Ginter, W.; Gentilini, M.V.; Nuñez, G.G.; Canellada, A.M.; Sparwasser, T.; Berod, L.; Castro, M.S.; et al. Immunostimulatory Effects Triggered by Enterococcus Faecalis CECT7121 Probiotic Strain Involve Activation of Dendritic Cells and Interferon-Gamma Production. PLoS ONE 2015, 10, e0127262. [Google Scholar] [CrossRef] [Green Version]
- Khalkhali, S.; Mojgani, N. Enterococcus Faecium; a Suitable Probiotic Candidate for Modulation of Immune Responses Against Pathogens. Int. J. Basic Sci. Med. 2017, 2, 77–82. [Google Scholar] [CrossRef] [Green Version]
- Scutera, S.; Riboldi, E.; Daniele, R.; Elia, A.R.; Fraone, T.; Castagnoli, C.; Giovarelli, M.; Musso, T.; Sozzani, S. Production and Function of Activin A in Human Dendritic Cells. Eur. Cytokine Netw. 2008, 19, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Krueger, F. Trim Galore: A Wrapper Tool Around Cutadapt and FastQC to Consistently Apply Quality and Adapter Trimming to FastQ Files. 2012. Available online: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 1 December 2021).
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Version 0.11.2. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 15 November 2020).
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile Genome Assembly Evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [Green Version]
- McGuffin, L.J.; Bryson, K.; Jones, D.T. The PSIPRED Protein Structure Prediction Server. Bioinformatics 2000, 16, 404–405. [Google Scholar] [CrossRef]
- Ko, J.; Park, H.; Heo, L.; Seok, C. GalaxyWEB Server for Protein Structure Prediction and Refinement. Nucleic Acids Res. 2012, 40, W294–W297. [Google Scholar] [CrossRef]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2016, 54, 5.6.1–5.6.37. [Google Scholar] [CrossRef] [Green Version]
- Benkert, P.; Künzli, M.; Schwede, T. QMEAN Server for Protein Model Quality Estimation. Nucleic Acids Res. 2009, 37, W510–W514. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- Rosay, T.; Bazire, A.; Diaz, S.; Clamens, T.; Blier, A.-S.; Mijouin, L.; Hoffmann, B.; Sergent, J.-A.; Bouffartigues, E.; Boireau, W.; et al. Pseudomonas Aeruginosa Expresses a Functional Human Natriuretic Peptide Receptor Ortholog: Involvement in Biofilm Formation. mBio 2015, 6, e01033-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elo, S.; Saxelin, M.; Salminen, S. Attachment of Lactobacillus Casei Strain GG to Human Colon Carcinoma Cell Line Caco-2: Comparison with Other Dairy Strains. Lett. Appl. Microbiol. 1991, 13, 154–156. [Google Scholar] [CrossRef]
- Chauviere, G.; Coconnier, M.-H.; Kerneis, S.; Fourniat, J.; Servin, A.L. Adhesion of Human Lactobacillus Acidophilus Strain LB to Human Enterocyte-like Caco-2 Cells. J. Gen. Microbiol. 1992, 138, 1689–1696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernet, M.F.; Brassart, D.; Neeser, J.R.; Servin, A.L. Lactobacillus acidophilus LA 1 Binds to Cultured Human Intestinal Cell Lines and Inhibits Cell Attachment and Cell Invasion by Enterovirulent Bacteria. Gut 1994, 35, 483–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nyrönen, T.; Pihlavisto, M.; Peltonen, J.M.; Hoffrén, A.-M.; Varis, M.; Salminen, T.; Wurster, S.; Marjamäki, A.; Kanerva, L.; Katainen, E.; et al. Molecular Mechanism for Agonist-Promoted α2A-Adrenoceptor Activation by Norepinephrine and Epinephrine. Mol. Pharmacol. 2001, 59, 1343–1354. [Google Scholar] [CrossRef]
- Lyte, M. Microbial endocrinology: A personal journey. In Microbial Endocrinology; Springer: Berlin/Heidelberg, Germany, 2010; pp. 1–16. [Google Scholar]
- Rimoldi, M.; Chieppa, M.; Salucci, V.; Avogadri, F.; Sonzogni, A.; Sampietro, G.M.; Nespoli, A.; Viale, G.; Allavena, P.; Rescigno, M. Intestinal Immune Homeostasis Is Regulated by the Crosstalk between Epithelial Cells and Dendritic Cells. Nat. Immunol. 2005, 6, 507–514. [Google Scholar] [CrossRef]
- Saarela, M.; Mogensen, G.; Fondén, R.; Mättö, J.; Mattila-Sandholm, T. Probiotic Bacteria: Safety, Functional and Technological Properties. J. Biotechnol. 2000, 84, 197–215. [Google Scholar] [CrossRef]
- Lyte, J.M.; Shrestha, S.; Wagle, B.R.; Liyanage, R.; Martinez, D.A.; Donoghue, A.M.; Daniels, K.M.; Lyte, M. Serotonin Modulates Campylobacter Jejuni Physiology and in Vitro Interaction with the Gut Epithelium. Poult. Sci. 2021, 100, 100944. [Google Scholar] [CrossRef]
- Vlisidou, I.; Lyte, M.; van Diemen, P.M.; Hawes, P.; Monaghan, P.; Wallis, T.S.; Stevens, M.P. The Neuroendocrine Stress Hormone Norepinephrine Augments Escherichia Coli O157:H7-Induced Enteritis and Adherence in a Bovine Ligated Ileal Loop Model of Infection. Infect. Immun. 2004, 72, 5446–5451. [Google Scholar] [CrossRef] [Green Version]
- Hegde, M.; Wood, T.K.; Jayaraman, A. The Neuroendocrine Hormone Norepinephrine Increases Pseudomonas Aeruginosa PA14 Virulence through the Las Quorum-Sensing Pathway. Appl. Microbiol. Biotechnol. 2009, 84, 763–776. [Google Scholar] [CrossRef]
- Dong, Y.; Liu, J.; Pang, M.; Du, H.; Wang, N.; Awan, F.; Lu, C.; Liu, Y. Catecholamine-Stimulated Growth of Aeromonas Hydrophila Requires the TonB2 Energy Transduction System but Is Independent of the Amonabactin Siderophore. Front. Cell. Infect. Microbiol. 2016, 6, 183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srinivasan, B.; Kolli, A.R.; Esch, M.B.; Abaci, H.E.; Shuler, M.L.; Hickman, J.J. TEER Measurement Techniques for in Vitro Barrier Model Systems. J. Lab. Autom. 2015, 20, 107–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obrenovich, M. Leaky Gut, Leaky Brain? Microorganisms 2018, 6, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneeberger, E.E.; Lynch, R.D. The Tight Junction: A Multifunctional Complex. Am. J. Physiol.-Cell Physiol. 2004, 286, C1213–C1228. [Google Scholar] [CrossRef] [PubMed]
- Lodemann, U.; Strahlendorf, J.; Schierack, P.; Klingspor, S.; Aschenbach, J.R.; Martens, H. Effects of the Probiotic Enterococcus Faecium and Pathogenic Escherichia Coli Strains in a Pig and Human Epithelial Intestinal Cell Model. Scientifica 2015, 2015, 235184. [Google Scholar] [CrossRef] [Green Version]
- Ohland, C.L.; MacNaughton, W.K. Probiotic Bacteria and Intestinal Epithelial Barrier Function. Am. J. Physiol.-Gastrointest. Liver Physiol. 2010, 298, G807–G819. [Google Scholar] [CrossRef] [Green Version]
- Resta-Lenert, S.; Barrett, K.E. Live Probiotics Protect Intestinal Epithelial Cells from the Effects of Infection with Enteroinvasive Escherichia Coli (EIEC). Gut 2003, 52, 988–997. [Google Scholar] [CrossRef] [Green Version]
- Anderson, R.C.; Cookson, A.L.; McNabb, W.C.; Park, Z.; McCann, M.J.; Kelly, W.J.; Roy, N.C. Lactobacillus Plantarum MB452 Enhances the Function of the Intestinal Barrier by Increasing the Expression Levels of Genes Involved in Tight Junction Formation. BMC Microbiol. 2010, 10, 316. [Google Scholar] [CrossRef] [Green Version]
- Zyrek, A.A.; Cichon, C.; Helms, S.; Enders, C.; Sonnenborn, U.; Schmidt, M.A. Molecular Mechanisms Underlying the Probiotic Effects of Escherichia Coli Nissle 1917 Involve ZO-2 and PKC? Redistribution Resulting in Tight Junction and Epithelial Barrier Repair. Cell. Microbiol. 2007, 9, 804–816. [Google Scholar] [CrossRef]
- Ewaschuk, J.B.; Diaz, H.; Meddings, L.; Diederichs, B.; Dmytrash, A.; Backer, J.; Looijer-van Langen, M.; Madsen, K.L. Secreted Bioactive Factors from Bifidobacterium Infantis Enhance Epithelial Cell Barrier Function. Am. J. Physiol.-Gastrointest. Liver Physiol. 2008, 295, G1025–G1034. [Google Scholar] [CrossRef] [Green Version]
- Cogan, T.A.; Thomas, A.O.; Rees, L.E.N.; Taylor, A.H.; Jepson, M.A.; Williams, P.H.; Ketley, J.; Humphrey, T.J. Norepinephrine Increases the Pathogenic Potential of Campylobacter Jejuni. Gut 2007, 56, 1060–1065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aroori, S.V.; Cogan, T.A.; Humphrey, T.J. Effect of Noradrenaline on the Virulence Properties of Campylobacter Species. Int. J. Microbiol. 2014, 2014, 279075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christensen, H.R.; Frøkiaer, H.; Pestka, J.J. Lactobacilli Differentially Modulate Expression of Cytokines and Maturation Surface Markers in Murine Dendritic Cells. J. Immunol. 2002, 168, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Meijerink, M.; van Hemert, S.; Taverne, N.; Wels, M.; de Vos, P.; Bron, P.A.; Savelkoul, H.F.; van Bilsen, J.; Kleerebezem, M.; Wells, J.M. Identification of Genetic Loci in Lactobacillus Plantarum That Modulate the Immune Response of Dendritic Cells Using Comparative Genome Hybridization. PLoS ONE 2010, 5, e10632. [Google Scholar] [CrossRef] [Green Version]
- Parvez, S.; Malik, K.A.; Ah Kang, S.; Kim, H.Y. Probiotics and Their Fermented Food Products Are Beneficial for Health. J. Appl. Microbiol. 2006, 100, 1171–1185. [Google Scholar] [CrossRef]
- Tuohy, K.M.; Probert, H.M.; Smejkal, C.W.; Gibson, G.R. Using Probiotics and Prebiotics to Improve Gut Health. Drug Discov. Today 2003, 8, 692–700. [Google Scholar] [CrossRef]
- Rescigno, M. Intestinal dendritic cells. In Advances in Immunology; Elsevier: Amsterdam, The Netherlands, 2010; Volume 107, pp. 109–138. ISBN 978-0-12-381300-8. [Google Scholar]
- Idriss, H.T.; Naismith, J.H. TNF Alpha and the TNF Receptor Superfamily: Structure-Function Relationship(s). Microsc. Res. Tech. 2000, 50, 184–195. [Google Scholar] [CrossRef]
- Moore, K.W.; de Waal Malefyt, R.; Coffman, R.L.; O’Garra, A. Interleukin-10 and the Interleukin-10 Receptor. Annu. Rev. Immunol. 2001, 19, 683–765. [Google Scholar] [CrossRef]
- Kühn, R.; Löhler, J.; Rennick, D.; Rajewsky, K.; Müller, W. Interleukin-10-Deficient Mice Develop Chronic Enterocolitis. Cell 1993, 75, 263–274. [Google Scholar] [CrossRef]
- de Moreno de Leblanc, A.; Del Carmen, S.; Zurita-Turk, M.; Santos Rocha, C.; van de Guchte, M.; Azevedo, V.; Miyoshi, A.; Leblanc, J.G. Importance of IL-10 Modulation by Probiotic Microorganisms in Gastrointestinal Inflammatory Diseases. ISRN Gastroenterol. 2011, 2011, 892971. [Google Scholar] [CrossRef] [Green Version]
- Fabersani, E.; Abeijon-Mukdsi, M.C.; Ross, R.; Medina, R.; González, S.; Gauffin-Cano, P. Specific Strains of Lactic Acid Bacteria Differentially Modulate the Profile of Adipokines In Vitro. Front. Immunol. 2017, 8, 266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreira, C.G.; Weinshenker, D.; Sperandio, V. QseC Mediates Salmonella Enterica Serovar Typhimurium Virulence In Vitro and In Vivo. Infect. Immun. 2010, 78, 914–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reading, N.C.; Rasko, D.A.; Torres, A.G.; Sperandio, V. The Two-Component System QseEF and the Membrane Protein QseG Link Adrenergic and Stress Sensing to Bacterial Pathogenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 5889–5894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, A.; Russell, R.M.; Pifer, R.; Menezes-Garcia, Z.; Cuesta, S.; Narayanan, S.; MacMillan, J.B.; Sperandio, V. The Serotonin Neurotransmitter Modulates Virulence of Enteric Pathogens. Cell Host Microbe 2020, 28, 41–53.e8. [Google Scholar] [CrossRef]
- Stock, A.M.; Robinson, V.L.; Goudreau, P.N. Two-Component Signal Transduction. Annu. Rev. Biochem. 2000, 69, 183–215. [Google Scholar] [CrossRef] [Green Version]
- Lyte, M.; Brown, D.R. Evidence for PMAT- and OCT-like Biogenic Amine Transporters in a Probiotic Strain of Lactobacillus: Implications for Interkingdom Communication within the Microbiota-Gut-Brain Axis. PLoS ONE 2018, 13, e0191037. [Google Scholar] [CrossRef] [Green Version]
- Lyte, M.; Arulanandam, B.P.; Frank, C.D. Production of Shiga-like Toxins by Escherichia Coli O157:H7 Can Be Influenced by the Neuroendocrine Hormone Norepinephrine. J. Lab. Clin. Med. 1996, 128, 392–398. [Google Scholar] [CrossRef]
- Ünal, C.M.; Singh, B.; Fleury, C.; Singh, K.; Chávez de Paz, L.; Svensäter, G.; Riesbeck, K. QseC Controls Biofilm Formation of Non-Typeable Haemophilus Influenzae in Addition to an AI-2-Dependent Mechanism. Int. J. Med. Microbiol. 2012, 302, 261–269. [Google Scholar] [CrossRef] [Green Version]
- Novak, E.A.; Shao, H.; Daep, C.A.; Demuth, D.R. Autoinducer-2 and QseC Control Biofilm Formation and In Vivo Virulence of Aggregatibacter Actinomycetemcomitans. Infect. Immun. 2010, 78, 2919–2926. [Google Scholar] [CrossRef] [Green Version]
- Dubrac, S.; Boneca, I.G.; Poupel, O.; Msadek, T. New Insights into the WalK/WalR (YycG/YycF) Essential Signal Transduction Pathway Reveal a Major Role in Controlling Cell Wall Metabolism and Biofilm Formation in Staphylococcus Aureus. J. Bacteriol. 2007, 189, 8257–8269. [Google Scholar] [CrossRef] [Green Version]
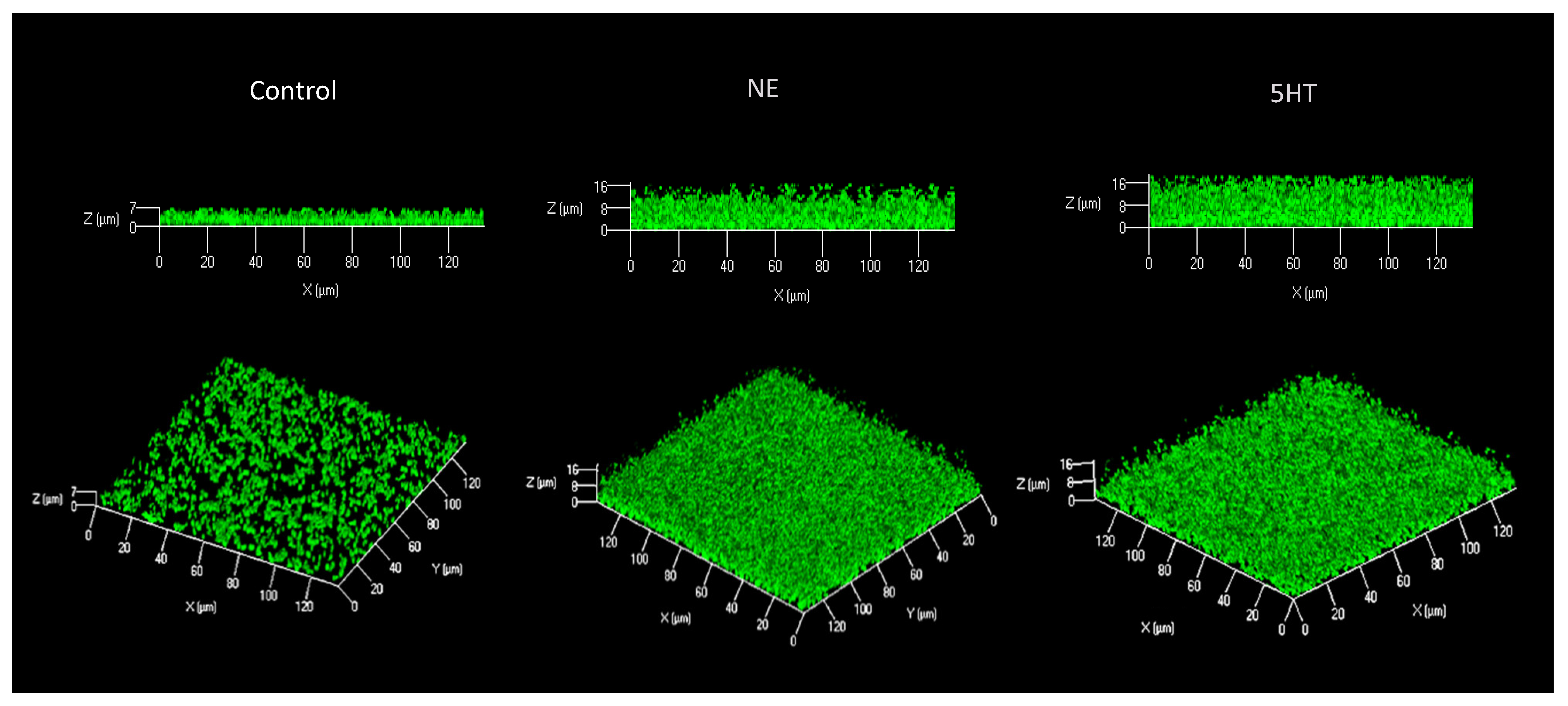





| Treatment | Biovolumes (µm3/µm2) | Average Thickness (µm) | Maximum Thickness (µm) |
|---|---|---|---|
| Control | 0.63 ± 0.27 | 1.4 ± 0.8 | 13.2 ± 2.3 |
| 100 µM NE | 0.98 ± 0.28 ** | 2.3 ± 1.0 * | 18.6 ± 4.1 *** |
| 50 µM 5HT | 1.24 ± 0.48 ** | 3.0 ± 1.2 *** | 18.4 ± 4.1 *** |
| Molecule | Binding Energy (kcal/mol) | Number of Conformations | Amino Acids Involved |
|---|---|---|---|
| NE | −6.35 | 7 | ILE63, ASP78, GLU82, ASN116, GLN118, THR127 |
| 5HT | −6.12 | 5 | ILE63, GLU64, ASP79, GLU82, ASN116, VAL117 |
| Phentolamine | −3.73 | 2 | ILE63, GLU64, ASP79, GLU82, VAL117, GLN118, PHE153, ALA155 |
| Propranolol | −5.78 | 4 | ILE63, GLU64, ARG66, GLU82, ASP97, ASN116, ILE129, PHE153, ALA155 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scardaci, R.; Bietto, F.; Racine, P.-J.; Boukerb, A.M.; Lesouhaitier, O.; Feuilloley, M.G.J.; Scutera, S.; Musso, T.; Connil, N.; Pessione, E. Norepinephrine and Serotonin Can Modulate the Behavior of the Probiotic Enterococcus faecium NCIMB10415 towards the Host: Is a Putative Surface Sensor Involved? Microorganisms 2022, 10, 487. https://doi.org/10.3390/microorganisms10030487
Scardaci R, Bietto F, Racine P-J, Boukerb AM, Lesouhaitier O, Feuilloley MGJ, Scutera S, Musso T, Connil N, Pessione E. Norepinephrine and Serotonin Can Modulate the Behavior of the Probiotic Enterococcus faecium NCIMB10415 towards the Host: Is a Putative Surface Sensor Involved? Microorganisms. 2022; 10(3):487. https://doi.org/10.3390/microorganisms10030487
Chicago/Turabian StyleScardaci, Rossella, Francesca Bietto, Pierre-Jean Racine, Amine M. Boukerb, Olivier Lesouhaitier, Marc G. J. Feuilloley, Sara Scutera, Tiziana Musso, Nathalie Connil, and Enrica Pessione. 2022. "Norepinephrine and Serotonin Can Modulate the Behavior of the Probiotic Enterococcus faecium NCIMB10415 towards the Host: Is a Putative Surface Sensor Involved?" Microorganisms 10, no. 3: 487. https://doi.org/10.3390/microorganisms10030487
APA StyleScardaci, R., Bietto, F., Racine, P.-J., Boukerb, A. M., Lesouhaitier, O., Feuilloley, M. G. J., Scutera, S., Musso, T., Connil, N., & Pessione, E. (2022). Norepinephrine and Serotonin Can Modulate the Behavior of the Probiotic Enterococcus faecium NCIMB10415 towards the Host: Is a Putative Surface Sensor Involved? Microorganisms, 10(3), 487. https://doi.org/10.3390/microorganisms10030487






