Distribution and Characteristics of Listeria spp. in Pigs and Pork Production Chains in Germany
Abstract
:1. Introduction
2. Materials and Methods
2.1. Origin of Isolates
2.2. Sequencing and Bioinformatic Analysis of Sequences
2.3. Antimicrobial Susceptibility Testing
3. Results
3.1. Presence of Listeria spp. in Pigs in Slaughterhouse A and B
3.2. Presence of Listeria spp. in Environmental Samples in Slaughterhouse A
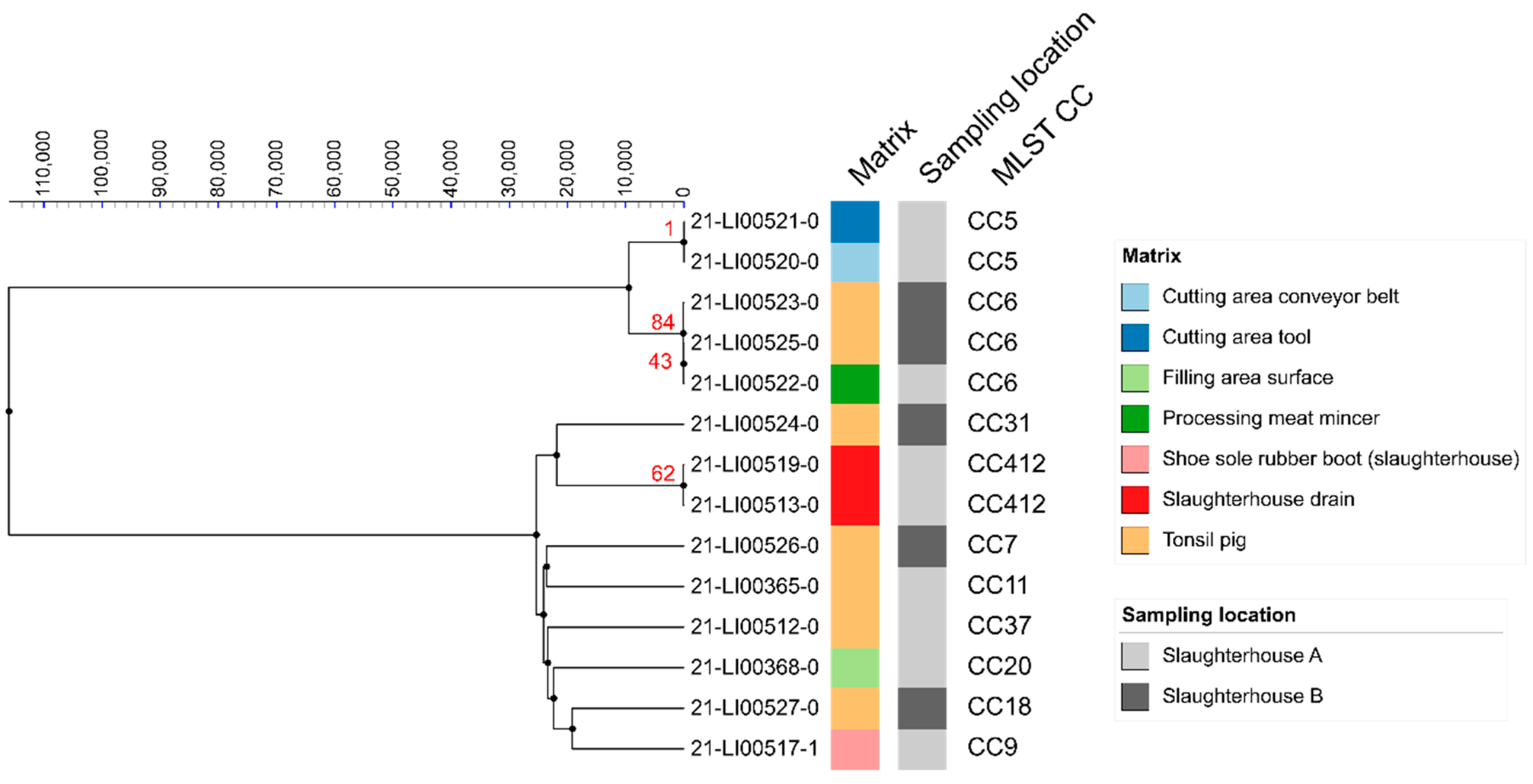
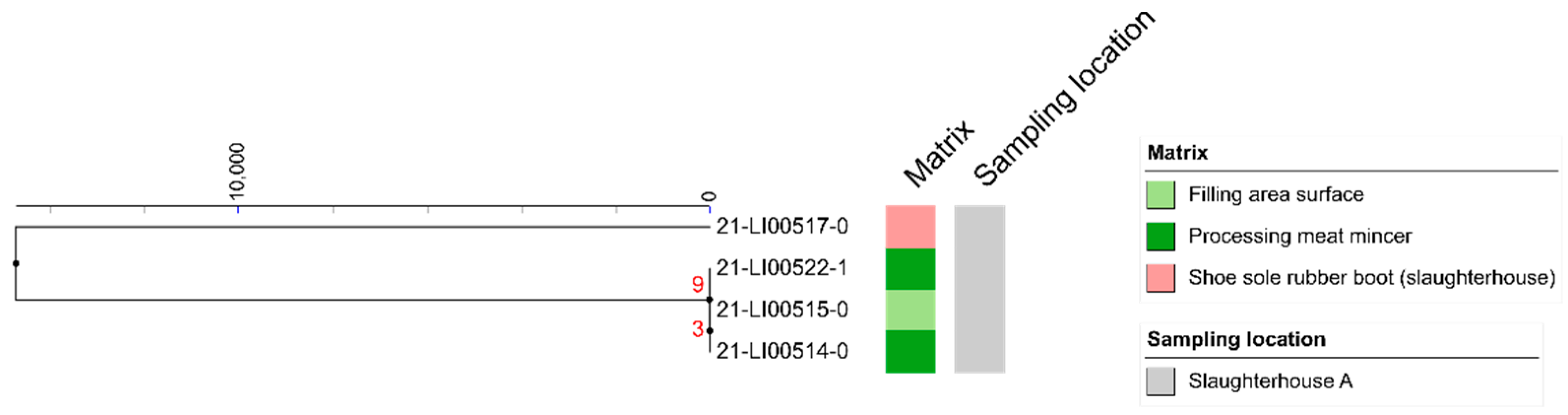
3.3. MLST Analyses
3.4. SNP Analyses
3.5. Antimicrobial Resistance Genes
3.6. Virulence Genes
3.7. Antimicrobial Susceptibility Testing Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Buchanan, R.L.; Gorris, L.; Hayman, M.M.; Jackson, T.C.; Whiting, R.C. A review of Listeria monocytogenes: An update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control 2017, 75, 1–13. [Google Scholar] [CrossRef]
- Leclercq, A.; Moura, A.; Vales, G.; Tessaud-Rita, N.; Aguilhon, C.; Lecuit, M. Listeria thailandensis sp. nov. Int. J. Syst. Evol. Microbiol. 2019, 69, 74–81. [Google Scholar] [CrossRef]
- Orsi, R.H.; den Bakker, H.C.; Wiedmann, M. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 2011, 301, 79–96. [Google Scholar] [CrossRef] [PubMed]
- Perrin, M.; Bemer, M.; Delamare, C. Fatal Case of Listeria innocua Bacteremia. J. Clin. Microbiol. 2003, 41, 5308–5309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Favaro, M.; Sarmati, L.; Sancesario, G.; Fontana, C. First case of Listeria innocua meningitis in a patient on steroids and eternecept. JMM Case Rep. 2014, 1, e003103. [Google Scholar] [CrossRef] [Green Version]
- Rocourt, J.; Hof, H.; Schrettenbrunner, A.; Malinverni, R.; Bille, J. Méningite purulente aiguë à Listeria seeligeri chez un adulte immunocompétent. Schweiz. Med. Wochenschr. 1986, 116, 248–251. (In French) [Google Scholar]
- Liu, D.; Lawrence, M.L.; Wiedmann, M.; Gorski, L.; Mandrell, R.E.; Ainsworth, A.J.; Austin, F.W. Listeria monocytogenes Subgroups IIIA, IIIB, and IIIC Delineate Genetically Distinct Populations with Varied Pathogenic Potential. J. Clin. Microbiol. 2006, 44, 4229–4233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, T.J.; Ducey, T.; Usgaard, T.; Dunn, K.; Bielawski, J. Multilocus Genotyping Assays for Single Nucleotide Polymorphism-Based Subtyping of Listeria monocytogenes Isolates. Appl. Environ. Microbiol. 2008, 74, 7629–7642. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, O.F.; Skouboe, P.; Dons, L.; Rossen, L.; Olsen, J.E. Listeria monocytogenes exists in at least three evolutionary lines: evidence from flagellin, invasive associated protein and listeriolysin O genes. Microbiology 1995, 141, 2053–2061. [Google Scholar] [CrossRef] [Green Version]
- Roberts, A.; Nightingale, K.; Jeffers, G.; Fortes, E.; Kongo, J.M.; Wiedmann, M. Genetic and phenotypic characterization of Listeria monocytogenes lineage III. Microbiology 2006, 152, 685–693. [Google Scholar] [CrossRef] [Green Version]
- Allerberger, F.; Huhulescu, S. Pregnancy related listeriosis: treatment and control. Expert Rev. Anti-Infec. Ther. 2014, 13, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Ragon, M.; Wirth, T.; Hollandt, F.; Lavenir, R.; Lecuit, M.; Le Monnier, A.; Brisse, S. A New Perspective on Listeria monocytogenes Evolution. PLOS Pathog. 2008, 4, e1000146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salcedo, C.; Arreaza, L.; Alcalá, B.; de la Fuente, L.; Vázquez, J.A.; Kazor, C.E.; Mitchell, P.M.; Lee, A.M.; Stokes, L.N.; Loesche, W.J.; et al. Development of a Multilocus Sequence Typing Method for Analysis of Listeria monocytogenes Clones. J. Clin. Microbiol. 2003, 41, 558–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chenal-Francisque, V.; Lopez, J.; Cantinelli, T.; Caro, V.; Tran, C.; Leclercq, A.; Lecuit, M.; Brisse, S. Worldwide Distribution of Major Clones of Listeria monocytogenes. Emerg. Infect. Dis. 2011, 17, 1110–1112. [Google Scholar] [CrossRef]
- Iida, T.; Kanzaki, M.; Nakama, A.; Kokubo, Y.; Maruyama, T.; Kaneuchi, C. Detection of Listeria monocytogenes in Humans, Animals and Foods. J. Veter- Med Sci. 1998, 60, 1341–1343. [Google Scholar] [CrossRef] [Green Version]
- Kanuganti, S.R.; Wesley, I.V.; Reddy, P.G.; Mckean, J.; Hurd, H.S. Detection of Listeria monocytogenes in Pigs and Pork. J. Food Prot. 2002, 65, 1470–1474. [Google Scholar] [CrossRef]
- Hellström, S.; Laukkanen-Ninios, R.; Siekkinen, K.-M.; Ranta, J.; Maijala, R.; Korkeala, H. Listeria monocytogenes Contamination in Pork Can Originate from Farms. J. Food Prot. 2010, 73, 641–648. [Google Scholar] [CrossRef]
- Oswaldi, V.; Dzierzon, J.; Thieme, S.; Merle, R.; Meemken, D. Slaughter pigs as carrier of Listeria monocytogenes in Germany. J. Consum. Prot. Food Saf. 2021, 16, 109–115. [Google Scholar] [CrossRef]
- Figueroa-López, A.M.; Maldonado-Mendoza, I.; López-Cervantes, J.; Verdugo-Fuentes, A.A.; Ruiz-Vega, D.A.; Cantú-Soto, E.U. Prevalence and characterization of Listeria monocytogenes isolated from pork meat and on inert surfaces. Braz. J. Microbiol. 2019, 50, 817–824. [Google Scholar] [CrossRef]
- Wang, Y.; Ji, Q.; Li, S.; Liu, M. Prevalence and Genetic Diversity of Listeria monocytogenes Isolated from Retail Pork in Wuhan, China. Front. Microbiol. 2021, 12, 459. [Google Scholar] [CrossRef]
- Duranti, A.; Sabbatucci, M.; Blasi, G.; Acciari, V.A.; Ancora, M.; Bella, A.; Busani, L.; Centorame, P.; Cammà, C.; Conti, F.; et al. A severe outbreak of listeriosis in central Italy with a rare pulsotype associated with processed pork products. J. Med Microbiol. 2018, 67, 1351–1360. [Google Scholar] [CrossRef] [PubMed]
- Giovannacci, I.; Ragimbeau, C.; Queguiner, S.; Salvat, G.; Vendeuvre, J.-L.; Carlier, V.; Ermel, G. Listeria monocytogenes in pork slaughtering and cutting plants: use of RAPD, PFGE and PCR–REA for tracing and molecular epidemiology. Int. J. Food Microbiol. 1999, 53, 127–140. [Google Scholar] [CrossRef]
- Boerlin, P.; Piffaretti, J.C. Typing of human, animal, food, and environmental isolates of Listeria monocytogenes by multilocus enzyme electrophoresis. Appl. Environ. Microbiol. 1991, 57, 1624–1629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Félix, B.; Feurer, C.; Maillet, A.; Guillier, L.; Boscher, E.; Kerouanton, A.; Denis, M.; Roussel, S. Population Genetic Structure of Listeria monocytogenes Strains Isolated from the Pig and Pork Production Chain in France. Front. Microbiol. 2018, 9, 684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demaître, N.; Rasschaert, G.; De Zutter, L.; Geeraerd, A.; De Reu, K. Genetic Listeria monocytogenes Types in the Pork Processing Plant Environment: from Occasional Introduction to Plausible Persistence in Harborage Sites. Pathogens 2021, 10, 717. [Google Scholar] [CrossRef]
- Deurenberg, R.H.; Bathoorn, E.; Chlebowicz, M.A.; Couto, N.; Ferdous, M.; García-Cobos, S.; Kooistra-Smid, A.M.; Raangs, E.C.; Rosema, S.; Veloo, A.C.; et al. Application of next generation sequencing in clinical microbiology and infection prevention. J. Biotechnol. 2016, 243, 16–24. [Google Scholar] [CrossRef]
- Maury, M.M.; Tsai, Y.-H.; Charlier, C.; Touchon, M.; Chenal-Francisque, V.; Leclercq, A.; Criscuolo, A.; Gaultier, C.; Roussel, S.; Brisabois, S.R.A.; et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 2016, 48, 308–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orsi, R.H.; Wiedmann, M. Characteristics and distribution of Listeria spp., including Listeria species newly described since 2009. Appl. Microbiol. Biotechnol. 2016, 100, 5273–5287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchrieser, C.; Rusniok, C.; Kunst, F.; Cossart, P.; Glaser, P. The Listeria Consortium Comparison of the genome sequences of Listeria monocytogenes and Listeria innocua: clues for evolution and pathogenicity. FEMS Immunol. Med. Microbiol. 2003, 35, 207–213. [Google Scholar] [CrossRef] [Green Version]
- Teuber, M. Spread of antibiotic resistance with food-borne pathogens. Exp. 1999, 56, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Troxler, R.; von Graevenitz, A.; Funke, G.; Wiedemann, B.; Stock, I. Natural antibiotic susceptibility of Listeria species: L. grayi, L. innocua, L. ivanovii, L. monocytogenes, L. seeligeri and L. welshimeri strains. Clin. Microbiol. Infect. 2000, 6, 525–535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poyart, C.; Carlier, C.; Trieu-Cuot, P.; Courvalin, P.; Courtieu, A.-L. Transferable plasmid-mediated antibiotic resistance in Listeria monocytogenes. Lancet 1990, 335, 1422–1426. [Google Scholar] [CrossRef]
- Olaimat, A.N.; Al-Holy, M.A.; Shahbaz, H.M.; Al-Nabulsi, A.A.; Abu Ghoush, M.H.; Osaili, T.M.; Ayyash, M.M.; Holley, R.A. Emergence of Antibiotic Resistance in Listeria monocytogenes Isolated from Food Products: A Comprehensive Review. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1277–1292. [Google Scholar] [CrossRef] [Green Version]
- Deneke, C.; Brendebach, H.; Uelze, L.; Borowiak, M.; Malorny, B.; Tausch, S. Species-Specific Quality Control, Assembly and Contamination Detection in Microbial Isolate Sequences with AQUAMIS. Genes 2021, 12, 644. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- CLSI. Methods for Antimicrobial Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria Isolated from Animals, VET06, 1st ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017. [Google Scholar]
- CLSI. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria, M45, 3rd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, M100, 31st ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2021. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, VET01S, 5th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Chesneau, O.; Ligeret, H.; Hosan-Aghaie, N.; Morvan, A.; Dassa, E. Molecular Analysis of Resistance to Streptogramin a Compounds Conferred by the Vga Proteins of Staphylococci. Antimicrob. Agents Chemother. 2005, 49, 973–980. [Google Scholar] [CrossRef] [Green Version]
- Crowe-McAuliffe, C.; Murina, V.; Turnbull, K.J.; Kasari, M.; Mohamad, M.; Polte, C.; Takada, H.; Vaitkevicius, K.; Johansson, J.; Ignatova, Z.; et al. Structural basis of ABCF-mediated resistance to pleuromutilin, lincosamide, and streptogramin A antibiotics in Gram-positive pathogens. Nat. Commun. 2021, 12, 1–14. [Google Scholar] [CrossRef]
- Berrang, M.E.; Frank, J.F. Generation of Airborne Listeria innocua from Model Floor Drains. J. Food Prot. 2012, 75, 1328–1331. [Google Scholar] [CrossRef]
- Maury, M.M.; Bracq-Dieye, H.; Huang, L.; Vales, G.; Lavina, M.; Thouvenot, P.; Disson, O.; Leclercq, A.; Brisse, S.; Lecuit, M. Hypervirulent Listeria monocytogenes clones’ adaption to mammalian gut accounts for their association with dairy products. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Martin, B.; Perich, A.; Gómez, D.; Yangüela, J.; Rodriguez, A.; Garriga, M.; Aymerich, T. Diversity and distribution of Listeria monocytogenes in meat processing plants. Food Microbiol. 2014, 44, 119–127. [Google Scholar] [CrossRef]
- Kubicová, Z.; Roussel, S.; Félix, B.; Cabanová, L. Genomic Diversity of Listeria monocytogenes Isolates from Slovakia (2010 to 2020). Front. Microbiol. 2021, 12, 3223. [Google Scholar] [CrossRef]
- Halbedel, S.; Prager, R.; Fuchs, S.; Trost, E.; Werner, G.; Flieger, A. Whole-Genome Sequencing of Recent Listeria monocytogenes Isolates from Germany Reveals Population Structure and Disease Clusters. J. Clin. Microbiol. 2018, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meloni, D.; Piras, F.; Mureddu, A.; Fois, F.; Consolati, S.G.; Lamon, S.; Mazzette, R. Listeria monocytogenes in Five Sardinian Swine Slaughterhouses: Prevalence, Serotype, and Genotype Characterization. J. Food Prot. 2013, 76, 1863–1867. [Google Scholar] [CrossRef]
- Lariviére-Gauthier, G.; Letellier, A.; Kérouanton, A.; Bekal, S.; Quessy, S.; Fournaise, S.; Fravalo, P. Analysis of Listeria monocytogenes Strain Distribution in a Pork Slaughter and Cutting Plant in the Province of Quebec. J. Food Prot. 2014, 77, 2121–2128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoller, A.; Stevens, M.J.A.; Stephan, R.; Guldimann, C. Characteristics of Listeria Monocytogenes Strains Persisting in a Meat Processing Facility over a 4-Year Period. Pathogens 2019, 8, 32. [Google Scholar] [CrossRef] [Green Version]
- Zakrzewski, A.J.; Chajęcka-Wierzchowska, W.; Zadernowska, A.; Podlasz, P. Virulence Characterization of Listeria monocytogenes, Listeria innocua, and Listeria welshimeri Isolated from Fish and Shrimp Using In Vivo Early Zebrafish Larvae Models and Molecular Study. Pathogens 2020, 9, 1028. [Google Scholar] [CrossRef]
- Lake, F.B.; van Overbeek, L.S.; Baars, J.J.; Koomen, J.; Abee, T.; Besten, H.M.D. Genomic characteristics of Listeria monocytogenes isolated during mushroom (Agaricus bisporus) production and processing. Int. J. Food Microbiol. 2021, 360, 109438. [Google Scholar] [CrossRef] [PubMed]
- Domann, E.; Wehland, J.; Rohde, M.; Pistor, S.; Hartl, M.; Goebel, W.; Leimeister-Wächter, M.; Wuenscher, M.; Chakraborty, T. A novel bacterial virulence gene in Listeria monocytogenes required for host cell microfilament interaction with homology to the proline-rich region of vinculin. EMBO J. 1992, 11, 1981–1990. [Google Scholar] [CrossRef] [PubMed]
- Volokhov, D.V.; Duperrier, S.; Neverov, A.A.; George, J.; Buchrieser, C.; Hitchins, A.D. The Presence of the Internalin Gene in Natural Atypically Hemolytic Listeria innocua Strains Suggests Descent from L. monocytogenes. Appl. Environ. Microbiol. 2007, 73, 1928–1939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno, L.Z.; Paixão, R.; Gobbi, D.D.; Raimundo, D.C.; Ferreira, T.P.; Hofer, E.; Matte, M.H.; Moreno, A.M. Characterization of atypical Listeria innocua isolated from swine slaughterhouses and meat markets. Res. Microbiol. 2012, 163, 268–271. [Google Scholar] [CrossRef] [PubMed]
- QS—Qualitätssicherung. 2. Statusbericht zum Antibiotikamonitoring im QS-System. 2019. Available online: https://www.q-s.de/services/files/mediencenter/publikationen/statusbericht-antibiotika/Statusbericht_QS-Antibiotikamonitoring_2019.pdf (accessed on 14 November 2021). (In German).
- Alonso-Hernando, A.; Prieto, M.; García-Fernández, C.; Alonso-Calleja, C.; Capita, R. Increase over time in the prevalence of multiple antibiotic resistance among isolates of Listeria monocytogenes from poultry in Spain. Food Control 2012, 23, 37–41. [Google Scholar] [CrossRef]

| Place of Sampling | Number of Samples Taken | Samples Positive for L. monocytogenes | Samples Positive for L. innocua | Samples Positive for L. welshimeri | ||||
|---|---|---|---|---|---|---|---|---|
| Date 3 | Date 4 | Date 3 | Date 4 | Date 3 | Date 4 | Date 3 | Date 4 | |
| Saws | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| Tonsil removal device | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| Floor drains | 5 | 6 | 1 | 1 | 0 | 0 | 0 | 0 |
| Rubber boots (sole) | 7 | 8 | 1 | 0 | 1 | 0 | 1 | 0 |
| Equipment in slaughter hall | 8 | 8 | 0 | 0 | 0 | 0 | 0 | 0 |
| Cutting plant (product contact surfaces/devices) | 5 | 6 | 0 | 2 | 0 | 0 | 0 | 0 |
| Processing plant (product contact surfaces/devices) | 5 | 7 | 0 | 2 | 0 | 1 | 2 | 1 |
| Total | 77 | 7 | 2 | 4 | ||||
| Sampled Matrix (Sample Number) | Slaughterhouse | Sampling Date | Lineage | MLST ST | MLST CC | Virulence Genes (Total Number) |
|---|---|---|---|---|---|---|
| Pig tonsil (21-LI00365-0) | A | 2 | II | 451 | 11 | actA, ami, aut, bsh, clpC, clpE, clpP, fbpA, gtcA, hly, hpt, iap/cwhA, inlA, inlB, inlC, inlF, inlJ, inlK, lap, lapB, lntA, lpeA, lpA1, lspA, mpl, oatA, pdgA, plcA, plcB, prfA, prsA2, vip (32) |
| Pig tonsil (21-LI00512-0) | A | 3 | II | 37 | 37 | actA, ami, aut, bsh, clpC, clpE, clpP, fbpA, gtcA, hly, hpt, iap/cwhA, inlA, inlB, inlC, inlF, inlJ, inlK, lap, lapB, lntA, lpeA, lpA1, lspA, mpl, oatA, pdgA, plcA, plcB, prfA, prsA2 (31) |
| Pig tonsil (21-LI00523-0) | B | 5 | I | 6 | 6 | actA, bsh, clpC, clpE, clpP, fbpA, gtcA, hly, hpt, iap/cwhA, inlA, inlB, inlC, inlF, inlK, lap, lapB, llsA, llsB, llsD, llsG, llsH, llsP, llsX, llsY, lntA, lpeA, lpA1, lspA, mpl, oatA, pdgA, plcA, plcB, prfA, prsA2, vip (37) |
| Pig tonsil (21-LI00524-0) | B | 5 | II | 325 | 31 | ami, aut, bsh, clpC, clpE, clpP, fbpA, gtcA, hly, hpt, iap/cwhA, inlA, inlB, inlC, inlF, inlJ, inlK, lap, lntA, lpeA, lpA1, lspA, mpl, oatA, pdgA, plcA, plcB, prfA, prsA2 (29) |
| Pig tonsil (21-LI00525-0) | B | 5 | I | 6 | 6 | actA, bsh, clpC, clpE, clpP, fbpA, gtcA, hly, hpt, iap/cwhA, inlA, inlB, inlC, inlF, inlK, lap, lapB, llsA, llsB, llsD, llsG, llsH, llsP, llsX, llsY, lntA, lpeA, lpA1, lspA, mpl, oatA, pdgA, plcA, plcB, prfA, prsA2, vip (37) |
| Pig tonsil (21-LI00526-0) | B | 5 | II | 7 | 7 | actA, ami, aut, bsh, clpC, clpE, clpP, fbpA, gtcA, hly, hpt, iap/cwhA, inlA, inlB, inlC, inlF, inlJ, inlK, lap, lapB, lntA, lpeA, lpA1, lspA, mpl, oatA, pdgA, plcA, plcB, prfA, prsA2 (31) |
| Pig tonsil (21-LI00527-0) | B | 5 | II | 18 | 18 | actA, ami, aut, bsh, clpC, clpE, clpP, fbpA, gtcA, hly, hpt, iap/cwhA, inlA, inlB, inlC, inlF, inlJ, inlK, lap, lapB, lntA, lpeA, lpA1, lspA, mpl, oatA, pdgA, plcA, plcB, prfA, prsA2, vip (32) |
| Floor drain (21-LI00513-0) | A | 3 | II | 412 | 412 | actA, ami, aut, bsh, clpC, clpE, clpP, fbpA, gtcA, hly, hpt, iap/cwhA, inlA, inlB, inlC, inlF, inlJ, inlK, lap, lntA, lpeA, lpA1, lspA, mpl, oatA, pdgA, plcA, plcB, prfA, prsA2 (30) |
| Sole of rubber boot (21-LI00517-1) | A | 3 | II | 9 | 9 | actA, ami, aut, bsh, clpC, clpE, clpP, fbpA, gtcA, hly, hpt, iap/cwhA, inlA, inlB, inlC, inlF, inlJ, inlK, lap, lapB, lntA, lpeA, lpA1, lspA, mpl, oatA, pdgA, plcA, plcB, prfA, prsA2, vip (32) |
| Floor drain (21-LI00519-0) | A | 4 | II | 412 | 412 | actA, ami, aut, bsh, clpC, clpE, clpP, fbpA, gtcA, hly, hpt, iap/cwhA, inlA, inlB, inlC, inlF, inlJ, inlK, lap, lntA, lpeA, lpA1, lspA, mpl, oatA, pdgA, plcA, plcB, prfA, prsA2 (30) |
| Cutting plant (21-LI00520-0) | A | 4 | I | 5 | 5 | ami, aut, bsh, clpC, clpE, clpP, fbpA, gtcA, hly, hpt, iap/cwhA, inlA, inlB, inlC, inlF, inlK, lap, lapB, lntA, lpeA, lpA1, lspA, mpl, oatA, pdgA, plcA, plcB, prfA, prsA2, vip (30) |
| Cutting plant (21-LI00521-0) | A | 4 | I | 5 | 5 | ami, aut, bsh, clpC, clpE, clpP, fbpA, gtcA, hly, hpt, iap/cwhA, inlA, inlB, inlC, inlF, inlK, lap, lapB, lntA, lpeA, lpA1, lspA, mpl, oatA, pdgA, plcA, plcB, prfA, prsA2, vip (30) |
| Processing plant (21-LI00522-0) | A | 4 | I | 6 | 6 | actA, bsh, clpC, clpE, clpP, fbpA, gtcA, hly, hpt, iap/cwhA, inlA, inlB, inlC, inlF, inlK, lap, lapB, llsA, llsB, llsD, llsG, llsH, llsP, llsX, llsY, lntA, lpeA, lpA1, lspA, mpl, oatA, pdgA, plcA, plcB, prfA, prsA2, vip (37) |
| Processing plant (21-LI00368-0) | A | 4 | II | 20 | 20 | actA, ami, aut, bsh, clpC, clpE, clpP, fbpA, gtcA, hly, hpt, iap/cwhA, inlA, inlB, inlC, inlF, inlJ, inlK, lap, lapB, lntA, lpeA, lpA1, lspA, mpl, oatA, pdgA, plcA, plcB, prfA, prsA2, vip (32) |
| Sampled Matrix (Laboratory Number) | Slaughterhouse | Sampling Date | Species | AMR Genes | Virulence Genes (Total Number) |
|---|---|---|---|---|---|
| Pig intestinal content (21-LI00510-0) | A | 1 | L. innocua | fosX; tet(S) | clpC, clpE, clpP, fbpA, gtcA, iap/cwhA, lap, lpeA, lpA1, lspA, oatA, pdgA, prsA2 (13) |
| Pig intestinal content (21-LI00511-0) | A | 2 | L. innocua | fosX | clpC, clpE, clpP, fbpA, gtcA, iap/cwhA, lap, llsA, llsG, llsH, llsX, lpeA, lpA1, lspA, oatA, pdgA, prsA2 (17) |
| Pig intestinal content (21-LI00518-0) | A | 4 | L. innocua | fosX; tet(M) | clpC, clpE, clpP, fbpA, gtcA, iap/cwhA, lap, lpeA, lpA1, lspA, oatA, pdgA, prsA2 (13) |
| Pig intestinal content (21-LI00367-0) | A | 4 | L. innocua | fosX | clpC, clpE, clpP, fbpA, gtcA, iap/cwhA, lap, llsA, llsG, llsH, llsX, lpeA, lpA1, lspA, oatA, pdgA, prsA2 (17) |
| Pig tonsil (21-LI00366-0) | A | 2 | L. innocua | fosX | clpC, clpE, clpP, fbpA, gtcA, iap/cwhA, lap, lpeA, lpA1, lspA, oatA, pdgA, prsA2 (13) |
| Pig tonsil (21-LI00528-0) | B | 6 | L. innocua | fosX; dfrG; tet(M); ant(6)-Ia | clpC, clpE, clpP, fbpA, gtcA, iap/cwhA, lap, lpeA, lpA1, lspA, oatA, pdgA, prsA2 (13) |
| Pig tonsil (21-LI00370-0) | B | 6 | L. innocua | fosX | clpC, clpE, clpP, fbpA, gtcA, iap/cwhA, lap, lpeA, lpA1, lspA, oatA, pdgA, prsA2 (13) |
| Pig tonsil (21-LI00371-0) | B | 6 | L. innocua | fosX | clpC, clpE, clpP, fbpA, gtcA, iap/cwhA, lap, llsA, llsG, llsH, llsX, lpeA, lpA1, lspA, oatA, pdgA, prsA2 (17) |
| Pig tonsil (21-LI00372-0) | B | 6 | L. innocua | fosX; tet(M); ant(6)-Ia | clpC, clpE, clpP, fbpA, gtcA, iap/cwhA, lap, lpeA, lpA1, lspA, oatA, pdgA, prsA2 (13) |
| Processing plant (21-LI00514-0) | A | 3 | L. welshimeri | fosX | clpC, clpE, clpP, fbpA, lap, lpeA, lpA1, lspA, prsA2 (9) |
| Processing plant (21-LI00515-0) | A | 3 | L. welshimeri | fosX | clpC, clpE, clpP, fbpA, lap, lpeA, lpA1, lspA, prsA2 (9) |
| Sole of rubber boot (21-LI00516-0) | A | 3 | L. innocua | fosX | clpC, clpE, clpP, fbpA, gtcA, iap/cwhA, lap, llsA, llsG, llsH, lpeA, lpA1, lspA, oatA, pdgA, prsA2 (16) |
| Sole of rubber boot (21-LI00517-0) | A | 3 | L. welshimeri | fosX | clpC, clpE, clpP, fbpA, lap, lpeA, lpA1, lspA, prsA2 (9) |
| Processing plant (21-LI00522-1) | A | 4 | L. welshimeri | fosX; vga(G) | clpC, clpE, clpP, fbpA, lap, lpeA, lpA1, lspA, prsA2 (9) |
| Processing plant (21-LI00369-0) | A | 4 | L. innocua | fosX | clpC, clpE, clpP, fbpA, gtcA, iap/cwhA, lap, llsA, llsG, llsH, llsX, lpeA, lpA1, lspA, oatA, pdgA, prsA2 (17) |
| Antimicrobial Agent(s) | Susceptible | Intermediate | Resistant | |||
|---|---|---|---|---|---|---|
| no. | % | no. | % | no. | % | |
| Penicillin | 14 | 100 | 0 | 0 | 0 | 0 |
| Ampicillin | 14 | 100 | 0 | 0 | 0 | 0 |
| Amoxicillin/clavulanic acid | 8 | 57 | 6 | 43 | 0 | 0 |
| Erythromycin | 14 | 100 | 0 | 0 | 0 | 0 |
| Clindamycin | 0 | 0 | 0 | 0 | 14 | 100 |
| Pirlimycin | 0 | 0 | 0 | 0 | 14 | 100 |
| Ciprofloxacin | 14 | 100 | 0 | 0 | 0 | 0 |
| Enrofloxacin | 13 | 93 | 1 | 7 | 0 | 0 |
| Marbofloxacin | 12 | 86 | 2 | 14 | 0 | 0 |
| Gentamicin | 14 | 100 | 0 | 0 | 0 | 0 |
| Streptomycin | 14 | 100 | 0 | 0 | 0 | 0 |
| Neomycin | 14 | 100 | 0 | 0 | 0 | 0 |
| Tetracycline | 14 | 100 | 0 | 0 | 0 | 0 |
| Doxycycline | 3 | 21 | 11 | 79 | 0 | 0 |
| Sulfamethoxazole/trimethoprim | 14 | 100 | 0 | 0 | 0 | 0 |
| Vancomycin | 14 | 100 | 0 | 0 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oswaldi, V.; Lüth, S.; Dzierzon, J.; Meemken, D.; Schwarz, S.; Feßler, A.T.; Félix, B.; Langforth, S. Distribution and Characteristics of Listeria spp. in Pigs and Pork Production Chains in Germany. Microorganisms 2022, 10, 512. https://doi.org/10.3390/microorganisms10030512
Oswaldi V, Lüth S, Dzierzon J, Meemken D, Schwarz S, Feßler AT, Félix B, Langforth S. Distribution and Characteristics of Listeria spp. in Pigs and Pork Production Chains in Germany. Microorganisms. 2022; 10(3):512. https://doi.org/10.3390/microorganisms10030512
Chicago/Turabian StyleOswaldi, Verena, Stefanie Lüth, Janine Dzierzon, Diana Meemken, Stefan Schwarz, Andrea T. Feßler, Benjamin Félix, and Susann Langforth. 2022. "Distribution and Characteristics of Listeria spp. in Pigs and Pork Production Chains in Germany" Microorganisms 10, no. 3: 512. https://doi.org/10.3390/microorganisms10030512
APA StyleOswaldi, V., Lüth, S., Dzierzon, J., Meemken, D., Schwarz, S., Feßler, A. T., Félix, B., & Langforth, S. (2022). Distribution and Characteristics of Listeria spp. in Pigs and Pork Production Chains in Germany. Microorganisms, 10(3), 512. https://doi.org/10.3390/microorganisms10030512







