Draft Genome Sequence of Lactococcus lactis Subsp. cremoris WA2-67: A Promising Nisin-Producing Probiotic Strain Isolated from the Rearing Environment of a Spanish Rainbow Trout (Oncorhynchus mykiss, Walbaum) Farm
Abstract
:1. Introduction
2. Materials and Methods
2.1. Growth Conditions and Genomic DNA Isolation
2.2. Draft Genome Sequencing, Assembly, and Mapping
2.3. Bioinformatic In Silico Analysis
2.3.1. Identification
2.3.2. Probiotic Traits
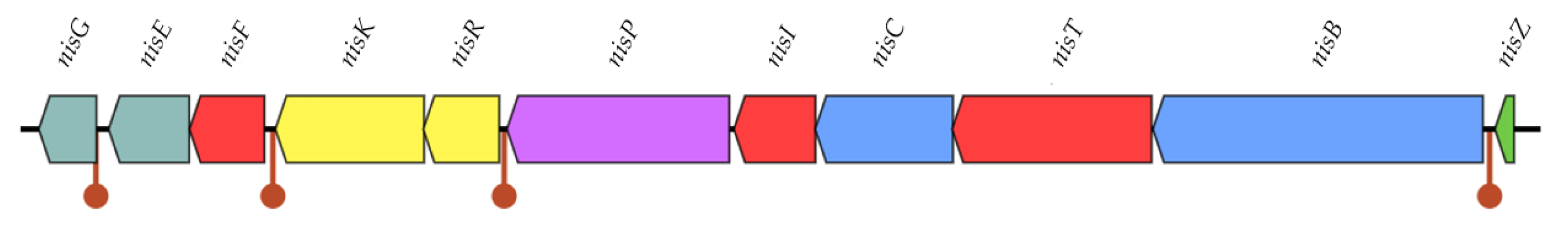
2.3.3. Bacteriocin Production
2.3.4. Mobile Genetic Elements (MGE)
Insertion Sequences (IS)
Plasmids
Prophages
2.3.5. CRISPR/CRISPR-Cas
2.3.6. Transferable Antibiotic Resistances
2.3.7. Virulence Factors
3. Results and Discussion
3.1. Draft Genome Sequencing, Assembly, and Mapping
3.2. Bioinformatic In Silico Analysis
3.2.1. Identification
3.2.2. Probiotic Traits
3.2.3. Bacteriocin Production
3.2.4. MGE (IS, Plasmids and Prophages)
3.2.5. CRISPR/CRISPR-Cas
3.2.6. Transferable Antibiotic Resistances
3.2.7. Virulence Factors
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Defoirdt, T.; Sorgeloos, P.; Bossier, P. Alternatives to antibiotics for the control of bacterial disease in aquaculture. Curr. Opin. Microbiol. 2011, 14, 251–258. [Google Scholar] [CrossRef]
- FAO. The State of World Fisheries and Aquaculture; FAO Fisheries and Aquaculture Department: Rome, Italy, 2020. [Google Scholar]
- Infante-Villamil, S.; Huerlimann, R.; Jerry, D.R. Microbiome diversity and dysbiosis in aquaculture. Rev. Aquac. 2021, 13, 1077–1096. [Google Scholar] [CrossRef]
- United Nations. World Population Prospects: The 2017 Revision, Key Findings and Advance Tables; Population Division, Working Paper No. ESA/P/WP/248; Department of Economic and Social Affairs: New York, NY, USA, 2017. [Google Scholar]
- Cabello, F.C.; Godfrey, H.P.; Buschmann, A.H.; Dölz, H.J. Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. Lancet Infect. Dis. 2016, 16, e127–e133. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, Q.E.; Zhou, X.; Wang, F.; Muurinen, J.; Virta, M.P.; Brandt, K.K.; Zhu, Y. Antibiotic resistome in the livestock and aquaculture industries: Status and solutions. Crit. Rev. Environ. Sci. 2021, 51, 2159–2196. [Google Scholar] [CrossRef]
- Pérez-Sánchez, T.; Ruiz-Zarzuela, I.; de Blas, I.; Balcázar, J.L. Probiotics in aquaculture: A current assessment. Rev. Aquac. 2014, 6, 133–146. [Google Scholar] [CrossRef]
- Gómez-Sala, B.; Feito, J.; Hernández, P.E.; Cintas, L.M. Lactic Acid Bacteria in aquatic environments and their applications. In Lactic Acid Bacteria: Microbiological and Functional Aspects, 5th ed.; Vinderola, G., Ouwehand, A.C., Salminen, S., von Wright, A., Eds.; CRC Press: Boca Raton, FL, USA, 2019; pp. 555–570. [Google Scholar]
- Wu, X.; Teame, T.; Hao, Q.; Ding, Q.; Liu, H.; Ran, C.; Yang, Y.; Zhang, Y.; Zhou, Z.; Duan, M.; et al. Use of a paraprobiotic and postbiotic feed supplement (HWF™) improves the growth performance, composition and function of gut microbiota in hybrid sturgeon (Acipenser baerii × Acipenser schrenckii). Fish Shellfish Immunol. 2020, 104, 36–45. [Google Scholar] [CrossRef]
- Rychen, G.; Aquilina, G.; Azimonti, G.; Bampidis, V.; Bastos, M.L.; Bories, G.; Chesson, A.; Cocconcelli, P.S.; Flachowsky, G.; Gropp, J.; et al. Guidance on the characterization of microorganisms used as feed additives or as production organisms. EFSA J. 2018, 16, e05206. [Google Scholar] [PubMed]
- EFSA (European Food Safety Authority). EFSA statement on the requirements for whole genome sequence analysis of microorganisms intentionally used in the food chain. EFSA J. 2021, 19, e06506. [Google Scholar]
- Fečkaninová, A.; Koščová, J.; Mudroňová, D.; Popelka, P.; Toropilová, J. The use of probiotic bacteria against Aeromonas infection in salmonid aquaculture. Aquaculture 2017, 469, 1–8. [Google Scholar] [CrossRef]
- Cintas, L.M.; Herranz, C.; Hernández, P.E. Natural and heterologous production of bacteriocins. In Prokaryotic Antimicrobial Peptides: From Genes to Applications, 1st ed.; Drider, D., Rebufatt, S., Eds.; Springer: Dordrecht, The Netherlands, 2011; pp. 115–143. [Google Scholar]
- Araújo, C.; Muñoz-Atienza, E.; Nahuelquín, Y.; Poeta, P.; Igrejas, G.; Hernández, P.E.; Herranz, C.; Cintas, L.M. Inhibition of fish pathogens by the microbiota from rainbow trout (Oncorhynchus mykiss, Walbaum) and rearing environment. Anaerobe 2015, 32, 7–14. [Google Scholar] [CrossRef]
- Araújo, C.; Muñoz-Atienza, E.; Ramírez, M.; Poeta, P.; Igrejas, G.; Hernández, P.E.; Herranz, C.; Cintas, L.M. Safety assessment, genetic relatedness and bacteriocin activity of potential probiotic Lactococcus lactis strains from rainbow trout (Oncorhynchus mykiss, Walbaum) and rearing environment. Eur. Food Res. Technol. 2015, 241, 647–662. [Google Scholar] [CrossRef]
- Araújo, C.; Muñoz-Atienza, E.; Pérez-Sánchez, T.; Poeta, P.; Igrejas, G.; Hernández, P.E.; Herranz, C.; Ruiz-Zarzuela, I.; Cintas, L.M. Nisin Z production by Lactococcus lactis subsp. cremoris WA2-67 of aquatic origin as a defense mechanism to protect rainbow trout (Oncorhynchus mykiss, Walbaum) against Lactococcus garvieae. Mar. Biotechnol. 2015, 17, 820–830. [Google Scholar] [CrossRef] [PubMed]
- Velásquez, J.E.; van der Donk, W. Genome mining for ribosomally synthesized natural products. Curr. Opin. Chem. Biol. 2011, 15, 11–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapse, N.G.; Engineer, A.S.; Gowdaman, V.; Wagh, S.; Dhakephalkar, P.K. Functional annotation of the genome unravels probiotic potential of Bacillus coagulans HS243. Genomics 2019, 111, 921–929. [Google Scholar] [CrossRef]
- Hussein, W.E.; Abdelhamid, A.G.; Rocha-Mendoza, D.; Garcia-Cano, I.; Yousef, A.E. Assessment of safety and probiotic traits of Enterococcus durans OSY-EGY, isolated from Egyptian artisanal cheese, using comparative genomics and phenotypic analysis. Front. Microbiol. 2020, 11, 3094. [Google Scholar] [CrossRef]
- Xiao, Y.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Mining genome traits that determine the different gut colonization potential of Lactobacillus and Bifidobacterium species. Microb. Genom. 2021, 7, 000581. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.A.; Korobeynikov, A.; Lapidus, A.; Prjibelski, A.D.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 2013, 20, 714–737. [Google Scholar] [CrossRef] [Green Version]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruit, K.D.; Borodovsky, M.; Ostell, J. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Stothard, P.; Wishart, D.S. Circular genome visualization and exploration using CGView. Bioinformatics 2005, 21, 537–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsen, M.V.; Cosentino, S.; Lukjancenko, O.; Saputra, D.; Rasmussen, S.; Hasman, H.; Sicheritz-Pontén, T.; Aarestrup, F.M.; Ussery, D.W.; Lund, O. Benchmarking of methods for genomic taxonomy. J. Clin. Microbiol. 2014, 52, 1529–1539. [Google Scholar] [CrossRef] [Green Version]
- Hasman, H.; Saputra, D.; Sicheritz-Ponten, T.; Lund, O.; Svendsen, C.A.; Frimodt-Møller, N.; Aarestrup, F.M. Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples. J. Clin. Microbiol. 2014, 52, 139–146. [Google Scholar] [CrossRef] [Green Version]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, 206–214. [Google Scholar] [CrossRef] [PubMed]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Song, W.; Sun, H.; Zhang, C.; Cheng, L.; Peng, Y.; Deng, Z.; Wang, D.; Wang, Y.; Hu, M.; Liu, W.; et al. Prophage Hunter: An integrative hunting tool for active prophages. Nucleic Acids Res. 2019, 47, W74–W80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef] [Green Version]
- Bortolaia, V.; Kaas, R.F.; Ruppe, E.; Roberts, M.C.; Scharwz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.R.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cosentino, S.; Larsen, M.V.; Aarestrup, F.M.; Lund, O. PathogenFinder—Distinguishing friend from foe using bacterial Whole Genome Sequence data. PLoS ONE 2013, 8, e77302. [Google Scholar] [CrossRef]
- Kapse, N.G.; Engineer, A.S.; Gowdaman, V.; Wagh, S.; Dhakephalkar, P.K. Genome profiling for health promoting and disease preventing traits unraveled probiotic potential of Bacillus clausii B106. Microbiol. Biotechnol. Lett. 2018, 46, 334–345. [Google Scholar] [CrossRef]
- Wang, W.; Sun, J.; Liu, C.; Xue, Z. Application of immunostimulants in aquaculture: Current knowledge and future perspectives. Aquac. Res. 2017, 48, 1–23. [Google Scholar] [CrossRef]
- Wang, C.; Chuprom, J.; Wang, Y.; Fu, L. Beneficial bacteria for aquaculture: Nutrition, bacteriostasis and immunoregulation. J. Appl. Microbiol. 2020, 128, 28–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, S.; Sinha, A.; Sahu, C. Effect of probiotic on reproductive performance in female livebearing ornamental fish. Aquac. Res. 2007, 38, 518–526. [Google Scholar] [CrossRef]
- Honeyfield, D.C.; Hinterkopf, J.P.; Fitzsimons, J.D.; Zajicek, J.L.; Brown, S.B. Development of thiamine deficiencies and Early Mortality Syndrome in lake trout by feeding experimental and feral fish diets containing thiaminase. J. Aquac. Anim. Health 2005, 17, 4–12. [Google Scholar] [CrossRef]
- Yossa, R.; Sarker, P.K.; Mock, D.M.; Lall, S.P. Vanderberg, G.W. Current knowledge on biotin nutrition in fish and research perspectives. Rev. Aquac. 2015, 7, 59–73. [Google Scholar] [CrossRef]
- Maeland, A.; Rønnestad, I.; Waagbø, R. Folate in eggs and developing larvae of Atlantic halibut, Hippoglossus hippoglossus, L. Aquac. Nutr. 2003, 9, 185–188. [Google Scholar] [CrossRef]
- Oliva-Teles, A. Nutrition and health of aquaculture fish. J. Fish Dis. 2012, 35, 83–108. [Google Scholar] [CrossRef] [PubMed]
- Hoseini, S.M.; Khan, M.A.; Yousefi, M.; Costas, B. Roles of arginine in fish nutrition and health: Insights for future research. Rev. Aquac. 2020, 12, 2091–2108. [Google Scholar] [CrossRef]
- Hoseini, S.M.; Pérez-Jiménez, A.; Costas, B.; Azeredo, R.; Gesto, M. Physiological roles of tryptophan in teleosts: Current knowledge and perspectives for future studies. Rev. Aquac. 2019, 11, 3–24. [Google Scholar] [CrossRef] [Green Version]
- Sarih, S.; Djellata, A.; Roo, J.; Hernández-Cruz, C.M.; Fontanillas, R.; Rosenlund, G.; Izquierdo, M.; Fernández-Palacios, H. Effects of increased protein, histidine and taurine dietary levels on egg quality of greater amberjack (Seriola dumerili, Risso, 1810). Aquaculture 2019, 499, 72–79. [Google Scholar] [CrossRef]
- Zhang, H.; HuangFu, H.; Wang, X.; Zhao, S.; Liu, Y.; Lv, H.; Qin, G.; Tan, Z. Antibacterial activity of lactic acid producing Leuconostoc mesenteroides QZ1178 against pathogenic Gallibacterium anatis. Front. Vet. Sci. 2021, 8, 630294. [Google Scholar] [CrossRef]
- Pessione, E. Lactic acid bacteria contribution to gut microbiota complexity: Lights and shadows. Front. Cell. Infect. Microbiol. 2012, 2, 86. [Google Scholar] [CrossRef] [Green Version]
- Chang, D.; Jung, H.; Rhee, J.; Pan, J. Homofermentative production of D- or L-lactate in metabolically engineered Escherichia coli RR1. Appl. Environ. Microbiol. 1999, 65, 1384–1389. [Google Scholar] [CrossRef] [Green Version]
- Lorenz, I. D-lactic acidosis in calves. Vet. J. 2009, 179, 197–203. [Google Scholar] [CrossRef]
- Wouters, J.A.; Frenkiel, H.; de Vos, W.M.; Kuipers, O.P.; Abee, T. Cold shock proteins of Lactococcus lactis MG1363 are involved in cryoprotection and in the production of cold-induced proteins. Appl. Environ. Microbiol. 2001, 67, 5171–5178. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Z.; Tang, H.; Wang, W.; Zhang, L.; Su, F.; Wu, Y.; Bai, L.; Li, S.; Sun, Y.; Tao, F.; et al. A cold shock protein promotes high-temperature microbial growth through binding to diverse RNA species. Cell Discov. 2021, 7, 15. [Google Scholar] [CrossRef]
- Ding, W.K.; Shah, N.P. Acid, bile, and heat tolerance of free and microencapsulated probiotic bacteria. J. Food Sci. 2007, 72, M446–M450. [Google Scholar] [CrossRef] [PubMed]
- Nag, A.; Das, S. Improving ambient temperature stability of probiotics with stress adaptation and fluidized bed drying. J. Funct. Foods. 2013, 5, 170–177. [Google Scholar] [CrossRef]
- Domínguez-Maqueda, M.; Cerezo, I.M.; Tapia-Paniagua, S.T.; de la Banda, I.G.; Moreno-Ventas, X.; Moriñigo, M.Á.; Balebona, M.C. A tentative study of the effects of heat-inactivation of the probiotic strain Shewanella putrefaciens Ppd11 on Senegalese sole (Solea senegalensis) intestinal microbiota and immune response. Microorganisms 2021, 9, 808. [Google Scholar] [CrossRef] [PubMed]
- Ventura, M.; Canchaya, C.; Zink, R.; Fitzgerald, G.F.; van Sinderen, D. Characterization of the groEL and groES loci in Bifidobacterium breve UCC 2003: Genetic, transcriptional, and phylogenetic analyses. Appl. Environ. Microbiol. 2004, 70, 6197–6209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duru, I.C.; Ylinen, A.; Belanov, S.; Pulido, A.A.; Paulin, L.; Auvinen, P. Transcriptomic time-series analysis of cold- and heat-shock response in psychrotrophic lactic acid bacteria. BMC Genom. 2021, 22, 28. [Google Scholar] [CrossRef]
- Naylor, R.L.; Hardy, R.W.; Buschmann, A.H.; Bush, S.R.; Cao, L.; Klinger, D.H.; Little, D.C.; Lubchenco, J.; Shumway, S.E.; Troell, M. A 20-year retrospective review of global aquaculture. Nature 2021, 591, 551–563. [Google Scholar] [CrossRef]
- Maas, R.M.; Verdegem, M.C.J.; Stevens, T.L.; Schrama, J.W. Effect of exogenous enzymes (phytase and xylanase) supplementation on nutrient digestibility and growth performance of Nile tilapia (Oreochromis niloticus) fed different quality diets. Aquaculture 2020, 529, 735723. [Google Scholar] [CrossRef]
- Hlophe-Ginindza, S.N.; Moyo, N.A.; Ng’ambi, J.W.; Ncube, I. The effect of exogenous enzyme supplementation on growth performance and digestive enzyme activities in Oreochromis mossambicus fed kikuyu-based diets. Aquac. Res. 2016, 47, 3777–3787. [Google Scholar] [CrossRef]
- Konkit, M.; Kim, W. Activities of amylase, proteinase, and lipase enzymes from Lactococcus chungangensis and its application in dairy products. J. Dairy Sci. 2016, 99, 4999–5007. [Google Scholar] [CrossRef] [Green Version]
- Padmavathi, T.; Bhargavi, R.; Priyanka, P.R.; Niranjan, N.R.; Pavitra, P.V. Screening of potential probiotic lactic acid bacteria and production of amylase and its partial purification. J. Genet. Eng. Biotechnol. 2018, 16, 357–362. [Google Scholar] [CrossRef]
- El-Haroun, E.R.; Goda, A.M.A.-S.; Kabir Chowdhury, M.A. Effect of dietary probiotic Biogen® supplementation as a growth promoter on growth performance and feed utilization of Nile tilapia Oreochromis niloticus (L.). Aquac. Res. 2006, 37, 1473–1480. [Google Scholar] [CrossRef]
- Arani, M.M.; Salati, A.P.; Safari, O.; Keyvanshokooh, S. Dietary supplementation effects of Pediococcus acidilactici as probiotic on growth performance, digestive enzyme activities and immunity response in zebrafish (Danio rerio). Aquacult. Nutr. 2019, 25, 854–861. [Google Scholar] [CrossRef]
- Nolasco-Soria, H. Amylase quantification in aquaculture fish studies: A revision of most used procedures and presentation of a new practical protocol for its assessment. Aquaculture 2021, 538, 736536. [Google Scholar] [CrossRef]
- Al-Tameemi, R.; Aldubaikul, A.; Salman, N.A. Comparative study of α-amylase activity in three Cyprinid species of different feeding habits from Southern Iraq. Turk. J. Fish. Aquat. Sci. 2010, 10, 411–414. [Google Scholar] [CrossRef]
- Siezen, R.J.; Kuipers, O.P.; de Vos, W.M. Comparison of lantibiotic gene clusters and encoded proteins. Antonie Van Leeuwenhoek 1996, 69, 171–184. [Google Scholar] [CrossRef] [Green Version]
- Cheigh, C.I.; Pyun, Y.R. Nisin biosynthesis and its properties. Biotechnol. Lett. 2005, 27, 1641–1648. [Google Scholar] [CrossRef]
- Fusieger, A.; Perin, L.M.; Teixeira, C.G.; de Carvalho, A.F.; Nero, L.A. The ability of Lactococcus lactis subsp. lactis bv. diacetylactis strains in producing nisin. Antonie Van Leeuwenhoek 2020, 113, 651–662. [Google Scholar] [CrossRef] [PubMed]
- de Vos, W.M.; Kuipers, O.P.; van der Meer, J.R.; Siezen, R.J. Maturation pathway of nisin and other lantibiotics: Post-translationally modified antimicrobial peptides exported by Gram-positive bacteria. Mol. Microbiol. 1995, 17, 427–437. [Google Scholar] [CrossRef] [Green Version]
- Qiao, M.; Saris, E.J. Evidence for a role of NisT in transport of the lantibiotic nisin produced by Lactococcus lactis N8. FEMS Microbiol. Lett. 1996, 144, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Zendo, T.; Yoneyama, F.; Sonomoto, K. Lactococcal membrane permeabilizing antimicrobial peptides. Appl. Microbiol. Biotechnol. 2010, 88, 1–9. [Google Scholar] [CrossRef]
- Tosukhowong, A.; Zendo, T.; Visessanguan, W.; Roytrakul, S.; Pumpuang, L.; Jaresitthikunchai, J.; Sonomoto, K. Garvieacin Q, a Novel Class II bacteriocin from Lactococcus garvieae BCC 43578. Appl. Environ. Microbiol. 2012, 78, 1619–1623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hölscher, T.; Görisch, H. Knockout and overexpression of pyrroloquinoline quinone biosynthetic genes in Gluconobacter oxydans 621H. J. Bacteriol. 2006, 188, 7668–7676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Nayyef, H.; Guyeux, C.; Bahi, J.M. A pipeline for insertion sequence detection and study for bacterial genome. arXiv 2017, arXiv:1706.08267. Available online: https://arxiv.org/pdf/1706.08267 (accessed on 25 December 2021).
- Argov, T.; Azulay, G.; Pasechnek, A.; Stadnyuk, O.; Ran-Sapir, S.; Borokov, I.; Sigal, N.; Herskovits, A.A. Temperate bacteriophages as regulators of host behavior. Curr. Opin. Microbiol. 2017, 38, 1–7. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary classification of CRISPR-Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Imperial, I.C.V.J.; Iban, J.A. Addressing the antibiotic resistance problem with probiotics: Reducing the risk of its double-edged sword effect. Front. Microbiol. 2016, 7, 1983. [Google Scholar] [CrossRef] [PubMed]
- Gueimonde, M.; Sánchez, B.; de los Reyes-Gavilán, C.G.; Margolles, A. Antibiotic resistance in probiotic bacteria. Front. Microbiol. 2013, 4, 202. [Google Scholar] [CrossRef] [Green Version]
- EFSA (European Food Safety Authority). Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 13: Suitability of taxonomic units notified to EFSA until September 2020. EFSA J. 2021, 18, 5965–6021. [Google Scholar]
- Muñoz-Atienza, E.; Gómez-Sala, B.; Araújo, C.; Campanero, C.; del Campo, R.; Hernández, P.E.; Herranz, C.; Cintas, L.M. Antimicrobial activity, antibiotic susceptibility and virulence factors of Lactic Acid Bacteria of aquatic origin intended for use as probiotics in aquaculture. BMC Microbiol. 2013, 13, 15. [Google Scholar] [CrossRef] [Green Version]
- Anokyewaa, M.A.; Amoah, K.; Li, Y.; Lu, Y.; Kuebutornye, F.K.A.; Asiedu, B.; Seidu, I. Prevalence of virulence genes and antibiotic susceptibility of Bacillus used in commercial aquaculture probiotics in China. Aquac. Rep. 2021, 21, 100784. [Google Scholar] [CrossRef]

| Analyzed Element | L. cremoris WA2-67 |
|---|---|
| IS | IS similar/family/origin/length (bp) |
| IS981/IS3/Lactococcus lactis/1224 IS-LL6/IS3/Lactococcus lactis/1254 | |
| Plasmids | ND a |
| Active prophages | ND a |
| CRISPR-cas systems b | CRISPR spacers/cas genes/contig |
| 4/ND/27 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feito, J.; Contente, D.; Ponce-Alonso, M.; Díaz-Formoso, L.; Araújo, C.; Peña, N.; Borrero, J.; Gómez-Sala, B.; del Campo, R.; Muñoz-Atienza, E.; et al. Draft Genome Sequence of Lactococcus lactis Subsp. cremoris WA2-67: A Promising Nisin-Producing Probiotic Strain Isolated from the Rearing Environment of a Spanish Rainbow Trout (Oncorhynchus mykiss, Walbaum) Farm. Microorganisms 2022, 10, 521. https://doi.org/10.3390/microorganisms10030521
Feito J, Contente D, Ponce-Alonso M, Díaz-Formoso L, Araújo C, Peña N, Borrero J, Gómez-Sala B, del Campo R, Muñoz-Atienza E, et al. Draft Genome Sequence of Lactococcus lactis Subsp. cremoris WA2-67: A Promising Nisin-Producing Probiotic Strain Isolated from the Rearing Environment of a Spanish Rainbow Trout (Oncorhynchus mykiss, Walbaum) Farm. Microorganisms. 2022; 10(3):521. https://doi.org/10.3390/microorganisms10030521
Chicago/Turabian StyleFeito, Javier, Diogo Contente, Manuel Ponce-Alonso, Lara Díaz-Formoso, Carlos Araújo, Nuria Peña, Juan Borrero, Beatriz Gómez-Sala, Rosa del Campo, Estefanía Muñoz-Atienza, and et al. 2022. "Draft Genome Sequence of Lactococcus lactis Subsp. cremoris WA2-67: A Promising Nisin-Producing Probiotic Strain Isolated from the Rearing Environment of a Spanish Rainbow Trout (Oncorhynchus mykiss, Walbaum) Farm" Microorganisms 10, no. 3: 521. https://doi.org/10.3390/microorganisms10030521
APA StyleFeito, J., Contente, D., Ponce-Alonso, M., Díaz-Formoso, L., Araújo, C., Peña, N., Borrero, J., Gómez-Sala, B., del Campo, R., Muñoz-Atienza, E., Hernández, P. E., & Cintas, L. M. (2022). Draft Genome Sequence of Lactococcus lactis Subsp. cremoris WA2-67: A Promising Nisin-Producing Probiotic Strain Isolated from the Rearing Environment of a Spanish Rainbow Trout (Oncorhynchus mykiss, Walbaum) Farm. Microorganisms, 10(3), 521. https://doi.org/10.3390/microorganisms10030521









