Production of Vitamin K by Wild-Type and Engineered Microorganisms
Abstract
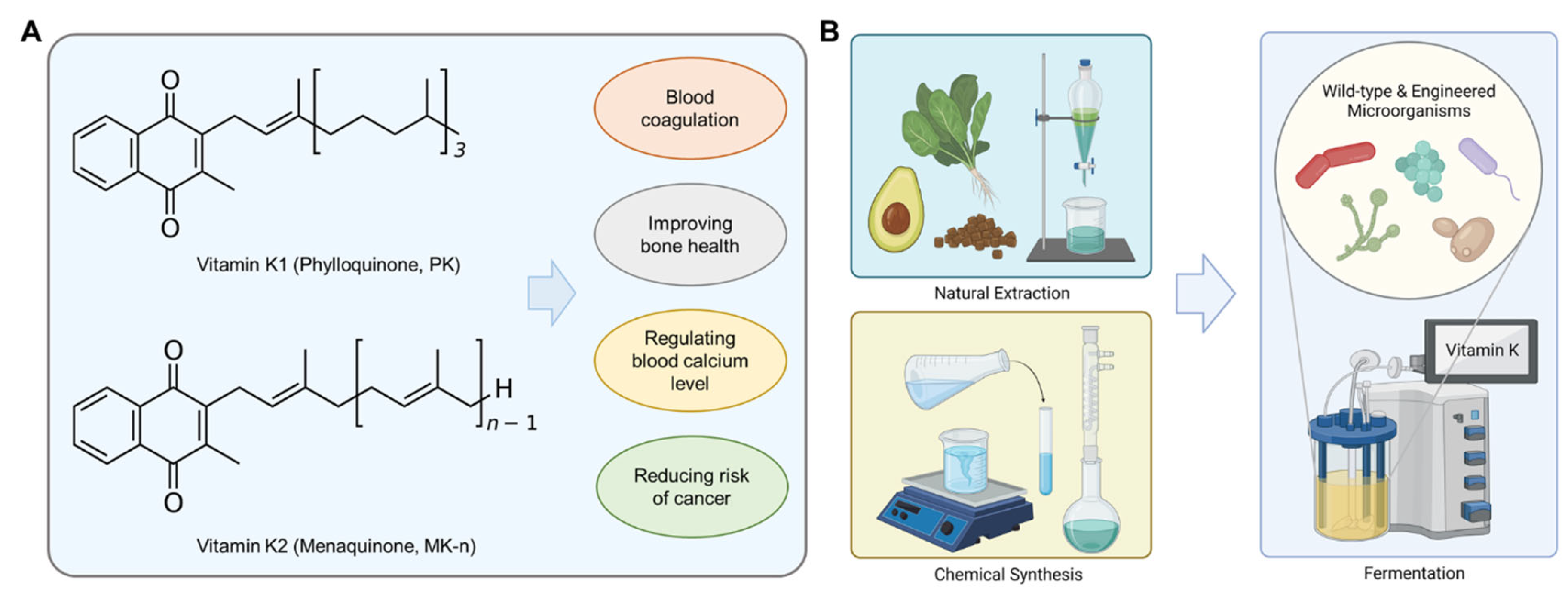
:1. Introduction
2. Production of Vitamin K by Wild-Type Microorganisms
2.1. Bacillus
2.2. Lactic Acid Bacteria
2.3. Other Microorganisms
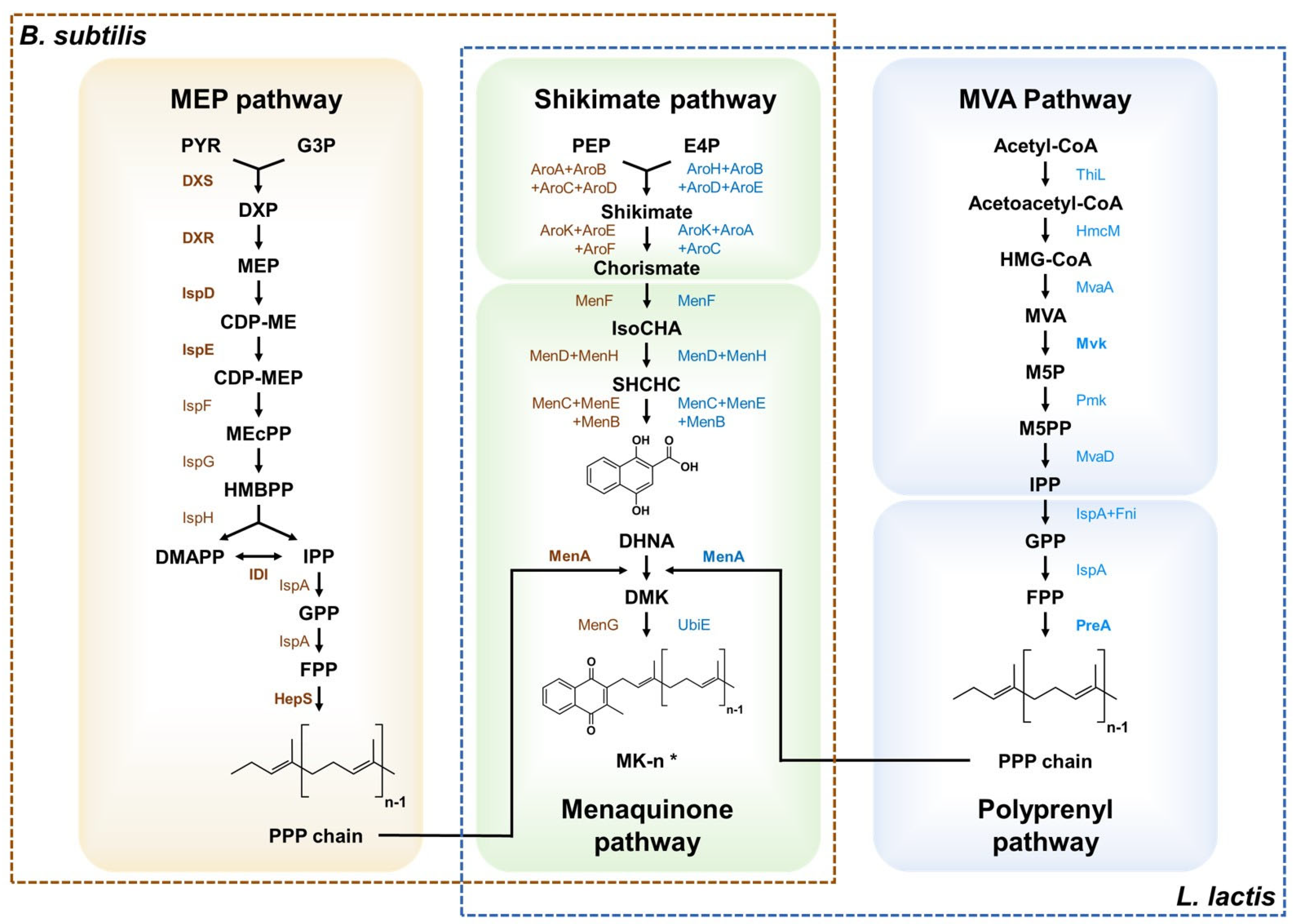
3. Production of Vitamin K Using Engineered Microorganisms
3.1. Engineered Bacillus
3.2. Engineered Lactic Acid Bacteria
3.3. Other Engineered Microorganisms
4. Conclusions
- Many studies for production of vitamin K using microorganisms have been conducted. Bacillus spp. are the most intensively studied microorganisms for vitamin K production. Many wild-type strains of Bacillus producing vitamin K have been isolated from soybean fermented foods, such as Natto and Cheonggukjang. Additionally, various LAB strains naturally producing different forms of vitamin K have been isolated and utilized for the manufacturing of vitamin K-enriched dairy products. Other than Bacillus and LAB, few microorganisms have been studied and characterized for vitamin K production. Most of them were isolated from gut microbiota, which contributes to vitamin K production in the human body. Several microorganisms producing vitamin K are not GRAS microorganisms, which limits the registration and commercialization of fermented products. To utilize the microbial-derived vitamin K as a food supplement, microbial production hosts are required to be safe and registered as GRAS microorganisms. The GRAS wild-type microorganisms are safe to use, but typically exhibit low yields of vitamin K. To improve the vitamin K production by the wild-type microorganisms, optimization of fermentation conditions and medium composition were mainly employed. Additionally, various chemical and physical methods, such as surfactant and ultrasound, have been used for increasing the biosynthesis and secretion of fat-soluble vitamin K out of the microbial cells.
- Random mutagenesis of the wild-type microorganisms can be used to increase vitamin K production, and vitamin K derived from the mutant strain is okay to use as a food ingredient. However, random mutagenesis strategy requires much time and effort to screen and select the mutants, which increased vitamin K production when compared to rational engineering. High-throughput screening methods may be utilized to accelerate the selection of vitamin K-overproducing strains from the mutant library.
- Microbial vitamin K production has also been improved by rational strain engineering strategies. These include overexpression of rate-limiting enzymes, deletion of competitive pathways, and fine-tuning of gene expression by modular system and quorum-sensing system. Although Bacillus, LAB, and other microbial cell factory platform strains have been engineered for overproduction of vitamin K, the current titers are still not high enough to support the commercialization of microbial-derived vitamin K by fermentation. The titer, yield, and productivity of vitamin K by microorganisms should be further improved by advanced genetic and fermentation technologies [117]. Systems metabolic engineering, which integrates various engineering tools of systems biology, synthetic biology, and evolutionary engineering, can facilitate the development of vitamin K-hyperproducing strains [118]. Moreover, in silico metabolic modeling and machine learning may help to develop the vitamin K-producing microbial hosts as industrially competitive [119].
- A recent trend suggests that most consumers prefer to take “natural” products than “synthetic” products. The microbial derived-vitamin K as a natural food supplement can satisfy the market demand and sustainable development goals. Even if the engineered host strains are considered as genetically modified organisms (GMO), recent advances in genome editing tools such as CRISPR/Cas9 system allow us to construct marker-free recombinant strains, facilitating safe and sustainable production of food ingredients [120].
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shah, K.; Gleason, L.; Villareal, D.T. Vitamin K and bone health in older adults. J. Nutr. Gerontol. Geriatr. 2014, 33, 10–22. [Google Scholar] [CrossRef]
- Kamao, M.; Suhara, Y.; Tsugawa, N.; Uwano, M.; Yamaguchi, N.; Uenishi, K.; Ishida, H.; Sasaki, S.; Okano, T. Vitamin K content of foods and dietary vitamin K intake in Japanese young women. J. Nutr. Sci. Vitaminol. 2007, 53, 464–470. [Google Scholar] [CrossRef] [Green Version]
- Basset, G.J.; Latimer, S.; Fatihi, A.; Soubeyrand, E.; Block, A. Phylloquinone (vitamin K1): Occurrence, biosynthesis and functions. Mini Rev. Med. Chem. 2017, 17, 1028–1038. [Google Scholar] [CrossRef]
- Bentley, R.; Meganathan, R. Biosynthesis of vitamin K (menaquinone) in bacteria. Microbiol. Rev. 1982, 46, 241–280. [Google Scholar] [CrossRef]
- Fusaro, M.; Mereu, M.C.; Aghi, A.; Iervasi, G.; Gallieni, M.J.; Metabolism, B. Vitamin K and bone. Clin. Cases Miner Bone Metab. 2017, 14, 200. [Google Scholar] [CrossRef]
- Schurgers, L.; Geleijnse, J.; Grobbee, D.; Pols, H.; Hofman, A.; Witteman, J.; Vermeer, C. Nutritional intake of vitamins K1 (phylloquinone) and K2 (menaquinone) in the Netherlands. J. Nutr. Environ. Med. 1999, 9, 115–122. [Google Scholar] [CrossRef]
- Van Oostende, C.; Widhalm, J.R.; Furt, F.; Ducluzeau, A.-L.; Basset, G.J. Vitamin K1 (phylloquinone): Function, enzymes and genes. Adv. Bot. Res. 2011, 59, 229–261. [Google Scholar]
- Mahdinia, E.; Demirci, A.; Berenjian, A. Production and application of menaquinone-7 (vitamin K2): A new perspective. World J. Microbiol. Biotechnol. 2017, 33, 2. [Google Scholar] [CrossRef]
- Beulens, J.W.; Booth, S.L.; van den Heuvel, E.G.; Stoecklin, E.; Baka, A.; Vermeer, C. The role of menaquinones (vitamin K2) in human health. Br. J. Nutr. 2013, 110, 1357–1368. [Google Scholar] [CrossRef] [Green Version]
- Vermeer, C.V. Vitamin K: The effect on health beyond coagulation—An overview. Food Nutr. Res. 2012, 56, 5329. [Google Scholar] [CrossRef]
- Bus, K.; Szterk, A. Relationship between Structure and Biological Activity of Various Vitamin K Forms. Foods 2021, 10, 3136. [Google Scholar] [CrossRef]
- DiNicolantonio, J.J.; Bhutani, J.; O’Keefe, J.H. The health benefits of vitamin K. Open Heart 2015, 2, e000300. [Google Scholar] [CrossRef]
- Dam, H.; Schønheyder, F.; Tage-Hansen, E. Studies on the mode of action of vitamin K. Biochem. J. 1936, 30, 1075–1079. [Google Scholar] [CrossRef]
- Olson, R.E.; Suttie, J.W. Vitamin K and gamma-carboxyglutamate biosynthesis. Vitam Horm. 1977, 35, 59–108. [Google Scholar] [CrossRef]
- Akiyama, Y.; Hara, K.; Matsumoto, A.; Takahashi, S.; Tajima, T. Comparison of intestinal absorption of vitamin K2 (menaquinone) homologues and their effects on blood coagulation in rats with hypoprothrombinaemia. Biochem. Pharmacol. 1995, 49, 1801–1807. [Google Scholar] [CrossRef]
- Halder, M.; Petsophonsakul, P.; Akbulut, A.C.; Pavlic, A.; Bohan, F.; Anderson, E.; Maresz, K.; Kramann, R.; Schurgers, L. Vitamin K: Double Bonds beyond Coagulation Insights into Differences between Vitamin K1 and K2 in Health and Disease. Int. J. Mol. Sci. 2019, 20, 896. [Google Scholar] [CrossRef] [Green Version]
- Weber, P. Vitamin K and bone health. Nutrition 2001, 17, 880–887. [Google Scholar] [CrossRef]
- Cockayne, S.; Adamson, J.; Lanham-New, S.; Shearer, M.J.; Gilbody, S.; Torgerson, D.J. Vitamin K and the prevention of fractures: Systematic review and meta-analysis of randomized controlled trials. Arch. Intern. Med. 2006, 166, 1256–1261. [Google Scholar] [CrossRef]
- Simes, D.C.; Viegas, C.S.B.; Araújo, N.; Marreiros, C. Vitamin K as a Powerful Micronutrient in Aging and Age-Related Diseases: Pros and Cons from Clinical Studies. Int. J. Mol. Sci. 2019, 20, 4150. [Google Scholar] [CrossRef] [Green Version]
- Iwamoto, J. Vitamin K2 Therapy for Postmenopausal Osteoporosis. Nutrients 2014, 6, 1971–1980. [Google Scholar] [CrossRef] [Green Version]
- Knapen, M.H.J.; Drummen, N.E.; Smit, E.; Vermeer, C.; Theuwissen, E. Three-year low-dose menaquinone-7 supplementation helps decrease bone loss in healthy postmenopausal women. Osteoporos. Int. 2013, 24, 2499–2507. [Google Scholar] [CrossRef]
- Shioi, A.; Morioka, T.; Shoji, T.; Emoto, M. The Inhibitory Roles of Vitamin K in Progression of Vascular Calcification. Nutrients 2020, 12, 583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lees, J.S.; Chapman, F.A.; Witham, M.D.; Jardine, A.G.; Mark, P.B. Vitamin K status, supplementation and vascular disease: A systematic review and meta-analysis. Heart 2019, 105, 938–945. [Google Scholar] [CrossRef]
- Zwakenberg, S.R.; de Jong, P.A.; Bartstra, J.W.; van Asperen, R.; Westerink, J.; de Valk, H.; Slart, R.; Luurtsema, G.; Wolterink, J.M.; de Borst, G.J.; et al. The effect of menaquinone-7 supplementation on vascular calcification in patients with diabetes: A randomized, double-blind, placebo-controlled trial. Am. J. Clin. Nutr. 2019, 110, 883–890. [Google Scholar] [CrossRef] [Green Version]
- Ingles, D.P.; Cruz Rodriguez, J.B.; Garcia, H. Supplemental Vitamins and Minerals for Cardiovascular Disease Prevention and Treatment. Curr. Cardiol. Rep. 2020, 22, 22. [Google Scholar] [CrossRef]
- Ferland, G. Vitamin K, an emerging nutrient in brain function. Biofactors 2012, 38, 151–157. [Google Scholar] [CrossRef]
- Vervoort, L.M.; Ronden, J.E.; Thijssen, H.H. The potent antioxidant activity of the vitamin K cycle in microsomal lipid peroxidation. Biochem. Pharmacol. 1997, 54, 871–876. [Google Scholar] [CrossRef]
- Li, J.; Lin, J.C.; Wang, H.; Peterson, J.W.; Furie, B.C.; Furie, B.; Booth, S.L.; Volpe, J.J.; Rosenberg, P.A. Novel role of vitamin k in preventing oxidative injury to developing oligodendrocytes and neurons. J. Neurosci. Off. J. Soc. Neurosci. 2003, 23, 5816–5826. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Wang, H.; Rosenberg, P.A. Vitamin K prevents oxidative cell death by inhibiting activation of 12-lipoxygenase in developing oligodendrocytes. J. Neurosci. Res. 2009, 87, 1997–2005. [Google Scholar] [CrossRef] [Green Version]
- Ren, L.; Peng, C.; Hu, X.; Han, Y.; Huang, H. Microbial production of vitamin K2: Current status and future prospects. Biotechnol. Adv. 2020, 39, 107453. [Google Scholar] [CrossRef]
- Krajewski, K.; Kutner, A.; Dzikowska, J.; Gutowska, R.; Napiórkowski, M.; Winiarski, J.; Kubiszewski, M.; Jedynak, Ł.; Morzycki, J.; Witkowski, S. Process for Preparation of MK-7 Type of Vitamin K2. Google Patents US9828323B2, 28 November 2017. [Google Scholar]
- Skattebol, L.; Aukrust, I.R.; Sandberg, M. Process for the Preparation of Vitamin K2. Google Patents EP2346806A1, 6 January 2016. [Google Scholar]
- Xu, Y.; Zhang, L.; Yang, R.; Yu, X.; Yu, L.; Ma, F.; Li, H.; Wang, X.; Li, P. Extraction and Determination of Vitamin K1 in Foods by Ultrasound-Assisted Extraction, SPE, and LC-MS/MS. Molecules 2020, 25, 839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarento, T.D.C.; McClure, D.D.; Talbot, A.M.; Regtop, H.L.; Biffin, J.R.; Valtchev, P.; Dehghani, F.; Kavanagh, J.M. A potential biotechnological process for the sustainable production of vitamin K1. Crit. Rev. Biotechnol. 2019, 39, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Shao, Z.; Zhao, H. Engineering microbial factories for synthesis of value-added products. J. Ind. Microbiol. Biotechnol. 2011, 38, 873–890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, E.T.; Schmidt-Dannert, C. Light-energy conversion in engineered microorganisms. Trends Biotechnol. 2008, 26, 682–689. [Google Scholar] [CrossRef]
- Lal, N.; Berenjian, A. Cis and trans isomers of the vitamin menaquinone-7: Which one is biologically significant? Appl. Microbiol. Biotechnol. 2020, 104, 2765–2776. [Google Scholar] [CrossRef]
- Szterk, A.; Zmysłowski, A.; Bus, K. Identification of cis/trans isomers of menaquinone-7 in food as exemplified by dietary supplements. Food Chem. 2018, 243, 403–409. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, L.; Jin, Z.; Zhang, D. Microbial Cell Factories for Green Production of Vitamins. Front. Bioeng. Biotechnol. 2021, 9, 661562. [Google Scholar] [CrossRef]
- Aguiar, T.Q.; Silva, R.; Domingues, L. Ashbya gossypii beyond industrial riboflavin production: A historical perspective and emerging biotechnological applications. Biotechnol. Adv. 2015, 33, 1774–1786. [Google Scholar] [CrossRef] [Green Version]
- Abbas, C.A.; Sibirny, A.A. Genetic control of biosynthesis and transport of riboflavin and flavin nucleotides and construction of robust biotechnological producers. Microbiol. Mol. Biol. Rev. 2011, 75, 321–360. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, J. Metabolic engineering. Appl. Microbiol. Biotechnol. 2001, 55, 263–283. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, Y.; Zhu, Y.; Li, Y. The importance of engineering physiological functionality into microbes. Trends Biotechnol. 2009, 27, 664–672. [Google Scholar] [CrossRef] [PubMed]
- Biedendieck, R.; Malten, M.; Barg, H.; Bunk, B.; Martens, J.-H.; Deery, E.; Leech, H.; Warren, M.J.; Jahn, D. Metabolic engineering of cobalamin (vitamin B12) production in Bacillus megaterium. Microb. Biotechnol. 2010, 3, 24–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LeBlanc, J.; Laiño, J.E.; del Valle, M.J.; Vannini, V.; van Sinderen, D.; Taranto, M.P.; de Valdez, G.F.; de Giori, G.S.; Sesma, F. B-Group vitamin production by lactic acid bacteria–current knowledge and potential applications. J. Appl. Microbiol. 2011, 111, 1297–1309. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Li, D.; Kang, J.; Jiang, P.; Sun, J.; Zhang, D. Metabolic engineering of Escherichia coli for de novo biosynthesis of vitamin B12. Nat. Commun. 2018, 9, 4917. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Liu, L.; Liu, C.; Sun, Y.; Zhang, D. New aspects of microbial vitamin K2 production by expanding the product spectrum. Microb. Cell Fact. 2021, 20, 84. [Google Scholar] [CrossRef]
- Westers, L.; Westers, H.; Quax, W.J.J.B.e.B.A.-M.C.R. Bacillus subtilis as cell factory for pharmaceutical proteins: A biotechnological approach to optimize the host organism. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2004, 1694, 299–310. [Google Scholar] [CrossRef]
- Berenjian, A.; Mahanama, R.; Talbot, A.; Biffin, R.; Regtop, H.; Valtchev, P.; Kavanagh, J.; Dehghani, F. Efficient media for high menaquinone-7 production: Response surface methodology approach. New Biotechnol. 2011, 28, 665–672. [Google Scholar] [CrossRef]
- Berenjian, A.; Mahanama, R.; Talbot, A.; Biffin, R.; Regtop, H.; Kavanagh, J.; Dehghani, F. The effect of amino-acids and glycerol addition on MK-7 production. Proc. World Congr. Eng. Comput. Sci. 2011, 11, 19–21. [Google Scholar]
- Berenjian, A.; Mahanama, R.; Talbot, A.; Regtop, H.; Kavanagh, J.; Dehghani, F. Advances in menaquinone-7 production by Bacillus subtilis natto: Fed-batch glycerol addition. Am. J. Biochem. Biotechnol. 2012, 8, 105–110. [Google Scholar]
- Berenjian, A.; Mahanama, R.; Talbot, A.; Regtop, H.; Kavanagh, J.; Dehghani, F. Designing of an intensification process for biosynthesis and recovery of menaquinone-7. Appl. Biochem. Biotechnol. 2014, 172, 1347–1357. [Google Scholar] [CrossRef]
- Hu, X.-C.; Liu, W.-M.; Luo, M.-M.; Ren, L.-J.; Ji, X.-J.; Huang, H. Enhancing Menaquinone-7 Production by Bacillus natto R127 Through the Nutritional Factors and Surfactant. Appl. Biochem. Biotechnol. 2017, 182, 1630–1641. [Google Scholar] [CrossRef] [PubMed]
- Mahdinia, E.; Demirci, A.; Berenjian, A. Strain and plastic composite support (PCS) selection for vitamin K (Menaquinone-7) production in biofilm reactors. Bioprocess Biosyst. Eng. 2017, 40, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
- Mahdinia, E.; Demirci, A.; Berenjian, A. Enhanced Vitamin K (Menaquinone-7) Production by Bacillus subtilis natto in Biofilm Reactors by Optimization of Glucose-based Medium. Curr. Pharm. Biotechnol. 2018, 19, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Mahdinia, E.; Demirci, A.; Berenjian, A. Utilization of glucose-based medium and optimization of Bacillus subtilis natto growth parameters for vitamin K (menaquinone-7) production in biofilm reactors. Biocatal. Agric. Biotechnol. 2018, 13, 219–224. [Google Scholar] [CrossRef]
- Mahdinia, E.; Demirci, A.; Berenjian, A. Implementation of fed-batch strategies for vitamin K (menaquinone-7) production by Bacillus subtilis natto in biofilm reactors. Appl. Microbiol. Biotechnol. 2018, 102, 9147–9157. [Google Scholar] [CrossRef]
- Mahdinia, E.; Demirci, A.; Berenjian, A. Optimization of Bacillus subtilis natto growth parameters in glycerol-based medium for vitamin K (Menaquinone-7) production in biofilm reactors. Bioprocess Biosyst. Eng. 2018, 41, 195–204. [Google Scholar] [CrossRef]
- Mahdinia, E.; Demirci, A.; Berenjian, A. Effects of medium components in a glycerol-based medium on vitamin K (menaquinone-7) production by Bacillus subtilis natto in biofilm reactors. Bioprocess Biosyst. Eng. 2019, 42, 223–232. [Google Scholar] [CrossRef]
- Mahdinia, E.; Demirci, A.; Berenjian, A. Evaluation of vitamin K (menaquinone-7) stability and secretion in glucose and glycerol-based media by Bacillus subtilis natto. Acta Aliment. 2019, 48, 405–414. [Google Scholar] [CrossRef]
- Novin, D.; van der Wel, J.; Seifan, M.; Berenjian, A. The effect of aeration and mixing in developing a dairy-based functional food rich in menaquinone-7. Bioprocess Biosyst. Eng. 2020, 43, 1773–1780. [Google Scholar] [CrossRef]
- Singh, R.; Puri, A.; Panda, B.P.P. Development of menaquinone-7 enriched nutraceutical: Inside into medium engineering and process modeling. J. Food Sci. Technol. 2015, 52, 5212–5219. [Google Scholar] [CrossRef] [Green Version]
- Ranmadugala, D.; Ebrahiminezhad, A.; Manley-Harris, M.; Ghasemi, Y.; Berenjian, A. Impact of 3–Aminopropyltriethoxysilane-Coated Iron Oxide Nanoparticles on Menaquinone-7 Production Using B. subtilis. Nanomaterials 2017, 7, 350. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.-J.; Ahn, B. Improved menaquinone (Vitamin K 2) production in cheonggukjang by optimization of the fermentation conditions. Food Sci. Biotechnol. 2011, 20, 1585–1591. [Google Scholar] [CrossRef]
- Zhao, C.; Wan, Y.; Tang, G.; Jin, Q.; Zhang, H.; Xu, Z. Comparison of different fermentation processes for the vitamin K2 (Menaquinone-7) production by a novel Bacillus velezensis ND strain. Process Biochem. 2021, 102, 33–41. [Google Scholar] [CrossRef]
- Morishita, T.; Tamura, N.; Makino, T.; Kudo, S. Production of menaquinones by lactic acid bacteria. J. Dairy Sci. 1999, 82, 1897–1903. [Google Scholar] [CrossRef]
- Lim, S.-D.; Kim, K.-S.; Do, J.-R. Physiological characteristics and production of vitamin K 2 by Lactobacillus fermentum LC272 isolated from raw milk. Food Sci. Anim. Resour. 2011, 31, 513–520. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; van Bennekom, E.O.; Zhang, Y.; Abee, T.; Smid, E.J. Long-chain vitamin K2 production in Lactococcus lactis is influenced by temperature, carbon source, aeration and mode of energy metabolism. Microb. Cell Fact. 2019, 18, 129. [Google Scholar] [CrossRef] [Green Version]
- Wei, H.; Wang, L.; Zhao, G.; Fang, Z.; Wu, H.; Wang, P.; Zheng, Z. Extraction, purification and identification of menaquinones from Flavobacterium meningosepticum fermentation medium. Process Biochem. 2018, 66, 245–253. [Google Scholar] [CrossRef]
- Fang, X.; Yang, Q.; Liu, H.; Wang, P.; Wang, L.; Zheng, Z.; Zhao, G. Effects of a combined processing technology involving ultrasound and surfactant on the metabolic synthesis of vitamin K2 by Flavobacterium sp. M1-14. Chem. Eng. Processing-Process Intensif. 2019, 135, 227–235. [Google Scholar] [CrossRef]
- Cooke, G.; Behan, J.; Costello, M. Newly identified vitamin K-producing bacteria isolated from the neonatal faecal flora. Microb. Ecol. Health Dis. 2006, 18, 133–138. [Google Scholar]
- Mahdinia, E.; Demirci, A.; Berenjian, A. Biofilm reactors as a promising method for vitamin K (menaquinone-7) production. Appl. Microbiol. Biotechnol. 2019, 103, 5583–5592. [Google Scholar] [CrossRef]
- Mahdinia, E.; Mamouri, S.J.; Puri, V.M.; Demirci, A.; Berenjian, A. Modeling of vitamin K (Menaquinoe-7) fermentation by Bacillus subtilis natto in biofilm reactors. Biocatal. Agric. Biotechnol. 2019, 17, 196–202. [Google Scholar] [CrossRef]
- Walther, B.; Karl, J.P.; Booth, S.L.; Boyaval, P. Menaquinones, Bacteria, and the Food Supply: The Relevance of Dairy and Fermented Food Products to Vitamin K Requirements. Adv. Nutr. 2013, 4, 463–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chollet, M.; Guggisberg, D.; Portmann, R.; Risse, M.-C.; Walther, B. Determination of menaquinone production by Lactococcus spp. and propionibacteria in cheese. Int. Dairy J. 2017, 75, 1–9. [Google Scholar] [CrossRef]
- Hojo, K.; Watanabe, R.; Mori, T.; Taketomo, N. Quantitative measurement of tetrahydromenaquinone-9 in cheese fermented by propionibacteria. J. Dairy Sci. 2007, 90, 4078–4083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LeBlanc, J.G.; Milani, C.; de Giori, G.S.; Sesma, F.; van Sinderen, D.; Ventura, M. Bacteria as vitamin suppliers to their host: A gut microbiota perspective. Curr. Opin. Biotechnol. 2013, 24, 160–168. [Google Scholar] [CrossRef]
- Ceyhan, M.; Celik, M. Elizabethkingia meningosepticum (Chryseobacterium meningosepticum) infections in children. Int. J. Pediatrics 2011, 2011, 215237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddy, R.M.; Reddy, P.G.; Seenayya, G. Enhanced production of thermostable β-amylase and pullulanase in the presence of surfactants by Clostridium thermosulfurogenes SV2. Process Biochem. 1999, 34, 87–92. [Google Scholar] [CrossRef]
- Ellis, J.L.; Karl, J.P.; Oliverio, A.M.; Fu, X.; Soares, J.W.; Wolfe, B.E.; Hernandez, C.J.; Mason, J.B.; Booth, S.L. Dietary vitamin K is remodeled by gut microbiota and influences community composition. Gut Microbes 2021, 13, 1887721. [Google Scholar] [CrossRef]
- Fernandez, F.; Collins, M.D. Vitamin K composition of anaerobic gut bacteria. FEMS Microbiol. Lett. 1987, 41, 175–180. [Google Scholar] [CrossRef]
- Yu, Q.; Li, Y.; Wu, B.; Hu, W.; He, M.; Hu, G. Novel mutagenesis and screening technologies for food microorganisms: Advances and prospects. Appl. Microbiol. Biotechnol. 2020, 104, 1517–1531. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Yamada, Y.; Ohtani, Y.; Mitsui, N.; Murasawa, H.; Araki, S. Production of Menaquinone (vitamin K2)-7 by Bacillus subtilis. J. Biosci. Bioeng. 2001, 91, 16–20. [Google Scholar] [CrossRef]
- Song, J.; Liu, H.; Wang, L.; Dai, J.; Liu, Y.; Liu, H.; Zhao, G.; Wang, P.; Zheng, Z. Enhanced Production of Vitamin K2 from Bacillus subtilis (natto) by Mutation and Optimization of the Fermentation Medium. Braz. Arch. Biol. Technol. 2014, 57, 606–612. [Google Scholar]
- Tsukamoto, Y.; Kasai, M.; Kakuda, H. Construction of a Bacillus subtilis (natto) with high productivity of vitamin K2 (menaquinone-7) by analog resistance. Biosci. Biotechnol. Biochem. 2001, 65, 2007–2015. [Google Scholar] [CrossRef]
- Wang, H.; Sun, X.; Wang, L.; Wu, H.; Zhao, G.; Liu, H.; Wang, P.; Zheng, Z. Coproduction of menaquinone-7 and nattokinase by Bacillus subtilis using soybean curd residue as a renewable substrate combined with a dissolved oxygen control strategy. Ann. Microbiol. 2018, 68, 655–665. [Google Scholar] [CrossRef]
- Puri, A.; Iqubal, M.; Zafar, R.; Panda, B.P. Influence of physical, chemical and inducer treatments on menaquinone-7 biosynthesis by Bacillus subtilis MTCC 2756. Songklanakarin J. Sci. Technol. 2015, 37, 283–289. [Google Scholar]
- Goodman, S.R.; Marrs, B.L.; Narconis, R.J.; Olson, R.E. Isolation and description of a menaquinone mutant from Bacillus licheniformis. J. Bacteriol. 1976, 125, 282–289. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.-Z.; Zhang, W. Menaquinone-7 production from maize meal hydrolysate by Bacillus isolates with diphenylamine and analogue resistance. J. Zhejiang Univ. Sci. B 2017, 18, 462–473. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Ruan, H.; Liu, L.; Zhang, W.; Xu, J. Temperature-induced mutagenesis-based adaptive evolution of Bacillus amyloliquefaciens for improving the production efficiency of menaquinone-7 from starch. J. Chem. Technol. Biotechnol. 2021, 96, 1040–1048. [Google Scholar] [CrossRef]
- Xu, J.-Z.; Yan, W.-L.; Zhang, W.-G. Enhancing menaquinone-7 production in recombinant Bacillus amyloliquefaciens by metabolic pathway engineering. RSC Adv. 2017, 7, 28527–28534. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; McClure, D.D.; Somerville, M.V.; Proschogo, N.W.; Dehghani, F.; Kavanagh, J.M.; Coleman, N.V. Metabolic Engineering of the MEP Pathway in Bacillus subtilis for Increased Biosynthesis of Menaquinone-7. ACS Synth. Biol. 2019, 8, 1620–1630. [Google Scholar] [CrossRef]
- Yang, S.; Cao, Y.; Sun, L.; Li, C.; Lin, X.; Cai, Z.; Zhang, G.; Song, H. Modular pathway engineering of Bacillus subtilis to promote de novo biosynthesis of menaquinone-7. ACS Synth. Biol. 2018, 8, 70–81. [Google Scholar] [CrossRef]
- Yuan, P.; Cui, S.; Liu, Y.; Li, J.; Lv, X.; Liu, L.; Du, G. Combinatorial engineering for improved menaquinone-4 biosynthesis in Bacillus subtilis. Enzym. Microb. Technol. 2020, 141, 109652. [Google Scholar] [CrossRef]
- Cui, S.; Lv, X.; Wu, Y.; Li, J.; Du, G.; Ledesma-Amaro, R.; Liu, L. Engineering a bifunctional Phr60-Rap60-Spo0A quorum-sensing molecular switch for dynamic fine-tuning of menaquinone-7 synthesis in Bacillus subtilis. ACS Synth. Biol. 2019, 8, 1826–1837. [Google Scholar] [CrossRef]
- Yuan, P.; Sun, G.; Cui, S.; Wu, Y.; Lv, X.; Liu, Y.; Li, J.; Du, G.; Liu, L. Engineering a ComA Quorum-Sensing circuit to dynamically control the production of Menaquinone-4 in Bacillus subtilis. Enzym. Microb. Technol. 2021, 147, 109782. [Google Scholar] [CrossRef]
- Cui, S.; Xia, H.; Chen, T.; Gu, Y.; Lv, X.; Liu, Y.; Li, J.; Du, G.; Liu, L. Cell membrane and electron transfer engineering for improved synthesis of menaquinone-7 in Bacillus subtilis. iScience 2020, 23, 100918. [Google Scholar] [CrossRef] [Green Version]
- Bøe, C.A.; Holo, H. Engineering Lactococcus lactis for Increased Vitamin K2 Production. Front. Bioeng. Biotechnol. 2020, 8, 191. [Google Scholar] [CrossRef] [Green Version]
- Kong, M.K.; Lee, P.C. Metabolic engineering of menaquinone-8 pathway of Escherichia coli as a microbial platform for vitamin K production. Biotechnol. Bioeng. 2011, 108, 1997–2002. [Google Scholar] [CrossRef]
- Gao, Q.; Chen, H.; Wang, G.; Yang, W.; Zhong, X.; Liu, J.; Huo, X.; Liu, W.; Huang, J.; Tao, Y.; et al. Highly Efficient Production of Menaquinone-7 from Glucose by Metabolically Engineered Escherichia coli. ACS Synth. Biol. 2021, 10, 756–765. [Google Scholar] [CrossRef]
- Liu, Y.; Ding, X.-M.; Xue, Z.-L.; Hu, L.-X.; Zhang, N.-J.; Wang, Z.; Yang, J.-W.; Cheng, Q.; Chen, M.-H.; Zhang, Z.-Z.; et al. The change of the state of cell membrane can enhance the synthesis of menaquinone in Escherichia coli. World J. Microbiol. Biotechnol. 2017, 33, 52. [Google Scholar] [CrossRef]
- Tani, Y.; Asahi, S.; Yamada, H. Menaquinone (vitamin K2)-6 production by mutants of Flavobacterium meningosepticum. J. Nutr. Sci. Vitaminol. 1986, 32, 137–145. [Google Scholar] [CrossRef] [Green Version]
- Moon, G.-S.; Pyun, Y.-R.; Park, M.S.; Ji, G.E.; Kim, W.J. Secretion of recombinant pediocin PA-1 by Bifidobacterium longum, using the signal sequence for bifidobacterial α-amylase. Appl. Environ. Microbiol. 2005, 71, 5630–5632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, M.; Seo, J.; Kim, J.; Ji, G. Heterologous gene expression and secretion in Bifidobacterium longum. Le Lait 2005, 85, 1–8. [Google Scholar] [CrossRef]
- Oh, J.-H.; van Pijkeren, J.-P. CRISPR–Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res. 2014, 42, e131. [Google Scholar] [CrossRef] [PubMed]
- Stout, E.A.; Sanozky-Dawes, R.; Goh, Y.J.; Crawley, A.B.; Klaenhammer, T.R.; Barrangou, R. Deletion-based escape of CRISPR-Cas9 targeting in Lactobacillus gasseri. Microbiology 2018, 164, 1098–1111. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.-J.; Seo, S.-O.; Kim, S.-A.; Li, L.; Kim, T.-J.; Kim, S.C.; Jin, Y.-S.; Han, N.S. Elimination of the cryptic plasmid in Leuconostoc citreum by CRISPR/Cas9 system. J. Biotechnol. 2017, 251, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Briner, A.E.; Lugli, G.A.; Milani, C.; Duranti, S.; Turroni, F.; Gueimonde, M.; Margolles, A.; Van Sinderen, D.; Ventura, M.; Barrangou, R. Occurrence and diversity of CRISPR-Cas systems in the genus Bifidobacterium. PLoS ONE 2015, 10, e0133661. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Yang, D.; Zhang, Z.; Liu, T.; Hu, G.; He, M.; Zhao, S.; Peng, N. High-Efficiency Genome Editing Based on Endogenous CRISPR-Cas System Enhances Cell Growth and Lactic Acid Production in Pediococcus acidilactici. Appl. Environ. Microbiol. 2021, 87, e0094821. [Google Scholar] [CrossRef]
- Yang, D.; Park, S.Y.; Park, Y.S.; Eun, H.; Lee, S.Y. Metabolic engineering of Escherichia coli for natural product biosynthesis. Trends Biotechnol. 2020, 38, 745–765. [Google Scholar] [CrossRef]
- Chen, X.; Zhou, L.; Tian, K.; Kumar, A.; Singh, S.; Prior, B.A.; Wang, Z. Metabolic engineering of Escherichia coli: A sustainable industrial platform for bio-based chemical production. Biotechnol. Adv. 2013, 31, 1200–1223. [Google Scholar] [CrossRef]
- Lee, S.Y.; Kim, H.U. Systems strategies for developing industrial microbial strains. Nat. Biotechnol. 2015, 33, 1061–1072. [Google Scholar] [CrossRef]
- Meganathan, R. Biosynthesis of the isoprenoid quinones menaquinone (vitamin K_2) and ubiquinone (coenzyme Q). In Escherichia Coli and Salmonella: Cellular and Molecular Biology; ASM Press: Washington, DC, USA, 1996; pp. 642–656. [Google Scholar]
- Lin, E.; Kuritzkes, D. Pathways for anaerobic electron transport. In Escherichia Coli and Salmonella Typhimurium: Cellular and Molecular Biology; American Society for Microbiology: Washington, DC, USA, 1987; pp. 201–221. [Google Scholar]
- Olson, R.E.; Rudney, H. Biosynthesis of ubiquinone. Vitam. Horm. 1983, 40, 1–43. [Google Scholar] [PubMed]
- Meganathan, R.; Kwon, O. Biosynthesis of Menaquinone (Vitamin K2) and Ubiquinone (Coenzyme Q). EcoSal Plus 2009, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, S.O.; Jin, Y.S. Next-Generation Genetic and Fermentation Technologies for Safe and Sustainable Production of Food Ingredients: Colors and Flavorings. Annu. Rev. Food Sci. Technol. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.R.; Jang, W.D.; Yang, D.; Cho, J.S.; Park, D.; Lee, S.Y. Systems Metabolic Engineering Strategies: Integrating Systems and Synthetic Biology with Metabolic Engineering. Trends Biotechnol. 2019, 37, 817–837. [Google Scholar] [CrossRef]
- Presnell, K.V.; Alper, H.S. Systems Metabolic Engineering Meets Machine Learning: A New Era for Data-Driven Metabolic Engineering. Biotechnol. J. 2019, 14, 1800416. [Google Scholar] [CrossRef]
- Hidalgo-Cantabrana, C.; O’Flaherty, S.; Barrangou, R. CRISPR-based engineering of next-generation lactic acid bacteria. Curr. Opin. Microbiol. 2017, 37, 79–87. [Google Scholar] [CrossRef]
| Class | Strain | Strategy | Carbon Source | Menaquinone Type | Fermentation Time | Titer | Extraction | Reference |
|---|---|---|---|---|---|---|---|---|
| Bacillus spp. | Bacillus subtilis natto | Optimization of medium and fermentation condition | Glycerol | MK-7 | 6 days | 62.32 mg/L | By aqueous medium | [49] |
| Bacillus subtilis natto | Carbon source addition during fermentation | Glycerol | MK-7 | 6 days | 68.6 mg/L | By aqueous medium | [50] | |
| Bacillus subtilis natto | Fed-batch fermentation | Glycerol | MK-7 | 6 days | 86.48 mg/L | By aqueous medium | [51] | |
| Bacillus subtilis natto | Optimization of stirrer speed and aeration rate | Glycerol | MK-7 | 100 h | 226 mg/L | By aqueous medium | [52] | |
| Bacillus natto R127 | Optimization of medium Supplementation of surfactant | Glycerol | MK-7 | 24 h | 40.96 mg/L | By aqueous medium | [53] | |
| Bacillus subtilis natto F2 | Static fermentation | Glycerol | MK-7 | 96 h | 35.5 mg/L | By fermentation broth | [54] | |
| Bacillus subtilis natto (NF1) | Biofilm reactors (Plastic composite support) Optimization of medium | Glucose | MK-7 | 144 h | 20.5 mg/L | By aqueous medium | [55] | |
| Bacillus subtilis natto (NF1) | Biofilm reactors (Plastic composite support) Optimization of fermentation condition | Glucose | MK-7 | 144 h | 18.45 mg/L | By aqueous medium | [56] | |
| Bacillus subtilis natto (NF1) | Biofilm reactors (Plastic composite support) Fed-batch fermentation | Glucose | MK-7 | 288 h | 28.7 mg/L | By aqueous medium | [57] | |
| Bacillus subtilis natto (NF1) | Biofilm reactors | Glycerol | MK-7 | 144 h | 12.09 mg/L | By aqueous medium | [58] | |
| Bacillus subtilis natto (NF1) | Biofilm reactors (Plastic composite support) Optimization of medium | Glycerol | MK-7 | 144 h | 14.7 mg/L | By aqueous medium | [59] | |
| Bacillus subtilis natto (NF1) | Fermentation in bottle Optimization of medium Static fermentation | Glycerol | MK-7 | 96 h | 32.5 mg/L | By aqueous medium | [60] | |
| Glucose | MK-7 | 96 h | 14.6 mg/L | By aqueous medium | ||||
| Bacillus subtilis natto | Optimization of aeration and agitation | Milk medium | MK-7 | 72 h | 3.54 mg/L | By aqueous medium | [61] | |
| Bacillus subtilis NCIM 2708 | Optimization of medium | Glycerol, mannitol | MK-7 | 24 h | 39.039 mg/g | By soybean samples | [62] | |
| Bacillus subtilis ATCC 6633 | Iron oxide nanoparticles coated | Glycerol | MK-7 | 108 h | 37.36 mg/L | By aqueous medium | [63] | |
| Bacillus amyloliquefaciens KCTC 11712BP | Optimization of medium and fermentation condition | Glycerol | MK-4 | 36 h | 0.76 mg/g | Fermented Cheonggukjang | [64] | |
| Glycerol | MK-7 | 36 h | 11.71 mg/g | Fermented Cheonggukjang | ||||
| Bacillus velezensis ND | Liquid-state fermentation | Glycerol | MK-7 | 168 h | 52.9 mg/L | By aqueous medium | [65] | |
| Biofilm-based fermentation | Glycerol | MK-7 | 144 h | 73.3 mg/L | By fermentation broth | |||
| Solid-state fermentation | Glycerol | MK-7 | 96 h | 150.02 mg/Kg | By fermentation broth | |||
| Lactic acid bacteria | Lactococcus lactis ssp. cremoris YIT 2011 | Batch fermentation | Glucose | MK-7, MK-8, MK-9 | 48 h | 534 nmol/L | By cell using chloroform and methanol | [66] |
| Lactococcus lactis ssp. lactis YIT 2027 | Batch fermentation | Glucose | MK-8, MK-9, MK-10 | 48 h | 717 nmol/L | By cell using chloroform and methanol | ||
| Leuconostoc lactis YIT 3001 | Soymilk fermentation | Soy milk | MK-7, MK-8, MK-9, MK-10 | 48 h | 2.60 nmol/L | By soymilk culture using chloroform and methanol | ||
| Lactobacillus fermentum LC272 | Batch fermentation | Glucose | MK-4 | 48 h | 0.18 mg/L | By cell using hexane and methanol | [67] | |
| Lactococcus lactis ssp. cremoris MG1363 | Optimization of temperature, carbon source, aeration, and mode of energy metabolism | Trehalose | MK-5, MK-6, MK-7, MK-8, MK-9, MK-10 | 48 h | 5.2-fold increase compared to the control (90 nmol/L medium) | By cell using hexane and 2-propanol | [68] | |
| Others | Flavobacterium meningosepticum | Changing extraction solvent | Glycerol | MK-4, MK-5, MK-6 | 6 days | 1.88 mg/g DCW | By cell using methanol | [69] |
| Flavobacterium sp. M1-14 | Using different surfactant with ultrasound | Glycerol | MK | 9 days | 30.03 mg/L | In aqueous medium | [70] | |
| Enterobacter agglomerans | Isolated from neonatal fecal flora | Tryptone soy broth powder | MK-4 | 72 h | ND | By cell using methanol and chloroform | [71] | |
| Serratia marcescens | ||||||||
| Enterococcus faecium |
| Class | Strain | Strategy | Carbon Source | Menaquinone Type | Fermentation Time | Titer | Extraction | Reference |
|---|---|---|---|---|---|---|---|---|
| Bacillus spp. | Bacillus subtilis natto OUV23481 | UV and analog resistance (HNA, pFP, mFP, β-TA) | Sucrose | MK-7 | 16 h | 3438 μg/100 g | By Natto | [85] |
| Bacillus subtilis D200-41 | Strain mutation (DPA) media optimization | Glycerol | MK-7 | 6 days | 60 mg/L | By aqueous medium | [83] | |
| B. subtilis(natto)-P15-11-1 | Strain mutation (NTG, HNA and N+ ion-beam) media optimization | Glycerol | MK-7 | 70 h | 3.593 mg/L | By cell using n-hexane | [84] | |
| B. subtilis(natto)-P15-11-1 | Strain mutation and media optimization | Lactose | MK-7 | 144 h | 91.25 mg/L | By aqueous medium | [86] | |
| Bacillus subtilis | Strain mutation (1-naphthol and Tween 80) | Glycerol | MK-7 | 24 h | 14.4 μg/mL | By cell and aqueous medium | [87] | |
| Bacillus licheniformis | Strain mutation (kanamycin and shikimate) | Glucose | MK-7 | 1 h | 0.3 nmol/mL | By cell using acetone | [88] | |
| Bacillus amyloliquefaciens H.β.D.R.-5 | Strain mutation (HNA, DPA and β-TA) | Corn starch hydrolysates | MK-7 | 6 days | 61.3 mg/L | By aqueous medium | [89] | |
| Bacillus amyloliquefaciens MK50-36 | Laboratory evolution at 50 °C | Corn starch | MK-7 | 144 h | 57 mg/L | By aqueous medium | [90] | |
| Bacillus subtilis | Strain mutation (1-naphthol and Tween 80) | Glycerol | MK-7 | 24 h | 14.4 μg/mL | By cell and aqueous medium | [87] | |
| Bacillus amyloliquefaciens Y-2 | Metabolic engineering (overexpression of hepS) | Glucose | MK-7 | 24 h | 273 μg/g DCW | By cell using n-hexane and 2-propanol mixture | [91] | |
| Bacillus subtilis | Metabolic engineering (overexpression of Dxs, Dxr, Idi, and MenA) | Glycerol | MK-7 | 6 days | 50 mg/L | By cell and aqueous medium | [92] | |
| Bacillus subtilis MK3-MEP123-Gly2-ΔdhbB | Metabolic engineering (overexpression of menA, dxs, dxr, yacM, yacN, and glpD, deletion of dhbB | Glycerol | MK-7 | 120 h | 69.5 mg/L | By aqueous medium | [93] | |
| Bacillus subtilis BY23 | Metabolic engineering (overexpression of menA, menG, crtE, dxs, dxr, and ispD-ispF, deletion of hepT) and introduction of MVA pathway | Glucose, glycerol | MK-4 | 144 h | 145 mg/L | By cell and aqueous medium | [94] | |
| Bacillus subtilis | Metabolic engineering (introduction of synthesis modules of MK-7) and Rap60-Spo0A quorum sensing system | Glucose | MK-7 | 6 days | 360 mg/L | By cell and aqueous medium | [95] | |
| Bacillus subtilis | Metabolic engineering (overexpressing ispH, crtE and menA) and PhrQ-RapQ-ComA quorum sensing system | Glucose | MK-4 | 82 h | 217 mg/L | By cell and aqueous medium | [96] | |
| Bacillus subtilis BS20-QT | Metabolic engineering (overexpression of TatAD-CD, QcrA-C) | Glucose, sucrose | MK-7 | 4 days | 310 mg/L | By cell and aqueous medium | [97] | |
| Lactic acid bacteria | Lactococcus lactis ssp. cremoris MG1363 | Metabolic engineering (Overexpression of mvk, preA, menA) | Glucose | MK-7, MK-8, MK-9 | Overnight | 680 nmol/L | By cell using heptane and 2-propanol | [98] |
| Others | Escherichia coli JM 109 | Metabolic engineering (Deletion of ubiC, ubiA, overexpression of MenA, MenD) | Glycerol | MK-8 | 28 h | 290 mg MK-8/g WCW | By cell using chloroform and methanol | [99] |
| Escherichia coli MK17 | Metabolic engineering (Overexpression of Idi, Metk, MenF MenA from E. coli, HepPPS, UbiE, from B. subtilis) | Glucose | MK-7 | 52 h | 1350 mg/L | By cell using hexane and propanol | [100] | |
| E. coli DH5α Fat B | Metabolic engineering (Overexpression of FatB from Umbellularia californica) | Glycerol | MK | 120 h | 10.71 ± 0.19 mg/L | By cell using methanol | [101] | |
| Flavobacterium meningosepticum | Mutagenesis (NTG, HNA) | Glycerol | MK | 72 h | 34 mg/L 5.5 mg/g DCW | By cell using acetone and ethyl ether | [102] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, M.-J.; Baek, K.-R.; Lee, Y.-R.; Kim, G.-H.; Seo, S.-O. Production of Vitamin K by Wild-Type and Engineered Microorganisms. Microorganisms 2022, 10, 554. https://doi.org/10.3390/microorganisms10030554
Kang M-J, Baek K-R, Lee Y-R, Kim G-H, Seo S-O. Production of Vitamin K by Wild-Type and Engineered Microorganisms. Microorganisms. 2022; 10(3):554. https://doi.org/10.3390/microorganisms10030554
Chicago/Turabian StyleKang, Min-Ji, Kwang-Rim Baek, Ye-Rim Lee, Geun-Hyung Kim, and Seung-Oh Seo. 2022. "Production of Vitamin K by Wild-Type and Engineered Microorganisms" Microorganisms 10, no. 3: 554. https://doi.org/10.3390/microorganisms10030554
APA StyleKang, M.-J., Baek, K.-R., Lee, Y.-R., Kim, G.-H., & Seo, S.-O. (2022). Production of Vitamin K by Wild-Type and Engineered Microorganisms. Microorganisms, 10(3), 554. https://doi.org/10.3390/microorganisms10030554







