Transfer of Human Microbiome to Drosophila Gut Model
Abstract
1. Introduction
2. Materials and Methods
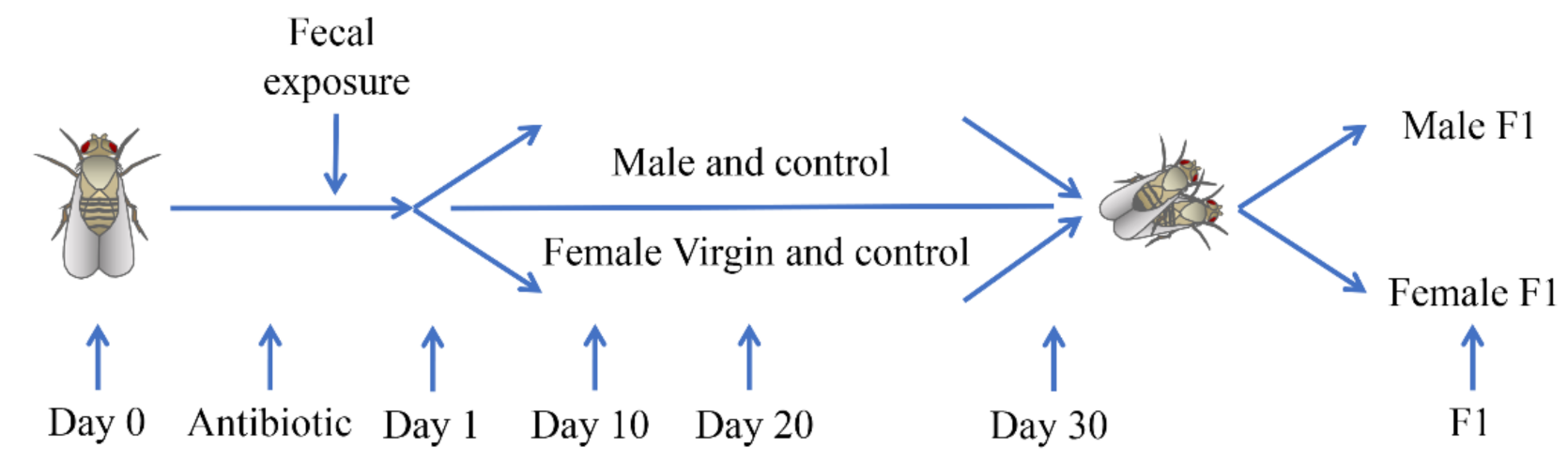
2.1. Collection and Processing of Fecal Samples
- Preliminary work: firstly, a suitable donor was screened according to the Brandt induction method and the donor questionnaire was filled out by the parents of the donor (as the donor was a two-year-old boy) [20]. Subsequently, the donor underwent comprehensive blood and fecal screening to detect potential transmissible pathogens. The donor’s parents were instructed not to administer any medications for at least one week before and during the fecal samples collection period as donors should not be treated by antibiotics for eight weeks prior to feces collection.
- Sample collection: we instructed the donor’s parents to collect the fecal samples in the collection cap according to the standard operating procedure for feces collection and then label the cap with the time and donor information after sealing. Urine, paper, or other waste were kept away from the sample during this process.
- Sample transportation and preservation: the fecal samples were maintained at 2–8 °C during transportation and then stored at −20 °C in the laboratory.
- Sample processing: weigh 50 g of uncontaminated fecal samples, add 300 mL of sterile water to it, stir well, filter with 4 × 4 sterile gauze, and then collect the filtrate to add to the drosophila culture medium. Details of the test process and the date of each sampling please see Figure 1.
2.2. Drosophila Husbandry
2.3. Lifespan Assay
2.4. Body Weight
2.5. Locomotor Activity
2.6. 16S rDNA Amplicon Sequencing and Analysis
2.7. Quantitative Real-Time PCR
2.8. Statistics
3. Results
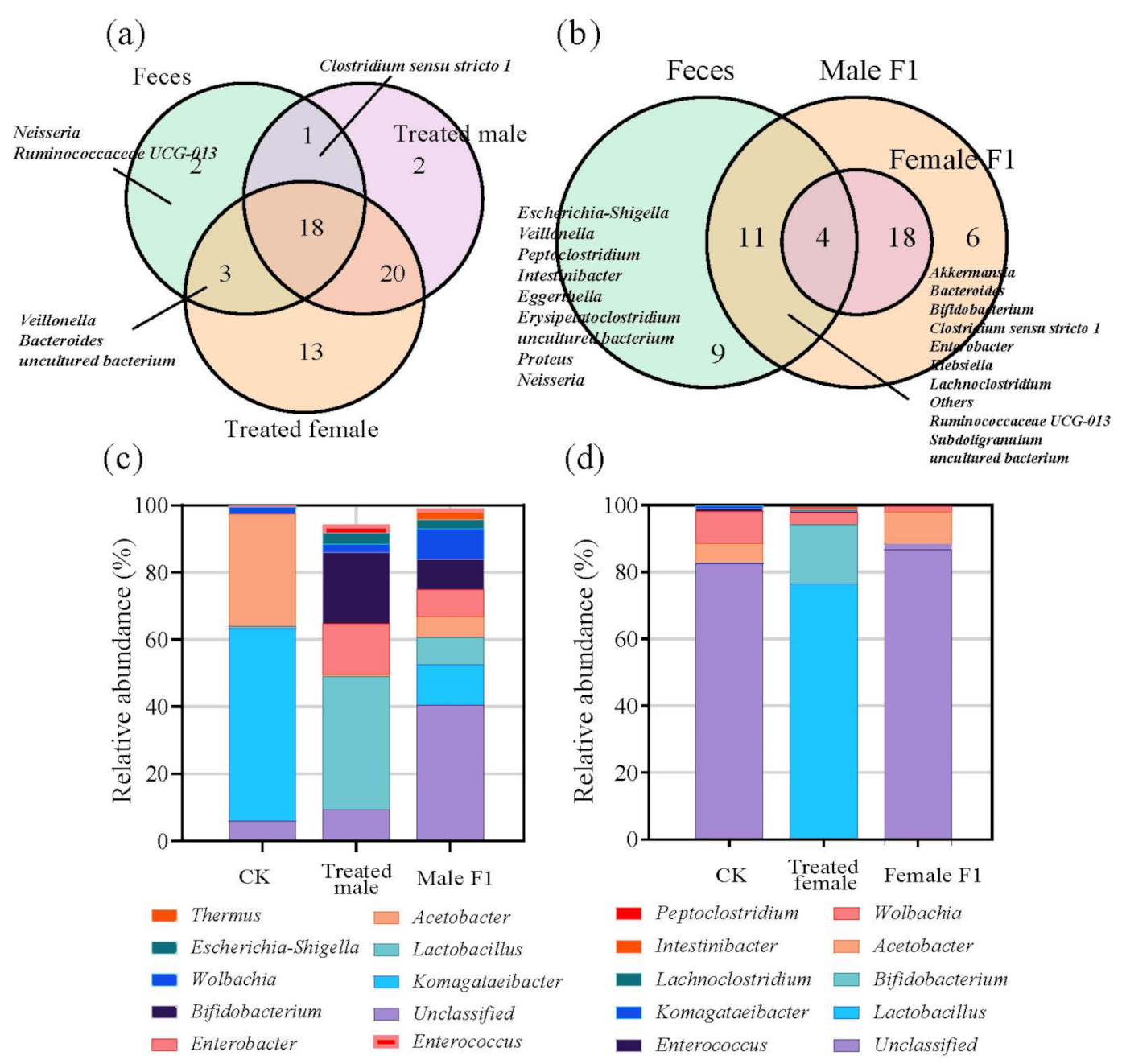
3.1. Dynamics of α-Diversity and Bacterial Composition of Gut Bacteria
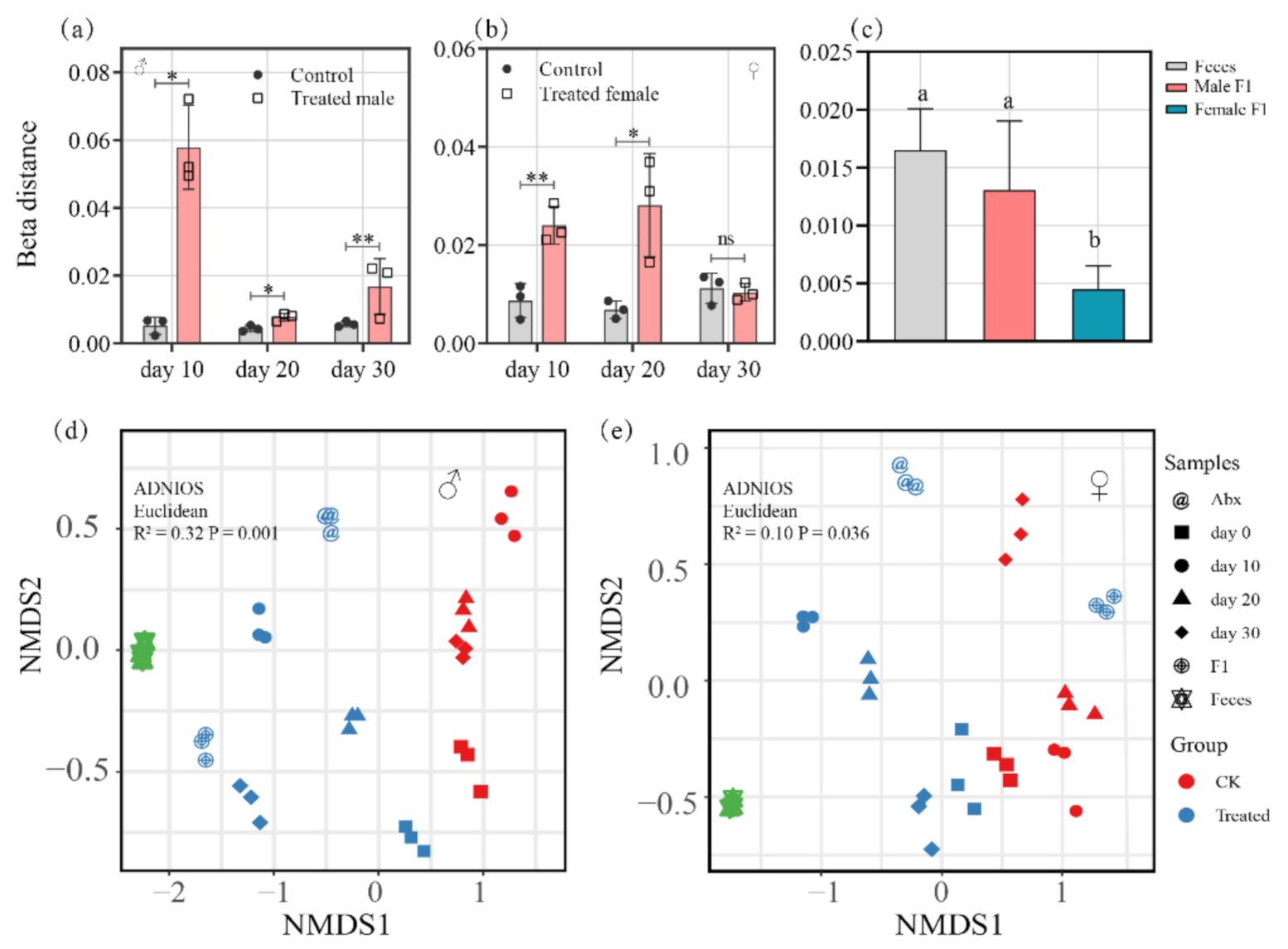
3.2. Analysis of Gut Bacterial Similarity
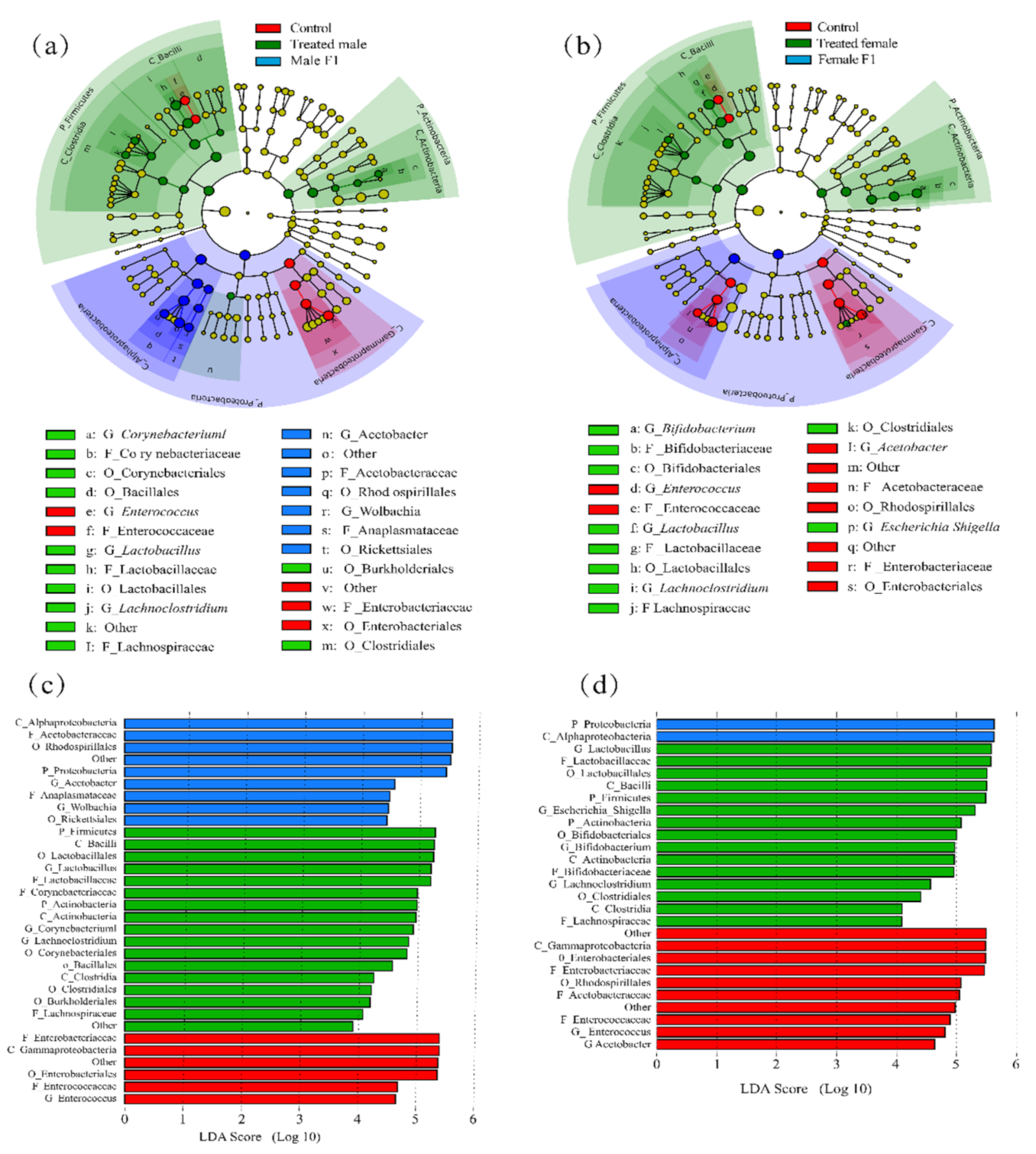
3.3. Effects of Fecal Microbiota Exposure on Species with Significant Differences in Abundance and Community Structure
3.4. Effects of Fecal Microbiota Exposure on Drosophila Lifespan and Body Weight
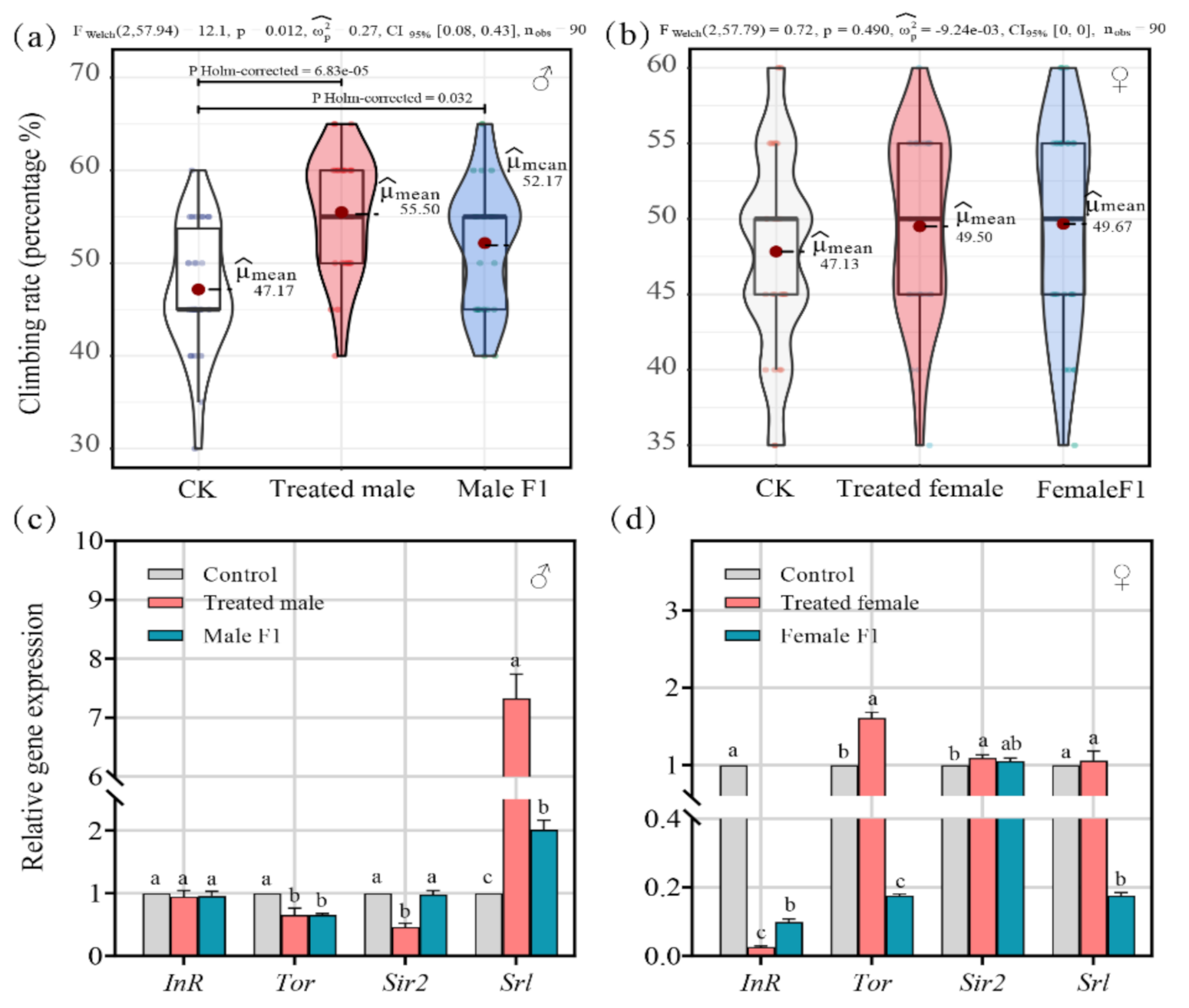
3.5. Effects of Fecal Microbiota Exposure on Vertical Climbing Ability and Expression of Age and Insulin-like Signaling-Related Genes of Drosophila
4. Discussion
4.1. Effects of Fecal Microbiota Exposure on Drosophila Gut Bacteria
4.2. Effects of Fecal Microbiota Exposure on Drosophila Lifespan and Body Weight
4.3. The Uses and Prospects of Anthropomorphic Drosophila Gut Model
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Hintze, K.J.; Cox, J.E.; Rompato, G.; Benninghoff, A.D.; Ward, R.E.; Broadbent, J.; Lefevre, M. Broad scope method for creating humanized animal models for animal health and disease research through antibiotic treatment and human fecal transfer. Gut Microbes 2014, 5, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Hrncir, T.; Hrncirova1, L.; Kverka, M.; Tlaskalova-Hogenova, H. The role of gut microbiota in intestinal and liver diseases. Lab. Anim. 2019, 53, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Hrncir, H.; Hrncirova, L.; Kverka, M.; Hromadka, R.; Machova, V.; Trckova, E.; Kostovcikova, K.; Kralickova, P.; Krejsek, J.; Tlaskalova-Hogenova, H. Gut Microbiota and NAFLD: Pathogenetic Mechanisms, Microbiota Signatures, and Therapeutic Interventions. Microorganisms 2021, 9, 957. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Knight, R.; Gordon, J.I. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 2009, 1, ra14. [Google Scholar] [CrossRef]
- Goodman, A.L.; Kallstrom, G.; Faith, J.J.; Reyes, A.; Moore, A.; Dantas, G.; Gordon, J.I. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc. Natl. Acad. Sci. USA 2011, 108, 6252–6257. [Google Scholar] [CrossRef]
- Gootenberg, D.B.; Turnbaugh, P.J. Companion animals symposium: Humanized animal models of the microbiome. J. Anim. Sci. 2011, 89, 1531–1537. [Google Scholar] [CrossRef][Green Version]
- Lundberg, R. Humanizing the gut microbiota of mice: Opportunities and challenges. Lab. Anim. 2019, 53, 244–251. [Google Scholar] [CrossRef]
- Adams, M.D.; Celniker, S.E.; Holt, R.A.; Evans, C.A.; Gocayne, J.D.; Amanatides, P.G.; Scherer, S.E.; Li, P.W.; Hoskins, R.A.; Galle, R.F.; et al. The genome sequence of Drosophila melanogaster. Science 2000, 287, 2185–2195. [Google Scholar] [CrossRef]
- Myers, E.W.; Sutton, G.G.; Delcher, A.L.; Dew, I.M.; Fasulo, D.P.; Flanigan, M.J.; Kravitz, S.A.; Mobarry, C.M.; Reinert, K.H.; Remington, K.A.; et al. A whole-genome assembly of Drosophila. Science 2000, 287, 2196–2204. [Google Scholar] [CrossRef]
- Pandy, U.B.; Nichols, C.D. Human disease models in Drosophila melanogaster and its role of the fly in therapeutic drug discovery. Pharmacol. Rev. 2011, 63, 411–436. [Google Scholar] [CrossRef] [PubMed]
- Erkosar, B.; Storelli, G.; Defaye, A.; Leulier, F. Host-intestinal microbiota mutualism: “Learning on the fly”. Cell Host Microbe 2013, 13, 8–14. [Google Scholar] [CrossRef]
- Ma, D.; Storelli, G.; Mitchell, M.; Leulier, F. Studying host-microbiota mutualism in Drosophila: Harnessing the power of gnotobiotic flies. Biomed. J. 2015, 38, 285–293. [Google Scholar] [CrossRef]
- Ryu, J.H.; Ha, E.M.; Lee, W.J. Innate immunity and gut–microbe mutualism in Drosophila. Dev. Comp. Immunol. 2010, 34, 369–376. [Google Scholar] [CrossRef]
- Storelli, G.; Defaye, A.; Erkosar, B.; Hols, P.; Royet, J.; Leulier, F. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 2011, 14, 403–414. [Google Scholar] [CrossRef]
- Broderick, N.A.; Lemaitre, B. Gut-associated microbes of Drosophila melanogaster. Gut Microbes 2012, 3, 307–321. [Google Scholar] [CrossRef]
- Blum, J.E.; Fischer, C.N.; Miles, J.; Handelsman, J. Frequent replenishment sustains the beneficial microbiome of Drosophila melanogaster. mBio 2013, 4, e00860-13. [Google Scholar] [CrossRef]
- Chaston, J.M.; Newell, P.D.; Douglas, A.E. Metagenome-wide association of microbial determinants of host phenotype in Drosophila melanogaster. mBio 2014, 5, e01631-14. [Google Scholar] [CrossRef]
- Storelli, G.; Strigini, M.; Grenier, T.; Bozonnet, L.; Schwarzer, M.; Daniel, C.; Matos, R.; Leulier, F. Drosophila Perpetuates Nutritional Mutualism by Promoting the Fitness of Its Intestinal Symbiont Lactobacillus plantarum. Cell Metab. 2018, 27, 362–377.e8. [Google Scholar] [CrossRef]
- Brandt, L.J.; Aroniadis, O.C. An overview of fecal microbiota transplantation: Techniques, indications, and outcomes. Gastrointest. Endosc. 2013, 78, 240–249. [Google Scholar] [CrossRef]
- Yuling, D.; Hao, S.; Weichao, Y.; Shuang, M.; Beibei, D.; Hui, X. The Effect of Inulin on Lifespan, Related Gene Expression and Gut Microbiota in InRp5545/TM3 Mutant Drosophila melanogaster: A Preliminary Study. Nutrients 2019, 11, 636. [Google Scholar]
- Oxenkrug, G.; Navrotskaya, V.; Vorobyova, L.; Summergrad, P. Minocycline effect on life and health span of Drosophila melanogaster. Aging Dis. 2012, 3, 352–359. [Google Scholar] [PubMed]
- Lebourg, E. The rate of living theory. Spontaneous locomotor activity, aging and longevity in Drosophila melanogaster. Exp. Gerontol. 1987, 22, 359–369. [Google Scholar] [CrossRef]
- Martinez, V.G.; Javadi, C.S.; Ngo, E.; Ngo, L.; Lagow, R.D.; Zhang, B. Age-Related changes in climbing behavior and neural circuit physiology in Drosophila. Dev. Neurobiol. 2007, 67, 778–791. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Shuang, M.; Hao, S.; Weichao, Y.; Mingfu, G.; Hui, X. Impact of Probiotic Combination in InR[E19]/TM2 Drosophila melanogaster on Longevity, Related Gene Expression, and Intestinal Microbiota: A Preliminary Study. Microorganisms 2020, 8, 1027. [Google Scholar]
- Wos-Oxley, M.; Bleich, A.; Oxley, A.P.; Kahl, S.; Janus, L.M.; Smoczek, A.; Nahrstedt, H.; Pils, M.C.; Taudien, S.; Platzer, M.; et al. Comparative evaluation of establishing a human gut microbial community within rodent models. Gut Microbes 2012, 3, 234–249. [Google Scholar] [CrossRef]
- Shin, S.C.; Kim, S.H.; You, H.; Kim, B.; Kim, A.C.; Lee, K.; Yoon, J.; Ryu, J.; Lee, W. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 2011, 334, 670–674. [Google Scholar] [CrossRef]
- Han, G.; Lee, H.J.; Jeong, S.E.; Jeon, C.O.; Hyun, S. Comparative Analysis of Drosophila melanogaster Gut Microbiota with Respect to Host Strain, Sex, and Age. Microb Ecol. 2017, 74, 207–216. [Google Scholar] [CrossRef]
- Buchon, N.; Broderick, N.A.; Lemaitre, B. Gut homeostasis in a microbial world: Insights from Drosophila melanogaster. Nat. Rev. Microbiol. 2013, 11, 615–626. [Google Scholar] [CrossRef]
- Yurkovetskiy, L.; Burrows, M.; Khan, A.A.; Graham, L.; Volchkov, P.; Becker, L.; Antonopoulos, D.; Umesaki, Y.; Chervonsky, A.V. Gender bias in autoimmunity is influenced by microbiota. Immunity 2013, 39, 400–412. [Google Scholar] [CrossRef]
- Org, E.; Mehrabian, M.; Parks, B.W.; Shipkova, P.; Liu, X.; Drake, T.A.; Lusis, A.J. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes 2016, 7, 313–322. [Google Scholar] [CrossRef]
- Pais, I.S.; Valente, R.S.; Sporniak, M.; Teixeira, L. Drosophila melanogaster establishes a species-specific mutualistic interaction with stable gut-colonizing bacteria. PLoS Biol. 2018, 16, e2005710. [Google Scholar] [CrossRef]
- Apidianakis, Y.; Rahme, L.G. Drosophila melanogaster as a model for human intestinal infection and pathology. Dis. Models Mech. 2011, 4, 21–30. [Google Scholar] [CrossRef]
- Miguel-Aliaga, I.; Jasper, H.; Lemaitre, B. Anatomy and Physiology of the Digestive Tract of Drosophila melanogaster. Genetics 2018, 210, 357–396. [Google Scholar] [CrossRef]
- Westfall, S.; Lomis, N.; Prakash, S. A novel polyphenolic prebiotic and probiotic formulation have synergistic effects on the gut microbiota influencing Drosophila melanogaster physiology. Artif. Cells Nanomed. Biotechnol. 2018, 46, 441–445. [Google Scholar] [CrossRef]
- Schriner, S.E.; Kuramada, S.; Lopez, T.E.; Truong, S.; Pham, A.; Jafari, M. Extension of Drosophila lifespan by cinnamon through a sex-specific dependence on the insulin receptor substrate chico. Exp. Gerontol. 2014, 60, 220–230. [Google Scholar] [CrossRef]
- Boyd, O.; Weng, P.; Sun, X.; Alberico, T.; Laslo, M.; Obenland, D.M.; Kern, B.; Zou, S. Nectarine promotes longevity in Drosophila melanogaster. Free Radic. Biol. Med. 2011, 50, 1669–1678. [Google Scholar] [CrossRef]
- Vaĭserman, A.M.; Koliada, A.K.; Zabuga, O.Z. Effect of dietary restriction during development on the level of expression of longevity-associated genes in Drosophila melanogaster. Adv. Gerontol. 2013, 26, 638–642. [Google Scholar] [CrossRef]
- Westfall, S.; Lomis, N.; Prakash, S. A polyphenol-rich prebiotic in combination with a novel probiotic formulation alleviates markers of obesity and diabetes in Drosophila. J. Funct. Foods 2018, 48, 374–386. [Google Scholar] [CrossRef]
- Kaeberlein, M.; Powers, R.W.; Steffen, K.K.; Westman, E.A.; Di, H.; Dang, N.; Kerr, E.O.; Kirkland, K.T.; Fields, S.; Kennedy, B.K. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 2005, 310, 1193–1196. [Google Scholar] [CrossRef]
- Vellai, T.; Takacs-Vellai, K.; Zhang, Y.; Kovacs, A.L.; Orosz, L.; Müller, F. Influence of TOR kinase on lifespan in C. elegans. Nature 2003, 426, 620. [Google Scholar] [CrossRef]
- Jia, K.; Chen, D.; Riddle, D.L. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development 2004, 131, 3897–3906. [Google Scholar] [CrossRef]
- Harrison, D.E.; Strong, R.; Sharp, Z.D.; Nelson, J.F.; Astle, C.M.; Flurkey, K.; Nadon, N.L.; Wilkinson, J.E.; Frenkel, K.; Carter, C.S.; et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 2009, 460, 392–395. [Google Scholar] [CrossRef]
- Kapahi, P.; Zid, B.M.; Harper, T.; Koslover, D.; Sapin, V.; Benzer, S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 2004, 14, 885–890. [Google Scholar] [CrossRef]
- Tiefenbock, S.K.; Baltzer, C.; Egli, N.A.; Frei, C. The Drosophila PGC-1 homologue Spargel coordinates mitochondrial activity to insulin signalling. EMBO J. 2010, 29, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Rogina, B.; Helfand, S.H. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad. Sci. USA 2004, 101, 15998–16003. [Google Scholar] [CrossRef] [PubMed]
- Tinkerhess, M.J.; Healy, L.; Morgan, M.; Sujkowski, A.; Matthys, E.; Zheng, L.; Wessells, R.J. The Drosophila PGC-1α homolog spargel modulates the physiological effects of endurance exercise. PLoS ONE 2012, 7, e31633. [Google Scholar] [CrossRef] [PubMed]
- Klünemann, M.; Andrejev, S.; Blasche, S.; Mateus, A.; Phapale, P.; Devendran, S.; Vappiani, J.; Simon, B.; Scott, T.A.; Kafkia, E.; et al. Bioaccumulation of therapeutic drugs by human gut bacteria. Nature 2021, 597, 533–538. [Google Scholar] [CrossRef]


| Total n. of Flies | Mean (%Change) | Median (%Change) | Logrank (vs. Control) | |
|---|---|---|---|---|
| Control | 158 | 29.90 | 30 | - |
| treated male | 159 | 30.73 (+2.78%) | 32 (+6.67%) | p < 0.05 |
| male F1 | 160 | 32.81 (+9.73%) | 30 (+0.00%) | p < 0.01 |
| Control | 158 | 32.97 | 33 | - |
| treated female | 159 | 28.58 (−13.32%) | 29 (−12.12%) | p < 0.05 |
| female F1 | 160 | 34.64 (+5.07%) | 32 (−3.03%) | p = 0.23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, D.; Sun, H.; Yang, W.; Gao, M.; Xu, H. Transfer of Human Microbiome to Drosophila Gut Model. Microorganisms 2022, 10, 553. https://doi.org/10.3390/microorganisms10030553
Ji D, Sun H, Yang W, Gao M, Xu H. Transfer of Human Microbiome to Drosophila Gut Model. Microorganisms. 2022; 10(3):553. https://doi.org/10.3390/microorganisms10030553
Chicago/Turabian StyleJi, Dongxu, Hao Sun, Weichao Yang, Mingfu Gao, and Hui Xu. 2022. "Transfer of Human Microbiome to Drosophila Gut Model" Microorganisms 10, no. 3: 553. https://doi.org/10.3390/microorganisms10030553
APA StyleJi, D., Sun, H., Yang, W., Gao, M., & Xu, H. (2022). Transfer of Human Microbiome to Drosophila Gut Model. Microorganisms, 10(3), 553. https://doi.org/10.3390/microorganisms10030553







